Abstract
Protein folding is an important problem in structural biology with significant medical implications, particularly for misfolding disorders like Alzheimer’s disease. Solving the folding problem will ultimately require a combination of theory and experiment, with theoretical models providing a comprehensive view of folding and experiments grounding these models in reality. Here we review progress towards this goal over the past decade, with an emphasis on recent theoretical advances that are empowering chemically-detailed models of folding and the new results these technologies are providing. In particular, we discuss new insights made possible by Markov state models (MSMs), including the role of non-native contacts and the hub-like character of protein folded states.
Introduction
Ever since Anfinsen’s discovery that denatured proteins can spontaneously self-assemble into their native conformations [1], researchers have tried to understand the molecular mechanisms by which proteins fold. Understanding these mechanisms has become increasingly important in light of many neurological disorders, such as Alzheimer’s disease, that have been directly linked to protein misfolding in the cell [2].
Ultimately, elucidating how proteins fold must involve a combination of theory and experiment, with theoretical models providing a complete picture of folding and experiments grounding these models in reality. Despite decades of research, however, it remains a challenge to predict how proteins fold to their native structures directly from their primary amino acid sequences.
Where are we in our ability to realize this goal? In this article, we summarize the transition from simplified, native-centric models to atomically-detailed, physics-based models that has occurred over the past decade and the factors that motivated this transition. We also describe recent technological and methodological advances that have enabled current theoretical approaches. Many advances in simulating protein folding have come from Markov State Model (MSM) approaches, a new paradigm in molecular simulation that uses large-scale statistical sampling to construct a comprehensive model of protein folding thermodynamics and kinetics on long timescales [3-16].
Much like the theory of quantum mechanics in its infancy, surprising experimental data has provided the impetus for new theoretical developments, and now in turn, theoretical models are providing a new set of surprising predictions. In particular, MSM methods, which can tame the otherwise overwhelming complexity of folding revealed by large-scale simulations, have yielded quantitative agreement with experiments [17-22] and led to new predictions about the importance of non-native contacts [18,19] as well as the resulting hub-like character of protein native states and slow dynamics within the unfolded ensemble [20,23].
Lastly, we describe some of the future challenges we face in making quantitative connections to experiment—including making direct predictions of experimental observables and interpreting the meaning of particular reaction coordinates in their larger context—and new directions for the protein folding community.
The native-centric view is unsatisfying
Ten years ago, all-atom molecular dynamics (MD) simulation was relegated to nanosecond to single microsecond timescales while folding experiments were typically capturing events on the millisecond timescale. As a result, many theorists turned to a native-centric view of folding, which had the potential to provide a unified picture of folding and computationally tractable models [24-26].
A variety of experimental results motivated the focus on native-centric models. Whereas chemically-detailed physical simulation of the folding process at that time appeared to be nearly intractable, experiments showed that, on a macroscopic scale, proteins fold very simply [27]. Fast-mixing studies of folding kinetics showed that small globular proteins are well described by a cooperative two-state mechanism, with an activation barrier separating unfolded and native states. φ-values measured in site-directed mutagenesis studies and high-temperature unfolding simulations suggested that the ensemble structure of these transition states were very native-like. The correlation between native-state topology and two-state folding rate further supported this view [28]. Thus, despite the huge complexity of the conformational search, many of the atomistic details of events preceding the folding transition appeared to be largely irrelevant.
This native-centric view inspired the widespread use of simplified models, called Go models, in which only native state interactions and self-exclusion of the protein chain are accounted for [29,30]. These were justified in part by spin-glass-like theories of folding that predicted that proteins should be minimally frustrated in order to avoid kinetic traps [24]. Go models (and other Go-like models capturing various levels of detail) speed folding dynamics, such that folding can be seen in nanosecond-scale simulations, and thus were considered to be sufficiently computationally efficient to capture aspects of the entire folding reaction and predict some mechanistic quantities [31].
Despite the promise of native-centric models in the field of protein folding, they represent an increasingly unsatisfying direction for further theoretical inquiry. Recent experiments have revealed details about folding that cannot be accounted for by native-state interactions alone. For example, non-native interactions in the unfolded state can be significant [32,33]. Native-centric models are not transferable, so they cannot address the fundamental question of how primary sequence dictates folding pathways, and in particular, how the chain is able to fold without an exhaustive search of conformational space. Moreover, they cannot be used when structural information is more limited or even unavailable, such as for protein misfolding and aggregation, or intrinsically unstructured proteins. For many other proteins, the two-state approximation breaks down [34,35]. Ultrafast downhill folders, for example, have apparent negative activation barriers, as their folding rate may be diffusion-controlled [36]. Additionally, some native-centric properties—such as the correlation between native topology and folding rate—may be more complicated than earlier studies have suggested [37].
