Abstract
An oncogenic mutation (G49A:E17K) in the AKT1 gene has been described recently in human breast, colon and ovarian cancers. The low frequency of this mutation and perhaps other selective pressures have prevented the isolation of human cancer cell lines that harbor this mutation thereby limiting functional analysis. Here we create a physiologic in vitro model to study the effects of this mutation by employing somatic cell gene targeting using the nontumorigenic human breast epithelial cell line, MCF10A. Surprisingly, knock in of E17K into the AKT1 gene had minimal phenotypic consequences and importantly, did not recapitulate the biochemical and growth characteristics seen with somatic cell knock in of PIK3CA hotspot mutations. These results suggest that mutations in critical genes within the PI3K pathway are not functionally equivalent, and that other cooperative genetic events may be necessary to achieve oncogenic PI3K pathway activation in cancers that contain the AKT1 E17K mutation.
Keywords: Akt1, oncogene, knock in, gene targeting, breast epithelial cells
Introduction
The PI3-kinase (PI3K) pathway is critical in cancer biology, as activation of this pathway by genomic mutations, deletions, or amplifications is frequent in a wide variety of human malignancies. Extensive preclinical and clinical efforts are underway to target this pathway for cancer therapy. Breast cancers have one of the highest frequencies of mutational activation of the PI3K pathway. Approximately one third of breast cancers harbor activating mutations or amplification of the PIK3CA gene, which encodes the catalytic p110α subunit of PI3K. Additional breast cancers have mutation or deletion of PTEN, a lipid phosphatase that negatively regulates PI3K activity. More recently a single hotspot mutation, G49A:E17K, in the pleckstrin homology domain of AKT1 was described, predominantly in human breast tumors (Carpten et al., 2007). The reported frequency of this mutation has ranged from 1.8%–8% in previous studies (Bleeker et al., 2008; Carpten et al., 2007; Kim et al., 2008; Stemke-Hale et al., 2008). Importantly, mutant AKT1 was shown to activate PI3K pathway signaling when overexpressed in NIH3T3 cells and to transform rodent fibroblasts and bone marrow cells (Carpten et al., 2007). Most studies have found PTEN, PIK3CA, and AKT1 mutations to be mutually exclusive in individual tumors (Abubaker et al., 2007; Bleeker et al., 2008; Carpten et al., 2007; Saal et al., 2005), suggesting that mutational activation of the PI3K pathway by any one of these means is biologically equivalent. However, some studies have found a small percentage of tumors with concurrent PTEN mutation/deletion and PIK3CA mutation (Perez-Tenorio et al., 2007; Stemke-Hale et al., 2008). The frequency of such events may be higher in particular tumor types, such as endometrial cancer, and may reflect differences in tumor biology (Cohen et al., 2009; Hayes et al., 2006; Oda et al., 2005; Shoji et al., 2009).
To date, there are no identified human cell lines with an AKT1 G49A:E17K mutation (hereafter referred to as AKT1 E17K), and the effects of this mutation have not been evaluated in human breast epithelial cells. Although overexpression studies have been performed, our laboratory and others have shown that physiologic expression of oncogenes from their endogenous promoters can produce distinct phenotypes from those seen with transgenic overexpression (Arena et al., 2007a; Konishi et al., 2007b). To this end, we and others recently reported the creation of isogenic human mammary epithelial cell lines with knock in of hotspot exon 9 and exon 20 PIK3CA mutations (Di Nicolantonio et al., 2008; Gustin et al., 2009a). In our system, PIK3CA mutations caused MCF10A cells to grow in the absence of epidermal growth factor (EGF), an effect which was independent of mTOR activation, and lowered the threshold for mTOR activation by exogenous growth factors. Here we report knock in of the AKT1 E17K hotspot mutation within the same human nontumorigenic mammary epithelial cell system. In contrast to PIK3CA mutations, the AKT1 mutation does not confer growth factor independence and does not alter the threshold for mTOR activation. Our analysis establishes a physiologically relevant in vitro model system to study the effects of the AKT1 mutation on breast tumorigenesis and reveals distinct phenotypes arising from mutations in the same signaling pathway. These differences could have potential implications for future targeted therapies.
