Abstract
Murine leukemia viruses (MuLVs) encode two forms of Gag polyprotein: the precursor for the viral core proteins (Pr65gag for Moloney MuLV [M-MuLV]) and a longer glycosylated form (glyco-gag, or gPr80gag). gPr80gag is translated from the same unspliced viral RNA as Pr65gag, from an upstream in-frame CUG initiation codon. As a result, gPr80gag contains 88 unique N-terminal amino acids that include a signal peptide that conducts gPr80gag into the rough endoplasmic reticulum, where it is glycosylated, exported to the cell surface, and cleaved into two proteins of 55 and 40 kDa. The amino-terminal 55-kDa protein remains cell associated with the 88 unique amino acids exposed to the cytosol. We previously showed that gPr80gag facilitates efficient M-MuLV release through lipid rafts. In this report, we found that the unique N-terminal domain of gPr80gag is sufficient to facilitate enhanced M-MuLV particle release from transfected 293T cells. A search for cellular proteins involved in gPr80gag function led to cellular La protein. Overexpression of mouse or human La enhanced M-MuLV particle release in the absence of glyco-gag, and the released virus had a reduced buoyant density characteristic of increased cholesterol content. Moreover, small interfering RNA (siRNA) knockdown of human La abolished glyco-gag enhancement of M-MuLV release. These results implicate La as a cellular protein involved in M-MuLV glyco-gag function. We also found that overexpression of mouse or human La could enhance HIV-1 release in the absence of gPr80gag. Therefore, M-MuLV and HIV-1 may share a pathway for release through lipid rafts involving La.
IMPORTANCE
Retroviruses cause diseases such as leukemia and AIDS. An important aspect of viral replication is how viruses are released from infected cells. We previously found that a unique protein encoded by murine leukemia viruses (MuLVs), glyco-gag (or gPr80gag), enhances efficient virus release through cholesterol-rich membrane subdomains called lipid rafts. In this study, we found that the N-terminal domain of gPr80gag is sufficient to enhance viral release. A search for cellular proteins that participate in gPr80gag function led to cellular La protein. Overexpression of La phenocopied glyco-gag in enhancing M-MuLV release, and knockdown of La abolished glyco-gag function. M-MuLV glyco-gag also enhanced release of HIV-1, as did overexpression La in the absence of glyco-gag. Thus, M-MuLV and HIV-1 may share a cellular pathway for release through lipid rafts involving La. These results may also be relevant for other viruses that are released through lipid rafts.
INTRODUCTION
Murine leukemia viruses (MuLVs) are prototypical simple retroviruses of the gammaretrovirus genus. One unique feature of MuLVs and many other gammaretroviruses is that they encode an alternate form of Gag polyprotein, gPr80gag (or glyco-gag), as well as the polyprotein precursor to Gag structural proteins, Pr65gag. gPr80gag is translated from unspliced viral mRNA via an upstream CUG initiation codon in the same reading frame as for Pr65gag (1–3). The N terminus of gPr80gag contains 88 unique amino acids, including a signal peptide that targets gPr80gag for transport into the rough endoplasmic reticulum, leading to its glycosylation and export to the cell surface (4). At the cell surface, mature gPr80gag is cleaved into two proteins of ca. 55 and 40 kDa (1, 5), and the 55-kDa amino-terminal portion is maintained in a type II integral membrane configuration, with the 88 unique amino acids in the cytosol (4).
Glycosylated Gag proteins are conserved among gammaretroviruses, but the molecular functions of these proteins have been unclear until recently. In mice, gPr80gag is a major pathogenic determinant for neuropathic MuLV (6–9). MuLV mutants of gPr80gag show substantial replication defects in mice, and there is strong selection for recovery of gPr80gag expression (10–12). We previously showed that gPr80gag plays a role in a late step in viral assembly or release. gPr80gag-negative MuLVs bud from cells from aberrant tube-like structures (12, 13). Expression of gPr80gag in mutant-infected cells increases virus particle release, and the tube-like structures are replaced by typical spherical particles (12). Recently, we found that there are two pathways for MuLV release from cells: interferon (IFN)-sensitive release through lipid rafts and interferon-resistant release through areas other than lipid rafts (14). gPr80gag facilitates viral release through lipid rafts, and this is apparently the more efficient pathway for release. We also found that Moloney MuLV (M-MuLV) gPr80gag can facilitate release of HIV-1 particles (14). It has also recently been reported that gPr80gag can complement a replication defect in human lymphocyte lines for Nef-deficient HIV-1, although in this study the effect of glyco-gag was on viral infectivity as opposed to viral release (15).
In this report, we show that the unique 88 amino acids at the N terminus of gPr80gag are sufficient for facilitating MuLV and HIV release through lipid rafts. Moreover, we have identified the cellular protein La/SSB (Sjogren’s syndrome autoantigen B) as being involved in the mechanism of gPr80gag action.
RESULTS
The N-terminal unique region of gPr80gag is sufficient for activity.
In our previous studies, we showed that expression of M-MuLV gPr80gag from the expression plasmid p8065-2 enhances M-MuLV particle release from NIH 3T3 fibroblasts and that this was done by directing release through lipid rafts, since the resulting particles had higher cholesterol content, reduced buoyant density, and enhanced association with cellular detergent-resistant membranes (DRMs) (14). Enhancement of virus release was also found in transiently transfected 293T cells (14). Since gPr80gag differs from Pr65gag by additional amino-terminal residues, we tested whether this unique region was sufficient for gPr80gag activity. An expression plasmid encoding the unique 88 amino-terminal sequences of gPr80gag fused to an N-terminal hemagglutinin (HA) epitope tag (HA-gg88) was generated (Fig. 1). As shown in Fig. 2A, when HA-gg88 or p8065-2 was cotransfected into 293T cells along with an M-MuLV gag-pol expression construct (AKAQ188), equivalent amounts of intracellular Gag protein were detected. Like p8065-2, HA-gg88 also facilitated virus release, in fact somewhat more effectively than p8065-2 (ca. 3-fold and 4.5-fold increases, respectively). In the absence of HA-gg88 or p8065-2, release of virions from AKAQ188-transfected cells was inefficient (but detectable), while the amount of cytoplasmic Gag protein produced was similar.