It is thus becoming increasingly clear that a comprehensive understanding of folding must require physics-based, transferrable molecular simulation models. Whereas these models were previously too computationally expensive, advances in hardware and simulation methodology have helped push simulations from nanosecond timescales into the millisecond regime, opening up new avenues in the field.
New simulation methodologies allow atomically-detailed models of folding on biologically relevant timescales
New advances in MD simulation software and hardware are now helping the protein folding field move beyond the native-centric view by allowing theorists to simulate biologically relevant timescales (e.g. microsecond to milliseconds timescales, see Figure 1) using physics-based simulations that can capture the balance between native and non-native interactions. For example, simulation packages like GROMACS [38] and OpenMM [39] are able to automatically select the best combination of hardware and algorithms on a given computer. When run on GPUs or multi-core CPUs, these codes allow the execution of simulations on tens of microseconds timescales on a routine basis, which is an impressive advance given that atomistic MD simulations had previously been limited to tens of nanosecond timescales. Packages like NAMD [40] and DESMOND [41] also allow simulations of tens of microseconds timescales by parallelizing the computation over many CPUs in addition to making very efficient use of individual CPUs. Therefore, the ability to simulate small, fast-folding proteins is widely available. Using more expensive, purpose-built hardware, it may even be possible to run individual simulations of millisecond timescales [42]. Another 2x to 10x speedup may be obtained by exploiting new algorithmic developments [43-46] and improvements to the force fields used to describe atomic interactions [47-51] are making these simulations a more accurate reflection of reality.
Figure 1.
The average folding time versus length for a number of fast-folding proteins from the experimental literature that are candidates for atomistic simulations and MSMs. Overlays of the crystal structure (cyan) and our predicted native state (dark blue) are shown for a number of systems highlighted in this review.
Directly simulating these timescales is an impressive feat (see http://www.youtube.com/watch?v=gFcp2Xpd29I for an example); however, long simulations on their own are not sufficient to provide better models of protein folding because protein dynamics are inherently stochastic and, therefore, demand a statistical description. For example, to predict an ensemble property like the average folding timescale of a protein, one must simulate numerous statistically-independent folding events. One could run a simulation that was hundreds or thousands of times longer than the average folding time to achieve this but such an approach is inherently inefficient. Instead, it is better to run many shorter simulations in parallel. Regardless of whether one runs a few long simulations or many shorter ones, however, analyzing the resulting data is still a significant challenge. Some methods based on non-equilibrium statistical-mechanics exist for extracting ensemble properties from numerous shorter simulations [52-54], but even these are limited in the types of systems and properties they can address.
MSMs are capable of providing a complete description of the equilibrium thermodynamics and kinetics of any simulated system, regardless of whether the specific dataset is composed of a few long simulations, many shorter ones, or a mixture thereof. MSMs are discrete-time master equation models for protein dynamics composed of a network of metastable states—or sets of rapidly mixing conformations that tend to persist for extended periods of time—and the probabilities (or, equivalently, the rates) of transitioning between pairs of states in some time interval called the lag time of the model [3-5]. Building and analyzing these models is challenging but now there are a number of software packages that can automate these tasks [10,16,19]. All of these approaches identify the states for an MSM by performing a kinetic clustering of an MD dataset. That is, conformations that can interconvert rapidly (indicating they are not separated by substantial free energy barriers) are grouped into the same state and ones that can only interconvert slowly (indicating that they are separated by substantial barriers) are grouped into separate states. One can then calculate the probabilities (or, equivalently, the rates) of transitioning between pairs of states from the number of transitions observed in the dataset. For details on constructing an appropriate set of states we recommend referring to Refs [17] and [19].
Because of the kinetic definition of states, an MSM is essentially a map of the underlying free energy landscape that determines a molecule’s thermodynamics and kinetics. As in a computer mapping program (such as Google or Yahoo maps), one can “zoom out” on an MSM to get a big picture view of processes like protein folding then “zoom in” on regions of conformational space of particular interest or calculate the average time for transitioning between two states. This “zooming in” or “zooming out” is achieved by varying the definition of what it means for two conformations to interconvert rapidly. For example, by requiring that two conformations be able to interconvert on a nanosecond timescale for them to be grouped in the same state, one can build a very high-resolution model with many small states that would be appropriate for making a quantitative comparison with experiments [17,19,55]. By only requiring that conformations in the same state be able to interconvert on microsecond timescales one can obtain a more coarse-grained model with fewer, larger states that would be appropriate for gaining human insight. Such human insight can be obtained by visualizing the model directly (i.e. extracting representative conformations from each state and examining the transition rates between states) or by using transition path theory (TPT) to visualize the highest flux pathways between some start and end state [19,56,57]. One can also use an MSM to generate new trajectories to mimic single molecule experiments or to model the relaxation of an ensemble of proteins to make a direct connection to bulk experiments.