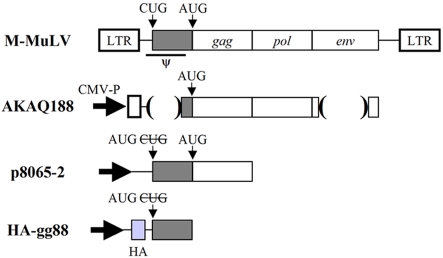
FIG 1 .
Expression plasmids used. Diagrams of the M-MuLV expression plasmids used in these experiments are shown. AKAQ188 has deletions in the region encoding the leader peptide (deletion of positions 215 to 561), containing both the packaging signal (positions 217 to 567) (46) and the CUG initiation codon for gPr80Gag (positions 357 to 359), and in the env gene (deletion of positions 5819 to 7197). The CUG start codon for gPr80gag was replaced by an AUG start codon in both p8065-2 and HA-gg88. In HA-gg88, viral sequences upstream of the CUG codon were also removed so that this plasmid would lack a functional packaging sequence (46). LTR, long terminal repeat; CMV-P, cytomegalovirus promotor.
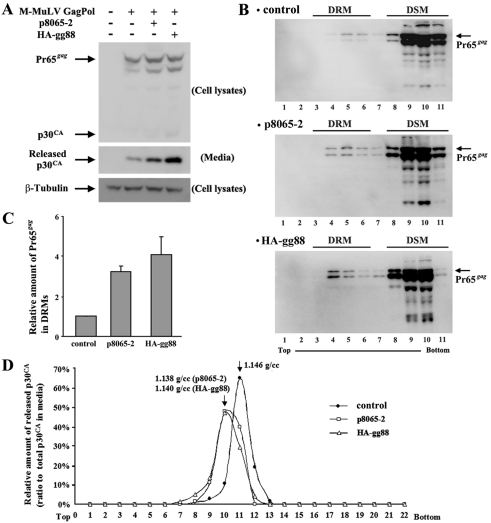
FIG 2 .
The 88 amino acids at the amino terminus in gPr80Gag facilitates M-MuLV particle release through lipid rafts. The M-MuLV Gag-Pol expression vector was transfected with p8065-2 and HA-gg88 into 293T cells. (A) The cells and media were harvested at 48 h posttransfection, and the same portion of cells and viruses were subjected to the Western blots with anti-p30CA and anti-β-Tubulin (loading control). (B) The cell lysates prepared from the transfected 293T cells with the lysis buffer containing 1% Triton X-100 were analyzed by membrane floatation in 5 to 30% sucrose gradients. Low-density fractions represented detergent-resistant membranes (DRM) and lipid rafts, while high-density fractions represented protein in detergent-soluble membranes (DSM) or cytosolic proteins. (C) The amount of Pr65gag was quantified by densitometry, and the relative amount of Pr65 in lipid rafts to control are shown (means ± standard deviations [SD]). (D) Viruses harvested from the transfected 293T cells were analyzed by density gradient centrifugation, and distribution of p30CA was analyzed by Western blotting and densitometry.
To test whether HA-gg88 also facilitates virus release through lipid rafts, we tested for the presence of Pr65gag in cellular DRMs that are cholesterol rich as described previously (14). As shown in Fig. 2B and C, p8065-2 resulted in a ca. 3-fold increase in Pr65gag in DRMs (as reported previously [14]), and HA-gg88 had an equivalent effect. The proportions of Pr65gag in DRMs for the transiently transfected 293T cells were less than those for NIH 3T3 fibroblasts infected with wild-type versus glyco-gag-negative M-MuLV (14), but the relative differences were similar. As a second measure of release through lipid rafts, we tested the buoyant densities of the resulting viruses in sucrose gradients. As shown in Fig. 2D, virions released from p8065-2 or HA-gg88-expressing cells both showed lower buoyant densities, characteristic of viruses with higher cholesterol content, than virus released from 293T cells in the absence of glyco-gag. The absolute values of the buoyant densities reported here were somewhat lower than those in our previous study (14), which was due to a change in the method for measurement of sucrose densities, but the relative differences between control virions and those released from glyco-gag-expressing cells were equivalent. Thus, HA-gg88 was as efficient as p8065-2 in directing virus release through lipid rafts, indicating that the N-terminal unique region of gPr80gag is necessary and sufficient for the activity of this protein.
Expression of HA-gg88 relocalizes cellular La protein to the cytoplasm.
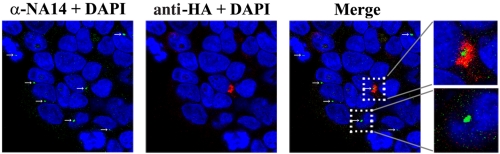
To investigate the mechanism of gPr80gag action, a yeast two-hybrid screen was conducted as described in Materials and Methods. The bait plasmid encoded the 88 amino-terminal unique amino acids fused to the LexA DNA binding domain. This plasmid was used to screen a mouse cDNA expression library fused to the B42 activation domain in Saccharomyces cerevisiae. Four cDNA fusion clones that showed strong interaction with the bait plasmid were identified, and sequencing of the inserts indicated that they all were cDNAs for Sjogren’s syndrome nuclear autoantigen 1 (SSNA1; also known as nuclear autoantigen 14 [NA14] [16, 17]). While the initial report described SSNA1/NA14 as a nuclear antigen, subsequent studies indicated that it predominantly binds to centrosomes in the cytoplasm (16–18). To test for potential in vivo interactions between NA14 and gPr80gag, we performed two-color immunofluorescence microscopy on 293T cells transfected with HA-gg88 (Fig. 3). Endogenous NA14 showed a cytoplasmic perinuclear location consistent with centrosomes reminiscent of previous studies (16, 18), regardless of whether the cells were expressing HA-gg88 or not. HA-gg88 was found in the cytoplasm and localized mainly in the perinuclear region surrounding the endogenous NA14 protein. However, these two proteins did not show actual colocalization. NIH 3T3 cells transiently transfected with an epitope-tagged Flag-NA14 protein showed sporadic localized signals in the cytoplasm as well as the condensed signals at perinuclear regions, but in cotransfections with HA-gg88, colocalization with Flag-NA14 was not observed (not shown). Glutathione S-transferase (GST) pulldown experiments with 293T cells cotransfected with a GST-tagged gg88 protein and Flag-NA14 also did not show evidence of direct binding between these two proteins (not shown). Thus, despite the two-hybrid results, gg88 does not appear to directly bind NA14 in mammalian cells.
FIG 3 .
Localization of HA-gg88, NA14, and Ro. The 293T cells were transfected with HA-gg88. The transfected cells were fixed at 36 to 48 h posttransfection with paraformaldehyde and then incubated with anti-HA and anti-NA14 antibodies. The antigens and nuclei were visualized by secondary antibodies conjugated with Alexa 488, Alexa 546, and DAPI (4′,6-diamidino-2-phenylindole). The panels for NA14-DAPI and HA-DAPI and a merge of the first two panels are shown. The inserts are enlarged images and are shown next to the merge picture.
Because the major Sjogren’s syndrome autoantigens are associated with immune responses and viral replication, we also tested whether glyco-gag might show any interactions (direct or indirect) with other Sjogren’s syndrome autoantigens (SSA [also known as Ro] and SSB [also known as La, or the lupus antigen]). Ro is a member of the tripartite motif (TRIM) protein family; this protein is an interferon-inducible E3 ligase that ubiquitinates interferon regulatory factor 3 (IRF-3) and IRF-8 (19–21). La is a predominantly nuclear RNA binding protein that binds certain RNA polymerase III (Pol III) transcripts, facilitating their processing and trafficking (22, 23); it also facilitates replication of some viruses (24–30). Two-color immunofluorescence microscopy for Ro and HA-gg88 was carried out with 293T cells. For these experiments, it was necessary to transfect cells with a Ro expression plasmid since endogenous Ro protein could not be detected by the anti-Ro antibody. 293T cells were cotransfected with the Ro expression plasmid and either pcDNA3.1 or HA-gg88. Ro protein was found in both the cytoplasm and the nucleus (more in the cytoplasm) (data not shown), and the distribution pattern was consistent with previously published studies (31, 32). However, coexpression of HA-gg88 did not change the cellular distribution of Ro, and colocalization of HA-gg88 and Ro was not observed (data not shown).
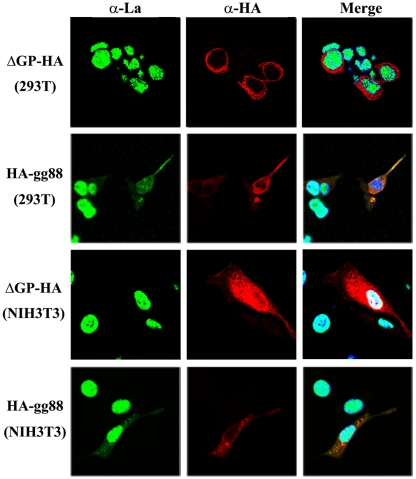
In contrast, expression of HA-gg88 affected the intracellular localization of La (Fig. 4). In 293T or NIH 3T3 cultures transfected with HA-gg88 (Fig. 4, second row and bottom row), La showed nuclear fluorescence in the cells that did not express HA-gg88, while the cells that were positive for HA-gg88 showed substantial cytoplasmic fluorescence for La. One possible explanation for the results is that transfection of any expression plasmid could lead to relocalization of La in those cells that took up the DNA. However, in cells transfected with the JSRV, jaagsiekte sheep retrovirus, Env expression vector, with ΔGP-HA (Fig. 4, top and third rows), or with the backbone pcDNA3 expression plasmids (not shown), La localization was largely nuclear. When the results were quantified, 43.5% of 293T cells from HA-gg88-transfected cultures showed cytoplasmic La staining, versus 18.6% showing cytoplasmic staining, after ΔGP-HA transfection. Likewise, NIH 3T3 cultures transfected with HA-gg88 showed cytoplasmic staining in 27.5% of cells, compared to 6.2% for ΔGP-HA-transfected cells. Taken together, these results indicated that HA-gg88 relocalizes La to the cytoplasm, which suggested that cytoplasmic La might play a role in the mechanism of glyco-gag action.
FIG 4 .
Relocalization of La by HA-gg88. The 293T and NIH 3T3 cells were transfected with HA-gg88 and the JSRV Env-expressing vector ΔGP-HA (transfection control). The transfected cells were fixed at 36 to 48 h posttransfection with paraformaldehyde and then incubated with anti-HA and anti-La antibodies. The antigens were visualized by the secondary antibodies conjugated with Alexa 488 and Alexa 546. The panels for La, HA, and a merge (La-HA-DAPI) are shown.
Overexpression of La phenocopies glycosylated gag.
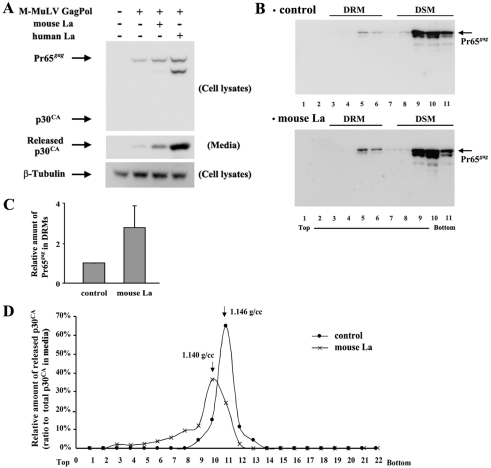
If M-MuLV glyco-gag facilitates virus release via La protein, one mechanism could be that when La is directed to the cytoplasm by glyco-gag, La plays a positive role in facilitating virus release through lipid rafts. We reasoned that overexpression of La might result in enhanced cytoplasmic levels of the protein, which might facilitate virus release in the absence of glyco-gag expression. As shown in Fig. 5A, cotransfection of 293T cells with expression plasmids for mouse or human La along with AKAQ188 resulted in significantly enhanced virus release, with human La more effective than mouse La (ca. 3-fold and 6-fold increases, respectively). Overexpression of mouse La also enhanced the appearance of Pr65gag in cellular DRMs (Fig. 5B and C), and virus released from cells overexpressing mouse La showed the characteristic shift to lighter buoyant density, indicative of higher cholesterol content. Thus, in the absence of glyco-gag, overexpression of La appears to result in the same enhancement of virus release through lipid rafts as that seen when glyco-gag is expressed.