Moreover, MSMs provide a way to exploit metastability—which is largely responsible for making long simulations inefficient—to build models with tremendous efficiency. For example, in a simple system with metastable states A↔B↔C (where B is an obligatory intermediate for transitioning between A and C) one can calculate the average timescale for the slow transition between states A and C even if no individual simulation starts off in state A and ends in state C so long as one sees the faster transitions between states A and B as well as between states B and C. Decomposing the problem in this manner is efficient because each step can be simulated in parallel. Adaptive sampling is an iterative algorithm that facilitates this parallelism by allowing one to use an initial MSM to decide where to run new simulations [58-60]. During each round of adaptive sampling, one calculates the contribution of each state to the statistical uncertainty in some property of interest (like the average folding rate), starts new simulations in the states causing the most uncertainty, and then rebuilds the model. By running N simulations per iteration of adaptive sampling one can reduce the time required to build a model by a factor of N and the total computational resources required to build that model by a factor of two [59]. Therefore, simulations that previously would have taken decades can now be performed in a matter of weeks or months.
MSMs have allowed us as a field to build more detailed models of protein folding and validate them against experiment. For example, many groups have successfully modeled the dynamics of small model systems for folding [9-12,14,15,61-64]. In one of the first forays into full protein systems, Jayachandran et al. built a model for the villin headpiece [65], which folds on a microsecond timescale. Bowman et al. have also modeled villin using new automated methods for constructing MSMs [17]. This model was built from hundreds of microsecond-timescale, atomistic simulations in explicit water [66] and, therefore, represents the state of the art in simulation accuracy. The native state of this model (i.e. lowest free energy state) was within 1.8 Å of the crystal structure, an important achievement given that all the simulations used to build the model started from unfolded conformations, and the average folding rate agreed with the experimental value within statistical uncertainty. In addition, MSMs for villin can predict triplet-triplet energy transfer (TTET) experiments for multiple probe locations and the metastable states that have been inferred from these experiments [22]. Noe et al. have also used MSMs to model the folding of another fast-folding protein, the PinWW domain (a 34-residue protein with a microsecond folding time) [19]. The ability of these models to predict structures, thermodynamics, and rates indicates they should be capable of predicting any experimental observable, since all are functions of these properties.
More recently, we have demonstrated that MSMs can be used to model the folding of larger, slower systems than one could hope to address with the traditional long simulation approach. For example, Voelz et al. have built an MSM for a 39-residue fragment of NTL9 [18], which has an average folding time of 1.5 milliseconds. This is an exciting achievement because long simulations can barely reach the millisecond timescale, much less capture the hundreds or thousands of milliseconds one would have to simulate to extract the average folding time for this system. Bowman et al. have also modeled ten millisecond timescales for an 80-residue fragment of the lambda repressor protein and made a number of hypotheses that suggest directions for further experimental investigation [21].
What you see is not always what you get
There is no doubt that folding is complex due to factors like the enormity of conformational space and the delicate balance between opposing forces. Despite this, protein folding looks quite simple in many experiments. For example, many folding experiments are dominated by single exponential relaxation and have sigmoidal melting curves. Therefore, based on Occam’s razor, it has been reasonable to conclude that protein folding is often a two-state process with a moderately populated unfolded basin separated from a well-populated native state by a single dominant barrier [27]. By the same logic, one can also reasonably conclude that multi-state systems fold via a single, linear pathway [67]. Using MSMs built from atomistic simulations and validated against experiments, we can finally hope to reconcile the known complexity of folding with this observed simplicity.