FIG 5 .
Overexpression of La facilitates M-MuLV particle release through lipid rafts. The M-MuLV gag-Pol expression vector (AKAQ188) was transfected with mouse La and human La expression vectors into 293T cells. The assessment for viral release (A), the distribution of Pr65gag in DRM (B and C), and the density of released viruses from the transfected cells (D) are described in the legend to Fig. 2.
Knockdown of La antagonizes the effects of HA-gg88.
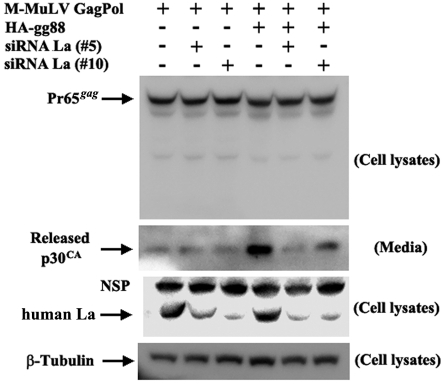
While the effects of overexpressing La were consistent with the idea that glyco-gag functions through La, we sought additional support for this model. We tested whether knockdown of human La would reduce HA-gg88-enhanced virus release from transfected 293T cells. As shown in Fig. 6, 293T cells were transiently transfected with AKAQ188, with or without HA-gg88, as well as two small interfering RNAs (siRNAs) against human La. The siRNAs were effective in reducing the amount of La present in the cells (Fig. 6, third panel from the top). As shown in the second panel, in the absence of the siRNAs, HA-gg88 substantially enhanced virus release; both siRNAs significantly reduced this enhancement. On the other hand, the lower levels of virus released in the absence of HA-gg88 were not affected by knockdown of La. Therefore, the effects of La knockdown were specific to glyco-gag-enhanced virus release. As shown in the top panel of Fig. 6, neither HA-gg88 nor anti-La siRNAs affected the amount of intracellular Pr65gag.
FIG 6 .
Interference of La expression antagonizes HA-gg88 in M-MuLV particle release. M-MuLV Gag-Pol expression vector is transfected into 293T cells with or without HA-gg88. At 24 hours posttransfection, siRNAs against human La (no. 5 and no. 10) were transfected. Two days after siRNA transfection, media were replaced, and then the cells and media were harvested after 8 h of further incubation. The same portion of cells and viruses were subjected to the Western blots with anti-p30CA, anti-La, and anti-β-Tubulin (loading control). NSP, non-specific signals.
Overexpression of La results in interferon-sensitive virus release.
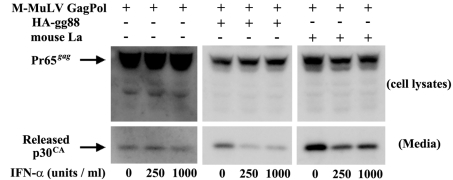
In our previous study, one of the first indications that gPr80gag directs M-MuLV release through lipid rafts was that wild-type-M-MuLV release was sensitive to mouse IFN-αA (IFN-α) but that gPr80gag-negative-virus release was largely resistant to it (14). Therefore, we tested whether the enhanced virus release resulting from overexpression of La is sensitive to IFN-α. As shown in Fig. 7, 293T cells were transiently transfected with AKAQ188 along with either HA-gg88 or the mouse La expression plasmid. The transfected cells were treated with different doses of IFN-α for 24 h, and viruses were gathered after 8 h of further incubation. As expected, the enhanced virus release resulting from cotransfection with HA-gg88 was inhibited by IFN-α treatment (Fig. 7, middle panels), while the smaller amounts of virus released from the control cells (left panels) were not affected by IFN-α. (Longer phosphorimager exposures were required for the left panels due to the lower levels of virus released.) As shown in the right panels, the enhanced virus release resulting from overexpression of mouse La was also inhibited by IFN-α treatment. Thus, the enhanced virus release mediated by La overexpression is IFN sensitive.
FIG 7 .
Effect of IFN-α on virus release from 293T cells overexpressing HA-gg88 and mouse La. The 293T cells were transiently transfected with M-MuLV Gag-Pol expression vector with HA-gg88 and a mouse La expression vector. The cells were treated with different concentrations of IFN-α for 24 h, after which media were replaced, and the cells and released viruses were collected after 6 h of further incubation. The same portions of cells and viruses were subjected to the Western blot analyses with anti-p30CA.
HA-gg88 and La overexpression both enhance release of HIV.
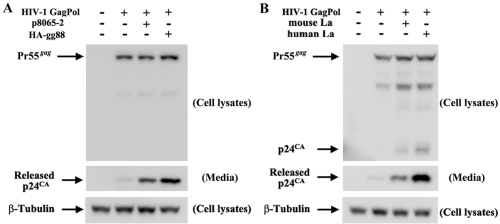
We previously showed that gPr80gag can also enhance release of another retrovirus, HIV-1, from transfected 293T cells (14). As shown in Fig. 8A, HA-gg88 also enhanced release of HIV-1 particles from 293T cells transiently transfected with an HIV-1 gag-pol expression vector, with HA-gg88 showing slightly higher efficiency than p8065-2 (ca. 3-fold and 4.5-fold increases, respectively). Overexpression of either mouse or human La also enhanced HIV-1 particle release (Fig. 8B), with human La again showing a stronger effect than mouse La in 293T cells (ca. 3.5-fold and 7-fold increases, respectively). Thus, the amino-terminal unique region of gPr80gag is also sufficient for enhancement of HIV-1 particle release, and glyco-gag enhancement of HIV-1 release is likely mediated through La.
FIG 8 .
Overexpression of HA-gg88 and La increases the efficiency of HIV-1 particle release. The HIV-1 Gag-Pol expression vector was transfected into 293T cells with p8065-2 and HA-gg88 (A) or mouse La and human La expression vectors (B). The cells and media were harvested at 48 h posttransfection, and the same portion of cells and viruses were analyzed by Western blotting with anti-HIV p24CΑ and anti-β-Tubulin (loading control).
DISCUSSION
In this report, we have obtained insights into the mechanism of action for MuLV-glycosylated Gag protein. We found that the N-terminal unique 88 amino acids of gPr80gag are necessary and sufficient for the ability of gPr80gag to target release through lipid rafts, as evidenced by the enhanced association of the Gag polyprotein precursor Pr65gag with detergent-resistant membranes, the lower buoyant density of released virions (reflective of a higher cholesterol content), and the sensitivity of virus release to interferon. A search for cellular proteins that interact with glyco-gag led indirectly to the cellular La protein, and HA-gg88 was found to relocalize La into the cytoplasm from the nucleus. Overexpression of La in transfected 293T cells phenocopied glyco-gag in all measures tested. Moreover, knockdown of La with siRNAs abrogated the ability of HA-gg88 to enhance MuLV virus particle release. These results strongly support a role for La in the mechanism of glyco-gag action, with La being a downstream effector of glyco-gag.
La protein is a highly abundant cellular protein, predominantly localized in the nucleus. The best known function of this protein is to bind the 3′ ends of RNA transcripts containing U residues, notably tRNA precursors and other RNA Pol III transcripts (22, 23). This binding protects pre-tRNAs from premature 3′ end processing, allowing for proper maturation. Substantial structural and functional studies have been conducted on La (22, 23). Other functions also have been reported for La. It binds to the genomic RNAs for several cytoplasmic RNA viruses, including poliovirus (33), coxsackievirus B3 (28) (picornaviruses), and hepatitis C virus (a flavivirus) (25), and the binding is important for the function of the viral internal ribosome entry sites (IRES). The latter observations make several points. First, La protein can function in the cytoplasm as well as in the nucleus. Second, it seems likely that La may interact with other cellular proteins in carrying out its various functions. Third, other viruses have also employed La protein to carry out key parts of their replication cycles. Thus, while at first surprising, the involvement of La in glyco-gag’s enhancement of MuLV release is reasonable.
While these studies have established a role for La in glyco-gag function, the mechanism of action remains to be determined. We have not detected direct interaction between HA-gg88 and La protein in pulldown assays from transfected cells (T. Nitta and R. Tam, unpublished), which might suggest that glyco-gag and La are interacting indirectly through another cellular protein(s) or particle. This notion is also consistent with the findings that neither gPr80gag nor La is found in DRMs from transfected cells, even though the DRMs from such cells contain enhanced levels of Pr65gag when either glyco-gag or La is expressed along with a gag-pol expression vector. Thus, we hypothesize that glyco-gag and La facilitate release through lipid rafts by interacting with other proteins or macromolecules that conduct Pr65gag to the DRMs/lipid rafts. It will be of substantial interest to identify such proteins or macromolecules. The results also indicated that human La was more efficient than mouse La in enhancing virus release from 293T cells. These cells are of human origin, which might suggest that there is species preference between La proteins and the cellular factors that they interact with for enhancing virus release.
Since La is an RNA binding protein, it will be interesting in the future to test whether the RNA binding activity is important for enhancement of virus release. When retroviruses are assembled, this involves interaction at membranes of the gag and gag-pol polyproteins, the genomic RNA, and the envelope protein. One possibility is that La is interacting with genomic RNA and facilitating incorporation into virus particles. However, the experiments presented here were with transiently transfected 293T cells expressing only Gag and Gag-pol polyproteins, with no Env protein or packageable viral RNA. Others have previously shown that MuLV particles consisting of Gag and Gag-pol proteins can be assembled and released from cells in the absence of viral genomic RNA or Env protein (34, 35). Thus, interaction of La with viral RNA is not involved in the release through lipid rafts shown here.
Previous reports by chemical analyses (4, 36) and computer predictions (not shown) indicated that gPr80gag is a type II integral membrane protein with a short amino-terminal cytoplasmic sequence. Here, we made HA-gg88, containing the initial 88 amino acids of gPr80Gag, and this truncated protein showed activity similar to that of the entire gPr80Gag protein in enhancing M-MuLV release through lipid rafts (Fig. 2), suggesting that neither the glycosylations on the p15MA and p30CA regions (36) nor any of the extracellular Gag residues are required for viral release enhancement. HA-gg88 distributes in the cytoplasm, mainly at perinuclear regions. Similar to the results reported here, a truncated gPr80gag containing the amino-terminal 189 amino acids showed strong localization at perinuclear regions (15). Thus, extracellular Gag residues, including those from position 89 to position 189, do not determine the perinuclear localization.
We previously showed that MuLV gPr80gag also enhances release of HIV particles through lipid rafts (14). Pizzato (15) has recently shown that MuLV glyco-gag complements a replication detect for Nef-negative HIV in lymphocytes, although in the latter study the major glyco-gag effect was on Nef-negative viral infectivity as opposed to release. The glyco-gag effects of Nef-negative HIV infectivity were found to be most pronounced for infected lymphocyte cell lines, while less pronounced for other cell lines, such as 293T, and even less for some fibroblast lines. The relationships of glyco-gag effects on viral infectivity from lymphocytes and particle release from 293T cells as described here and from NIH 3T3 cells in our previous study (14) remain to be elucidated. The results shown here also demonstrate that overexpression of La can phenocopy the enhanced virus release for HIV particles. Thus, La appears to be involved in facilitating HIV release through lipid rafts as well. It will interesting to test whether more-distantly related viruses that exit cells through lipid rafts employ a similar mechanism.