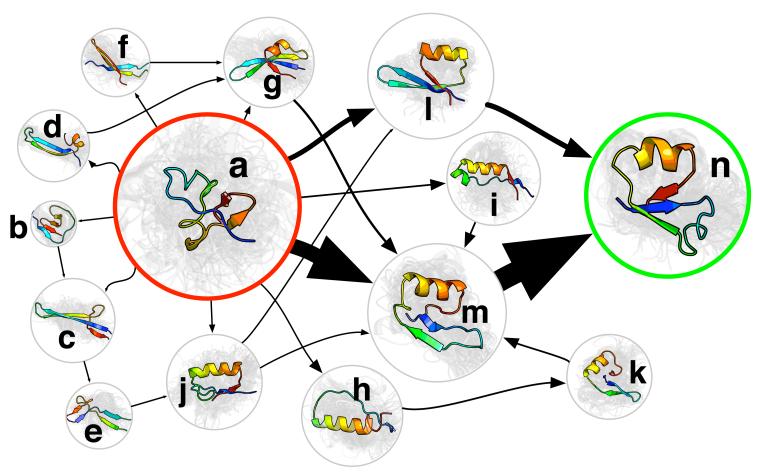
MSMs for a number of systems suggest that many proteins may fold via parallel paths [17-20]. For instance, great parallelism was found in early MSMs for the villin headpiece [65], and this observation has been confirmed by more recent models with more thorough sampling [17,20,22]. Parallel paths have been suggested in a number of previous theoretical and experimental studies [24,36,66,68,69] but with MSMs we can better map out these paths. For example, Noe et al. developed an algorithm for examining the highest flux paths between two states in a model and showed that it takes 3-5 pathways to account for 50% of the folding flux for the PinWW domain and up to 20 pathways to account for 90% of the flux [19]. Despite this, the relaxation of the model is dominated by a single-exponential with a timescale in remarkable agreement with the experimentally measured folding rate. Using similar methods, Voelz et al. have mapped out a number of parallel folding pathways for NTL9 [18]. This is a landmark study because it is the first demonstration that MSMs built from atomistic simulations can capture, with statistical significance, millisecond timescale events that could not be addressed by conventional simulation methodologies. The results of this NTL9 study demonstrate that there is a good deal of mixing between these parallel pathways (Figure 2). In addition, many of the intermediate states along these pathways have significant non-native contacts that appear to help the protein fold. For example, state L in Figure 2 has an extended beta-hairpin that is not found in the native state (state N). While direct tests of the specific non-native interactions found in our models have yet to be performed, these predictions certainly seem reasonable given a large body of recent experimental work demonstrating complex structures in the unfolded ensembles of proteins [32,70,71] and even within the native basin [72].
Figure 2.
The highest flux folding pathways from a 2000-state MSM for NTL9 taken from Ref [18]. These pathways account for only ~25% of the total flux and transit only 14 of the 2000 metastable states (labeled a-n). The size of each state is proportional to the logarithm of its equilibrium population and arrow sizes are proportional to the logarithm of the interstate flux.
Taking the idea of parallel pathways a step further, Bowman et al. recently discovered that the native states of many simulated systems appear to act as kinetic hubs [20]. As a result, many folding pathways have distinct starting points, as shown in Figure 3, though there is still mixing between pathways. In addition, transitioning between different non-native states is typically quite slow compared to folding due to the strength of non-native contacts, the enormity of conformational space, and greater evolutionary pressure to fold than to transition rapidly between unfolded conformations. This result is in direct contradiction of the two- and three-state models typically used to describe folding, which assume that dynamics within the unfolded ensemble are fast relative to folding. Despite their unexpected topology, these hub-like models still produce the simple single and biexponential relaxation seen in many protein folding experiments [20,21]. Therefore, while the simplest explanation for the relaxation seen in experiments is often a two- or three-state model, it would appear that these kinetics may result from the convolution of different rates for crossing a large number of free energy barriers of different heights. Once again, direct experimental tests of this prediction have not yet been performed but there is a growing body of work that corroborates our models. For example, recent microfluidic mixing experiments have identified extremely slow dynamics in the unfolded ensembles of proteins [73]. If further experiments validate this model, then the folding field may have to re-evaluate much of the existing literature, where a two-state assumption is often made in interpreting both theoretical and experimental results.
Figure 3.
Schematic of a native hub model with a native state (N) and non-native states (nn) taken from Ref [20]. The size of each node is correlated with its equilibrium probability and the connectivity falls off as one moves away from the native state.
While the hub model is more complex than the two- and three-state models that have dominated the protein folding field for some time now, these new models still have their own elegant simplicity. Pande has shown that one can analytically derive a hub-like model by assuming that the transition state between states A and B only has residue-residue contacts that are formed in both the start and end state [23].
Conclusions & future directions
We are now entering an era when theory could take the lead in protein folding by explaining the origins of experimental observables and generating new hypotheses, both about specific systems and general principles of folding. Making this advance has required moving from native-centric models to transferrable, physics-based simulation models. It has also required moving from an anecdotal description of simulations to a statistical description using Markov state models—highly parallelizable, multi-scale methods that can address long timescale events by drawing on information from shorter timescale simulations.
Three important results from MSMs are: (1) many proteins fold via parallel paths [17-19], (2) non-native contacts can play important roles in folding [18,20,23], and (3) the native states of many proteins may be kinetic hubs [20,21,23]. Despite these complexities, these models are still capable of predicting the apparently simple behavior of many experimental probes. They have also driven the development of new models for protein folding [23].