Extensive research has been conducted on HIV and MuLV assembly in infected cells. The interaction of Gag L domains (through motifs such as PS/TAP or YPDL) with components of the cellular vesicular trafficking machinery (e.g., ESCRT proteins) results in transport of Gag polyprotein to the plasma membrane or maybe internal sites of assembly (e.g., multivesicular bodies) (37–40). It does not seem likely that L domains of Gag polyprotein and the resulting association with the vesicular trafficking machinery are sufficient for targeting virus release through lipid rafts. In fact, the Gag polyprotein produced in a gPr80gag mutant MuLV is identical to that of wild-type virus, with the same L domains (41, 42). Thus, transport of Gag polyprotein (Pr65gag) to the plasma membrane should not be affected by gPr80gag status. Rather, once Gag polyprotein arrives at the plasma membrane MuLV, glyco-gag, working through La, may then conduct it to DRMs and lipid rafts, where efficient virus release occurs.
MATERIALS AND METHODS
Cells.
Human 293T and mouse NIH 3T3 fibroblasts were grown in Dulbecco’s modified Eagle’s medium supplemented with penicillin (100 U/ml), streptomycin (100 mg/ml), and 10% fetal bovine serum (293T cells) or 10% calf serum (NIH 3T3 cells).
DNA constructs.
The plasmid p8065-2, expressing MuLV gPr80gag, was described previously (Fig. 1) (12). To make the plasmid HA-gg88, which expresses an HA epitope tag and the 88 amino acids at the N terminus of gPr80gag, the tetherin-coding sequence was removed by digesting HA-tetherin (43) with EcoRI and NotI. The region encoding the N-terminal 88 amino acids was amplified from p8065-2 by PCR with primers 5′-TCCCCGGAATTCCTCGAGGCCACCATGGGAGACGTCCCAGGGAC and 5′-TCCCCGGCGGCCGCCTAATTCTCAGACAAATACAGAAAC. The amplified PCR product was digested with EcoRI and NotI and was ligated with the backbone of the digested HA-tetherin plasmid. For assessment of viral release from cells transiently transfected with retroviral constructs carrying gag and pol, the gPr80gag-negative M-MuLV Gag-Pol expression vector AKAQ188 (Fig. 1) and the HIV-1-based packaging vector pCMV-dR8.74 (http://www.lentiweb.com/) were used. The plasmids pcDNA3 (Invitrogen) and pEGFP-N1 (Clontech) were used as negative controls and also added to equalize the total amounts of DNA transfected. cDNAs for human SSNA1/NA14, SSA/Ro, and SSB/La and mouse SSB/La were obtained from OpenBiosystems. For the bait plasmid in yeast two-hybrid screening, the gPr80gag unique region at the N terminus of gPr80gag was amplified with the primers 5′-GCCGAATTCATGGGAGACGTCCCAGGGACTTCG and 5′-GCCGTCGACTCAATTCTCAGACAAATACAGAAACAC. The PCR product was digested with EcoRI and SalI and ligated with similarly digested pEG202-LexA to express gg88 fused to LexA DNA binding protein. The plasmid ΔGP-HA, expressing JSRV Env with the HA epitope tag from the cytomegalovirus promoter, was described previously (44).
Antibodies and chemicals.
Rabbit polyclonal anti-MuLV p30CA antiserum was described previously (45). Mouse monoclonal anti-HIV-1 p24CA antibody (YDHIVgp24) was purchased from MyBioSource. For detection of epitope tags, mouse and rabbit anti-HA antibodies (Cell Signaling), anti-HA antibodies conjugated with horseradish peroxidase (HRP; GenScript), and anti-Flag antibodies (Cell Signaling) were used. The antibodies against Ro (D-12) and NA14 were purchased from Santa Cruz Biotechnology and ProteinTech Group. To detect La, mouse monoclonal anti-La antibodies (312B; Santa Cruz Biotechnology) and rabbit polyclonal anti-La antibodies (Abgent) were used. Beta-Tubulin was used for the loading control in Western blots and was detected by rabbit anti-beta-Tubulin (Cell Signaling). For Western blots, we used anti-mouse IgG conjugated with horseradish peroxidase (Thermo Scientific) and anti-rabbit IgG conjugated with horseradish peroxidase (GE Healthcare). For indirect fluorescence microscopy, anti-mouse and anti-rabbit IgGs conjugated with Alexa 488 or 546 (Invitrogen) were used. Mouse alpha A interferon (IFN-αA) was obtained from Calbiochem.
Indirect immunofluorescence microscopy.
The 293T and NIH 3T3 cells were plated on glass coverslips 1 day before transfection. The cells were transfected with HA-gg88, a Ro-expressing vector, and pcDNA3 by Lipofectamine 2000 (Invitrogen) or BioT (Bioland) and then were incubated for 36 to 48 h. The cells were fixed with 4% paraformaldehyde for 30 min at room temperature. After the cells were washed and blocked, antibodies were added. The cells were stained with fluorescence-conjugated secondary antibodies, followed by mounting with Vectashield mounting medium (Vector Laboratories). The images were analyzed with Axiovert200 and LSM510 microscopes (Carl Zeiss).
Yeast two-hybrid screening.
To find cellular proteins binding to the gPr80gag-unique amino acids at the N terminus of gPr80gag, we conducted the yeast two-hybrid screen using pLexA-gg88 and a mouse liver cDNA library. The bacterial strain XL-1-Blue (supE44 hsdR17 recA1 endA1 gyrA46 thi relA1 lac) and the Saccharomyces cerevisiae strain EGY48 [MATα his3 trp1 ura3-52 leu::pLeu-LexAop6/Psh18-34 (LexA-op-lacZ reporter)] were used. The Saccharomyces cerevisiae strain EGY48 was sequentially cotransformed with pLexA-gg88 and the cDNA library and then plated on medium lacking uracil, histidine, and tryptophan (SD/ −U, −H, −W). After the resulting colonies were harvested and pooled, approximately 5 × 106 colonies were replated in selection medium lacking uracil, histidine, tryptophan, and leucine (SD/ −U, −H, −W, −L) and containing 2% galactose to induce the expression of cDNAs. The resulting colonies were streaked on the X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-containing medium (SD/ −U, −H, −W) to identify and eliminate false positives.
Flotation of DRM domains.
Flotation of DRMs was performed as previously described (14), with slight modifications. 293T cells transiently transfected with AKAQ188, p8065-2, HA-gg88, a mouse La expressing-vector, and control vectors (pCDNA3 or pEGFP-N1) were treated on the dish with lysis buffer (0.8 ml/10-cm dish) containing 25 mM Tris-HCl (pH 8.0), 140 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM Na3VO4, and a protease inhibitor cocktail (Roche) for 20 min at 4°C. The lysates were harvested and then centrifuged at 10,000 × g for 5 min at 4°C. The postnuclear lysate was adjusted to 40% (wt/vol) sucrose, and then a 5 to 30% discontinuous sucrose gradient was layered on the top. Samples were centrifuged at 100,000 × g for 18 to 24 h at 4°C. The fractions were collected from the top of the gradient tube. The densities of the fractionated samples were determined by use of a refractometer.
Knockdown of La expression by siRNA.
Interference of La expression was conducted by siRNAs. The 293T cells on 6-well plates were transfected with AKAQ188 and HA-gg88 or control vectors by use of a CalPhos mammalian transfection kit (Clontech). At 24 hours posttransfection, two different siRNAs against human La (no. 5 and no. 10; Qiagen) were transfected at 10 µM by use of Lipofectamine RNAiMAX (Invitrogen). Two days after siRNA transfection, media were replaced, and then the cells and media were harvested after 8 h of further incubation.
IFN treatment and detection of viruses.
Treatment of IFN-α and detection of released viruses and viruses in cells by anti-p30CA antibodies were described previously (14).
ACKNOWLEDGMENTS
We thank Audrey Low, Christopher Smith, Edward Paz, and Guopei Luo for assistance and participation in experiments and the Optical Biology Shared Resource of the Chao Family Comprehensive Cancer Center for confocal microscopy. The helpful discussions with John Rossi are appreciated. We thank Paul Bieniasz and Lorraine Albritton for the plasmids HA-tetherin (P.B.) and AKAQ188 (L.A.).
This work was supported by NIH grant CA94188 (to H.F.)
Footnotes
Citation Nitta, T., R. Tam, J. W. Kim, and H. Fan. 2011. The cellular protein La functions in enhancement of virus release through lipid rafts facilitated by murine leukemia virus glycosylated Gag. mBio 2(1):e00341-10. doi:10.1128/mBio.00341-10.
REFERENCES
- 1. Edwards S. A., Fan H. 1979. Gag-related polyproteins of Moloney murine leukemia virus: evidence for independent synthesis of glycosylated and unglycosylated forms. J. Virol. 30:551–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buetti E., Diggelmann H. 1980. Murine leukemia virus proteins expressed on the surface of infected cells in culture. J. Virol. 33:936–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Prats A. C., De Billy G., Wang P., Darlix J. L. 1989. CUG initiation codon used for the synthesis of a cell surface antigen coded by the murine leukemia virus. J. Mol. Biol. 205:363–372 [DOI] [PubMed] [Google Scholar]
- 4. Fujisawa R., McAtee F. J., Zirbel J. H., Portis J. L. 1997. Characterization of glycosylated Gag expressed by a neurovirulent murine leukemia virus: identification of differences in processing in vitro and in vivo. J. Virol. 71:5355–5360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ledbetter J., Nowinski R. C., Emery S. 1977. Viral proteins expressed on the surface of murine leukemia cells. J. Virol. 22:65–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fujisawa R., McAtee F. J., Wehrly K., Portis J. L. 1998. The neuroinvasiveness of a murine retrovirus is influenced by a dileucine-containing sequence in the cytoplasmic tail of glycosylated Gag. J. Virol. 72:5619–5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Portis J. L., Fujisawa R., McAtee F. J. 1996. The glycosylated gag protein of MuLV is a determinant of neuroinvasiveness: analysis of second site revertants of a mutant MuLV lacking expression of this protein. Virology 226:384–392 [DOI] [PubMed] [Google Scholar]
- 8. Portis J. L., Spangrude G. J., McAtee F. J. 1994. Identification of a sequence in the unique 5′ open reading frame of the gene encoding glycosylated Gag which influences the incubation period of neurodegenerative disease induced by a murine retrovirus. J. Virol. 68:3879–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Munk C., Prassolov V., Rodenburg M., Kalinin V., Lohler J., Stocking C. 2003. 10A1-MuLV but not the related amphotropic 4070A MuLV is highly neurovirulent: importance of sequences upstream of the structural Gag coding region. Virology 313:44–55 [DOI] [PubMed] [Google Scholar]
- 10. Chun R., Fan H. 1994. Recovery of glycosylated gag virus from mice infected with a glycosylated gag-negative mutant of Moloney murine leukemia virus. J. Biomed. Sci. 1:218–223 [DOI] [PubMed] [Google Scholar]
- 11. Corbin A., Prats A. C., Darlix J. L., Sitbon M. 1994. A nonstructural gag-encoded glycoprotein precursor is necessary for efficient spreading and pathogenesis of murine leukemia viruses. J. Virol. 68:3857–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Low A., Datta S., Kuznetsov Y., Jahid S., Kothari N., McPherson A., Fan H. 2007. Mutation in the glycosylated gag protein of murine leukemia virus results in reduced in vivo infectivity and a novel defect in viral budding or release. J. Virol. 81:3685–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuznetsov Y. G., Datta S., Kothari N. H., Greenwood A., Fan H., McPherson A. 2002. Atomic force microscopy investigation of fibroblasts infected with wild-type and mutant murine leukemia virus (MuLV). Biophys. J. 83:3665–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nitta T., Kuznetsov Y., McPherson A., Fan H. 2010. Murine leukemia virus glycosylated Gag (gPr80gag) facilitates interferon-sensitive virus release through lipid rafts. Proc. Natl. Acad. Sci. U. S. A. 107:1190–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pizzato M. 2010. MLV glycosylated-Gag is an infectivity factor that rescues Nef-deficient HIV-1. Proc. Natl. Acad. Sci. U. S. A. 107:9364–9369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pfannenschmid F., Wimmer V. C., Rios R. M., Geimer S., Krockel U., Leiherer A., Haller K., Nemcova Y., Mages W. 2003. Chlamydomonas DIP13 and human NA14: a new class of proteins associated with microtubule structures is involved in cell division. J. Cell Sci. 116:1449–1462 [DOI] [PubMed] [Google Scholar]