In the coming years, making quantitative comparisons between theory and experiment should further deepen our understanding of processes like folding and allow us to refine simulation methodologies and parameterizations. Some important future directions for theory and simulation are (1) making a more direct connection with experiments by more closely mimicking experimental procedures and probes, (2) comparing mutants to dissect parts of the folding process, as is often done in the experimental literature, and (3) developing a novel analytic theory which can explain and predict the behavior seen in both detailed simulations and new experiments. This endeavor will also benefit from the development of experiments that can probe folding in greater detail and that can be more easily understood and modeled (e.g. do not require invoking quantum mechanics to predict fluorescence). We can then hope to address considerably larger systems, design folding pathways, and understand folding in cellular contexts using atomistic simulations and MSMs.
Thus, with the ability to predict experimentally relevant timescales (milliseconds to seconds) with experimentally relevant systems for folding (e.g. on the ~100 amino acid length scale), we are beginning to realize the dream of marrying theory and experiment to finally solve the protein folding problem. Looking to the future, it will perhaps be in the application of this knowledge to numerous related problems, such as protein misfolding (relevant for numerous diseases [2]) and protein dynamics associated with function (such as enzymatic activity [74]), where advances in protein folding will continue to yield insight and impact for many years to come.
Acknowledgements
This work was funded by NIH R01-GM062868, NSF-MCB-0954714, and NSF EF-0623664. GRB was supported by the Berry Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anfinsen CB, Haber E, Sela M, White FH., Jr. The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain. Proc Natl Acad Sci USA. 1961;47:1309–1314. doi: 10.1073/pnas.47.9.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uversky VN. Intrinsic disorder in proteins associated with neurodegenerative diseases. Front Biosci. 2009;14:5188–5238. doi: 10.2741/3594. [DOI] [PubMed] [Google Scholar]
- 3.Schütte C, Fischer A, Huisinga W, Deuflhard P. A direct approach to conformational dynamics based on hybrid Monte Carlo. J Comput Phys. 1999;151:146–168. [Google Scholar]
- 4.Bowman GR, Huang X, Pande VS. Network models for molecular kinetics and their initial applications to human health. Cell Res. 2010;20:622–630. doi: 10.1038/cr.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noe F, Fischer S. Transition networks for modeling the kinetics of conformational change in macromolecules. Curr Opin Struct Biol. 2008;18:154–162. doi: 10.1016/j.sbi.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Singhal N, Snow CD, Pande VS. Using path sampling to build better Markovian state models: predicting the folding rate and mechanism of a tryptophan zipper beta hairpin. J. Chem. Phys. 2004;121:415–425. doi: 10.1063/1.1738647. [DOI] [PubMed] [Google Scholar]
- 7.Elmer SP, Park S, Pande VS. Foldamer dynamics expressed via Markov state models. I. Explicit solvent molecular-dynamics simulations in acetonitrile, chloroform, methanol, and water. J Chem Phys. 2005;123:114902. doi: 10.1063/1.2001648. [DOI] [PubMed] [Google Scholar]
- 8.Swope WC, Pitera JW, Suits F. Describing protein folding kinetics by molecular dynamics simulations. 1. Theory. J Phys Chem B. 2004;108:6571–6581. [Google Scholar]
- 9.Noe F, Horenko I, Schutte C, Smith JC. Hierarchical analysis of conformational dynamics in biomolecules: transition networks of metastable states. J Chem Phys. 2007;126:155102. doi: 10.1063/1.2714539. [DOI] [PubMed] [Google Scholar]
- 10.Chodera JD, Singhal N, Pande VS, Dill KA, Swope WC. Automatic discovery of metastable states for the construction of Markov models of macromolecular conformational dynamics. J Chem Phys. 2007;126:155101. doi: 10.1063/1.2714538. [DOI] [PubMed] [Google Scholar]
- 11.Buchete NV, Hummer G. Coarse master equations for peptide folding dynamics. J Phys Chem B. 2008;112:6057–6069. doi: 10.1021/jp0761665. [DOI] [PubMed] [Google Scholar]
- 12.Pan AC, Roux B. Building Markov state models along pathways to determine free energies and rates of transitions. J Chem Phys. 2008;129:064107. doi: 10.1063/1.2959573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang S, Banavali NK, Roux B. Mapping the conformational transition in Src activation by cumulating the information from multiple molecular dynamics trajectories. Proc Natl Acad Sci U S A. 2009;106:3776–3781. doi: 10.1073/pnas.0808261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrec M, Felts AK, Gallicchio E, Levy RM. Protein folding pathways from replica exchange simulations and a kinetic network model. Proc Natl Acad Sci U S A. 2005;102:6801–6806. doi: 10.1073/pnas.0408970102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gfeller D, De Los Rios P, Caflisch A, Rao F. Complex network analysis of free-energy landscapes. Proc Natl Acad Sci U S A. 2007;104:1817–1822. doi: 10.1073/pnas.0608099104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowman GR, Huang X, Pande VS. Using generalized ensemble simulations and Markov state models to identify conformational states. Methods. 2009;49:197–201. doi: 10.1016/j.ymeth.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowman GR, Beauchamp KA, Boxer G, Pande VS. Progress and challenges in the automated construction of Markov state models for full protein systems. J Chem Phys. 2009;131:124101. doi: 10.1063/1.3216567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voelz VA, Bowman GR, Beauchamp KA, Pande VS. Molecular simulation of ab initio protein folding for a millisecond folder NTL9(1-39) J Am Chem Soc. 2010;132:1526–1528. doi: 10.1021/ja9090353. ••The first atomically-detailed model of folding on millisecond timescales. This study points to the importance of non-native contacts and parallel pathways in folding.
- 19.Noe F, Schutte C, Vanden-Eijnden E, Reich L, Weikl TR. Constructing the equilibrium ensemble of folding pathways from short off-equilibrium simulations. Proc Natl Acad Sci U S A. 2009;106:19011–19016. doi: 10.1073/pnas.0905466106. ••Demonstrates the use of flux analysis, called transition path theory (TPT), to map out protein folding pathways. This study emphasizes that many pathways must be considered to account for the full folding flux.
- 20.Bowman GR, Pande VS. Protein folded states are kinetic hubs. Proc Natl Acad Sci U S A. 2010;107:10890–10895. doi: 10.1073/pnas.1003962107. ••The first observation of the hub-like character of native states in atomically-detailed MSMs for multiple proteins. This study highlights parallel pathways and slow dynamics in the unfolded ensemble (relative to folding) due to factors like non-native contacts and the enormity of conformational space. It also demonstrates how this can be reconciled with existing experimental data that has been interpreted as being two-state.
- 21.Bowman GR, Voelz VA, Pande VS. Atomistic folding simulations of the five helix bundle protein λ6-85. 2010. submitted. •A landmark simulation study demonstrating the ability of MSMs to capture ten millisecond timescales for relatively large systems (80 residues). This study shows that even large systems have a native hub and provides an alternative explanation for apparent “downhill” folding in addition to yielding a number of other hypotheses that warrant further experimental investigation.
- 22.Beauchamp KA, Ensign DL, Das R, Pande VS. Fine Structure in Protein Folding: Quantitative Comparison of HP35 Simulations and Triplet-Triplet Energy Transfer Experiments. 2010. submitted. [DOI] [PMC free article] [PubMed]
- 23.Pande VS. A simple theory of protein folding kinetics. Phys Rev Lett. 2010 doi: 10.1103/PhysRevLett.105.198101. in submssion. •Derivation of a simple theoretical model for protein folding that captures the native hub observed in simulation studies by accounting for non-native contacts.
- 24.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein folding: a synthesis. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 25.Leopold PE, Montal M, Onuchic JN. Protein folding funnels: a kinetic approach to the sequence-structure relationship. Proc Natl Acad Sci U S A. 1992;89:8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat Struct Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 27.Jackson SE, Fersht AR. Folding of chymotrypsin inhibitor 2. 1. Evidence for a two-state transition. Biochemistry. 1991;30:10428–10435. doi: 10.1021/bi00107a010. [DOI] [PubMed] [Google Scholar]
- 28.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J Mol Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 29.Hills RD, Jr., Brooks CL., 3rd Insights from coarse-grained go models for protein folding and dynamics. Int J Mol Sci. 2009;10:889–905. doi: 10.3390/ijms10030889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 31.Clementi C. Coarse-grained models of protein folding: toy models or predictive tools? Current Opinion in Structural Biology. 2008;18:10–15. doi: 10.1016/j.sbi.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Shan B, Eliezer D, Raleigh DP. The unfolded state of the C-terminal domain of the ribosomal protein L9 contains both native and non-native structure. Biochemistry. 2009;48:4707–4719. doi: 10.1021/bi802299j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Chan HS. Competition Between Native Topology and Nonnative Interactions in Simple and Complex Folding Kinetics of Natural and Designed Proteins. Proc. Natl. Acad. Sci. 2010;107:2920–2925. doi: 10.1073/pnas.0911844107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spudich GM, Miller EJ, Marqusee S. Destabilization of the Escherichia coli RNase H kinetic intermediate: switching between a two-state and three-state folding mechanism. J Mol Biol. 2004;335:609–618. doi: 10.1016/j.jmb.2003.10.052. [DOI] [PubMed] [Google Scholar]
- 35.Kim PS, Baldwin RL. Intermediates in the folding reactions of small proteins. Annu Rev Biochem. 1990;59:631–660. doi: 10.1146/annurev.bi.59.070190.003215. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh K, Ozkan SB, Dill KA. The Ultimate Speed Limit to Protein Folding Is Conformational Searching. J. Am. Chem. Soc. 2007;129:11920–11927. doi: 10.1021/ja066785b. [DOI] [PubMed] [Google Scholar]