- 17. Ramos-Morales F., Infante C., Fedriani C., Bornens M., Rios R. M. 1998. NA14 is a novel nuclear autoantigen with a coiled-coil domain. J. Biol. Chem. 273:1634–1639 [DOI] [PubMed] [Google Scholar]
- 18. Errico A., Claudiani P., D’Addio M., Rugarli E. I. 2004. Spastin interacts with the centrosomal protein NA14, and is enriched in the spindle pole, the midbody and the distal axon. Hum. Mol. Genet. 13:2121–2132 [DOI] [PubMed] [Google Scholar]
- 19. Yang K., Shi H. X., Liu X. Y., Shan Y. F., Wei B., Chen S., Wang C. 2009. TRIM21 is essential to sustain IFN regulatory factor 3 activation during antiviral response. J. Immunol. 182:3782–3792 [DOI] [PubMed] [Google Scholar]
- 20. Higgs R., Ni Gabhann J., Ben Larbi N., Breen E. P., Fitzgerald K. A., Jefferies C. A. 2008. The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J. Immunol. 181:1780–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yoshimi R., Chang T. H., Wang H., Atsumi T., Morse H. C., III, Ozato K. 2009. Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-kappaB-dependent cytokine expression in fibroblasts. J. Immunol. 182:7527–7538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wolin S. L., Cedervall T. 2002. The La protein. Annu. Rev. Biochem. 71:375–403 [DOI] [PubMed] [Google Scholar]
- 23. Maraia R. J., Intine R. V. 2001. Recognition of nascent RNA by the human La antigen: conserved and divergent features of structure and function. Mol. Cell. Biol. 21:367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raha T., Pudi R., Das S., Shaila M. S. 2004. Leader RNA of Rinderpest virus binds specifically with cellular La protein: a possible role in virus replication. Virus Res. 104:101–109 [DOI] [PubMed] [Google Scholar]
- 25. Ali N., Siddiqui A. 1997. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc. Natl. Acad. Sci. U. S. A. 94:2249–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meerovitch K., Svitkin Y. V., Lee H. S., Lejbkowicz F., Kenan D. J., Chan E. K., Agol V. I., Keene J. D., Sonenberg N. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 67:3798–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Svitkin Y. V., Pause A., Sonenberg N. 1994. La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J. Virol. 68:7001–7007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ray P. S., Das S. 2002. La autoantigen is required for the internal ribosome entry site-mediated translation of Coxsackievirus B3 RNA. Nucleic Acids Res. 30:4500–4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bitko V., Musiyenko A., Bayfield M. A., Maraia R. J., Barik S. 2008. Cellular La protein shields nonsegmented negative-strand RNA viral leader RNA from RIG-I and enhances virus growth by diverse mechanisms. J. Virol. 82:7977–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vashist S., Anantpadma M., Sharma H., Vrati S. 2009. La protein binds the predicted loop structures in the 3′ non-coding region of Japanese encephalitis virus genome: role in virus replication. J. Gen. Virol. 90:1343–1352 [DOI] [PubMed] [Google Scholar]
- 31. Tanaka M., Tanji K., Niida M., Kamitani T. 2010. Dynamic movements of Ro52 cytoplasmic bodies along microtubules. Histochem. Cell Biol. 133:273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Espinosa A., Oke V., Elfving A., Nyberg F., Covacu R., Wahren-Herlenius M. 2008. The autoantigen Ro52 is an E3 ligase resident in the cytoplasm but enters the nucleus upon cellular exposure to nitric oxide. Exp. Cell Res. 314:3605–3613 [DOI] [PubMed] [Google Scholar]
- 33. Craig A. W., Svitkin Y. V., Lee H. S., Belsham G. J., Sonenberg N. 1997. The La autoantigen contains a dimerization domain that is essential for enhancing translation. Mol. Cell. Biol. 17:163–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Levin J. G., Grimley P. M., Ramseur J. M., Berezesky I. K. 1974. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J. Virol. 14:152–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Muriaux D., Mirro J., Harvin D., Rein A. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. U. S. A. 98:5246–5251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pillemer E. A., Kooistra D. A., Witte O. N., Weissman I. L. 1986. Monoclonal antibody to the amino-terminal L sequence of murine leukemia virus glycosylated gag polyproteins demonstrates their unusual orientation in the cell membrane. J. Virol. 57:413–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Demirov D. G., Freed E. O. 2004. Retrovirus budding. Virus Res. 106:87–102 [DOI] [PubMed] [Google Scholar]
- 38. Pelchen-Matthews A., Kramer B., Marsh M. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162:443–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sherer N. M., Lehmann M. J., Jimenez-Soto L. F., Ingmundson A., Horner S. M., Cicchetti G., Allen P. G., Pypaert M., Cunningham J. M., Mothes W. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4:785–801 [DOI] [PubMed] [Google Scholar]
- 40. Morita E., Sundquist W. I. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395–425 [DOI] [PubMed] [Google Scholar]
- 41. Yuan B., Li X., Goff S. P. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700–4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Segura-Morales C., Pescia C., Chatellard-Causse C., Sadoul R., Bertrand E., Basyuk E. 2005. Tsg101 and Alix interact with murine leukemia virus Gag and cooperate with Nedd4 ubiquitin ligases during budding. J. Biol. Chem. 280:27004–27012 [DOI] [PubMed] [Google Scholar]
- 43. Neil S. J., Zang T., Bieniasz P. D. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430 [DOI] [PubMed] [Google Scholar]
- 44. Maeda N., Inoshima Y., Fruman D. A., Brachmann S. M., Fan H. 2003. Transformation of mouse fibroblasts by Jaagsiekte sheep retrovirus envelope does not require phosphatidylinositol 3-kinase. J. Virol. 77:9951–9959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mueller-Lantzsch N., Fan H. 1976. Monospecific immunoprecipitation of murine leukemia virus polyribosomes: identification of p30 protein-specific messenger RNA. Cell 9:579–588 [DOI] [PubMed] [Google Scholar]
- 46. Mann R., Mulligan R. C., Baltimore D. 1983. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell 33:153–159 [DOI] [PubMed] [Google Scholar]