- 37.Ouyang Z, Liang J. Predicting protein folding rates from geometric contact and amino acid sequence. Protein Sci. 2008;17:1256–1263. doi: 10.1110/ps.034660.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: Fast, flexible, and free. Journal of computational chemistry. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 39.Friedrichs MS, Eastman P, Vaidyanathan V, Houston M, Legrand S, Beberg AL, Ensign DL, Bruns CM, Pande VS. Accelerating molecular dynamic simulation on graphics processing units. J Comput Chem. 2009;30:864–872. doi: 10.1002/jcc.21209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowers K, Chow E, Xu H, Dror R, Eastwood M, Gregersen B, Klepeis J, Kolossvary I, Moraes M, Sacerdoti F, et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. Proceedings of the 2006 ACM/IEEE conference on Supercomputing.2006. [Google Scholar]
- 42.Shaw D, Dror R, Salmon J, Grossman J, Mackenzie K, Bank J, Young C, Deneroff M, Batson B, Bowers K, et al. Millisecond-scale molecular dynamics simulations on Anton. Proceedings of the Conference on High Performance Computing Networking, Storage and Analysis.2009. [Google Scholar]
- 43.Huang X, Bowman GR, Pande VS. Convergence of folding free energy landscapes via application of enhanced sampling methods in a distributed computing environment. J Chem Phys. 2008;128:205106. doi: 10.1063/1.2908251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laio A, Gervasio F. Metadynamics: a method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science. Reports on Progress in Physics. 2008;71 [Google Scholar]
- 45.Sweet CR, Petrone P, Pande VS, Izaguirre JA. Normal mode partitioning of Langevin dynamics for biomolecules. J Chem Phys. 2008;128:145101. doi: 10.1063/1.2883966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamelberg D, Mongan J, McCammon JA. Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. J Chem Phys. 2004;120:11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- 47.Wang JM, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comp Chem. 2000;21:1049–1074. [Google Scholar]
- 48.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sorin EJ, Pande VS. Exploring the helix-coil transition via all-atom equilibrium ensemble simulations. Biophys J. 2005;88:2472–2493. doi: 10.1529/biophysj.104.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mittal J, Best RB. Tackling force-field bias in protein folding simulations: folding of Villin HP35 and Pin WW domains in explicit water. Biophys J. 2010;99:L26–28. doi: 10.1016/j.bpj.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ponder JW, Wu C, Ren P, Pande VS, Chodera JD, Schnieders MJ, Haque I, Mobley DL, Lambrecht DS, DiStasio RA, Jr., et al. Current status of the AMOEBA polarizable force field. J Phys Chem B. 114:2549–2564. doi: 10.1021/jp910674d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolhuis PG, Chandler D, Dellago C, Geissler PL. Transition path sampling: throwing ropes over rough mountain passes, in the dark. Annu Rev Phys Chem. 2002;53:291–318. doi: 10.1146/annurev.physchem.53.082301.113146. [DOI] [PubMed] [Google Scholar]
- 53.Faradjian AK, Elber R. Computing time scales from reaction coordinates by milestoning. J Chem Phys. 2004;120:10880–10889. doi: 10.1063/1.1738640. [DOI] [PubMed] [Google Scholar]
- 54.Pande VS, Baker I, Chapman J, Elmer SP, Khaliq S, Larson SM, Rhee YM, Shirts MR, Snow CD, Sorin EJ, et al. Atomistic protein folding simulations on the submillisecond time scale using worldwide distributed computing. Biopolymers. 2003;68:91–109. doi: 10.1002/bip.10219. [DOI] [PubMed] [Google Scholar]
- 55.Sarich M, Noe F, Schutte C. On the approximation quality of Markov state models. SIAM Multiscale Model Simul. 2010 in press. [Google Scholar]
- 56.Vanden Eijnden E. Toward a theory of transition paths. J Stat Phys. 2006;123:503–523. [Google Scholar]
- 57.Berezhkovskii A, Hummer G, Szabo A. Reactive flux and folding pathways in network models of coarse-grained protein dynamics. J Chem Phys. 2009;130:205102. doi: 10.1063/1.3139063. •Derivation of a method for Markovpping out the highest flux pathways in protein folding.
- 58.Hinrichs NS, Pande VS. Calculation of the distribution of eigenvalues and eigenvectors in Markovian state models for molecular dynamics. J Chem Phys. 2007;126:244101. doi: 10.1063/1.2740261. [DOI] [PubMed] [Google Scholar]
- 59.Bowman GR, Ensign DL, Pande VS. Enhanced modeling via network theory: adaptive sampling of Markov state models. J Chem Theory Comput. 2010;6:787–794. doi: 10.1021/ct900620b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang X, Bowman GR, Bacallado S, Pande VS. Rapid equilibrium sampling initiated from nonequilibrium data. Proc Natl Acad Sci U S A. 2009;106:19765–19769. doi: 10.1073/pnas.0909088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swope WC, Pitera JW, Suits F, Pitman M, Eleftheriou M. Describing protein folding kinetics by molecular dynamics simulations. 2. Example applications to alanine dipeptide and beta-hairpin peptide. J Phys Chem B. 2004;108:6582–6594. [Google Scholar]
- 62.Elmer SP, Pande VS. Foldamer simulations: novel computational methods and applications to poly-phenylacetylene oligomers. J Chem Phys. 2004;121:12760–12771. doi: 10.1063/1.1812272. [DOI] [PubMed] [Google Scholar]
- 63.Schultheis V, Hirschberger T, Carstens H, Tavan P. Extracting Markov Models of Peptide Conformational Dynamics from Simulation Data. JCTC. 2005;1:515–526. doi: 10.1021/ct050020x. [DOI] [PubMed] [Google Scholar]
- 64.Rao F, Caflisch A. The protein folding network. J Mol Biol. 2004;342:299–306. doi: 10.1016/j.jmb.2004.06.063. •An early study of a small peptide that points to parallel pathways, a power-law degree distribution, and high connectivity to the native state.
- 65.Jayachandran G, Vishal V, Pande VS. Folding Simulations of the Villin Headpiece in All-Atom Detail. J. Chem. Phys. 2006;124:164902. doi: 10.1063/1.2186317. [DOI] [PubMed] [Google Scholar]
- 66.Ensign DL, Kasson PM, Pande VS. Heterogeneity even at the speed limit of folding: large-scale molecular dynamics study of a fast-folding variant of the villin headpiece. J Mol Biol. 2007;374:806–816. doi: 10.1016/j.jmb.2007.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krishna MM, Maity H, Rumbley JN, Lin Y, Englander SW. Order of steps in the cytochrome C folding pathway: evidence for a sequential stabilization mechanism. J Mol Biol. 2006;359:1410–1419. doi: 10.1016/j.jmb.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 68.Radford SE, Dobson CM, Evans PA. The folding of hen lysozyme involves partially structured intermediates and multiple pathways. Nature. 1992;358:302–307. doi: 10.1038/358302a0. [DOI] [PubMed] [Google Scholar]
- 69.Kamagata K, Sawano Y, Tanokura M, Kuwajima K. Multiple parallel-pathway folding of proline-free Staphylococcal nuclease. J Mol Biol. 2003;332:1143–1153. doi: 10.1016/j.jmb.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 70.Waldauer SA, Bakajin O, Ball T, Chen Y, Decamp SJ, Kopka M, Jager M, Singh VR, Wedemeyer WJ, Weiss S, et al. Ruggedness in the folding landscape of protein L. HFSP J. 2008;2:388–395. doi: 10.2976/1.3013702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Voelz VA, Singh VR, Wedemeyer WJ, Lapidus LJ, Pande VS. Unfolded state dynamics and structure of protein L characterized by simulation and experiment. J Am Chem Soc. 2010;132:4702–4709. doi: 10.1021/ja908369h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reiner A, Henklein P, Kiefhaber T. An unlocking/relocking barrier in conformational fluctuations of villin headpiece subdomain. Proc Natl Acad Sci U S A. 2010;107:4955–4960. doi: 10.1073/pnas.0910001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Waldauer SA, Bakajin O, Lapidus LJ. Extremely slow intramolecular diffusion in unfolded protein L. Proc Natl Acad Sci U S A. 2010;107:13713–13717. doi: 10.1073/pnas.1005415107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fraser JS, Clarkson MW, Degnan SC, Erion R, Kern D, Alber T. Hidden alternative structures of proline isomerase essential for catalysis. Nature. 2009;462:669–673. doi: 10.1038/nature08615. [DOI] [PMC free article] [PubMed] [Google Scholar]