Results
Identification of AKT1 mutations in primary human breast cancers
To confirm the frequency of AKT1 E17K mutations in human breast cancers, we sequenced this region via PCR of genomic DNA and direct sequencing using 100 primary human breast cancers. Our analysis identified 3 tumors with somatic AKT1 E17K mutations, indicating an overall mutational frequency of 3% (Figure 1a and data not shown). This frequency is consistent with previously published reports (Bleeker et al., 2008; Carpten et al., 2007; Kim et al., 2008; Stemke-Hale et al., 2008). Similar to other studies, AKT1 E17K mutations were not found concurrently with oncogenic hotspot PIK3CA or PTEN mutations, suggesting that activation of the PI3K pathway by mutational activation of either AKT1 or PIK3CA is functionally equivalent. As in previous studies, AKT1 mutations were found exclusively in breast cancers that were positive for estrogen receptor α (ER) expression, demonstrating a potential selection bias for PI3K pathway activation in breast cancers that express ER.
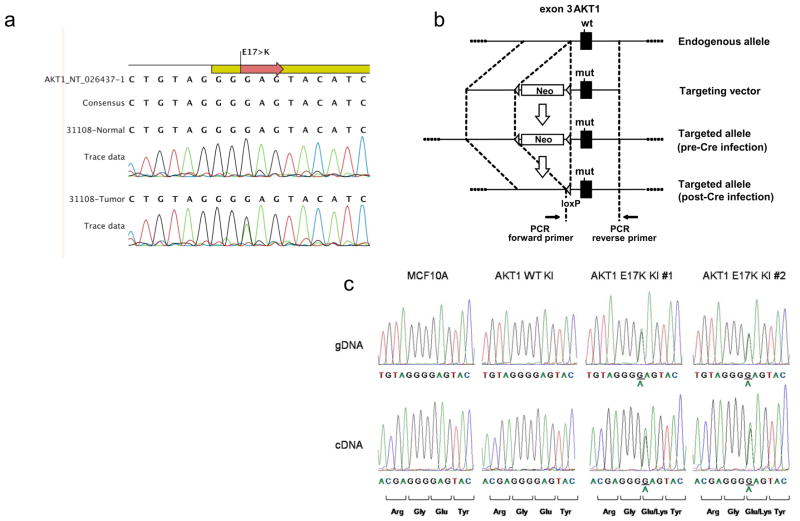
Figure 1. Resequencing for the AKT1 G49A:E17K mutation in primary human breast cancers and knock in of the AKT1 G49A:E17K mutation in MCF10A human breast epithelial cells.
(a) Representative sequencing traces from gDNA templates of matched tumor and normal tissue. (b) Design of the targeting construct. The AKT1 G49A:E17K mutation is on the 3′ homology arm, which includes all of exon 3. “Neo” signifies a cassette containing a short synthetic intron, splice acceptor, internal ribosome entry site, neomycin resistance gene, and polyadenylation signal. (c) Sequence analysis of gDNA and cDNA from parental MCF10A cells and targeted subclones.
Targeted knock in of the AKT1 E17K hotspot mutation in nontumorigenic human breast epithelial cells
Because there are no known cell lines that harbor the AKT1 E17K mutation, in vitro study of this mutation has been limited. To further analyze the effects of the AKT1 E17K mutation, we used gene targeting to create isogenic pairs of human mammary epithelial cell lines, similar to our previous work (Gustin et al., 2009b; Konishi et al., 2007a). Using the nontumorigenic MCF10A cell line, we have demonstrated that knock in of oncogenic PIK3CA mutations results in properties of transformation in vitro (Gustin et al., 2009b). Thus, in a similar fashion, we created two independent MCF10A clones with targeted knock in of the AKT1 E17K mutation. An additional clone was identified that was positive by PCR screening for gene targeting but sequencing of gDNA and cDNA showed that this clone preserved the wild-type AKT1 sequence in both alleles (Figure 1b). This presumably occurs because the DNA break during homologous recombination takes place proximal to the mutation present within the homology arm, as we have previously described (Konishi et al., 2007a). This clone serves as an ideal control since it has undergone the same process of gene targeting and single cell dilution as the two AKT1 E17K knock in clones. Sequencing of gDNA and cDNA verified that the two E17K knock in clones are heterozygous for the mutation and express the two alleles equally (Figure 1b). We also verified that there were no additional randomly integrated copies of the targeting construct in these clones via PCR and direct sequencing of genomic DNA, using primers located within the homology arm containing the E17K mutation (data not shown).
Effect of mutant Akt1 on downstream signaling pathways
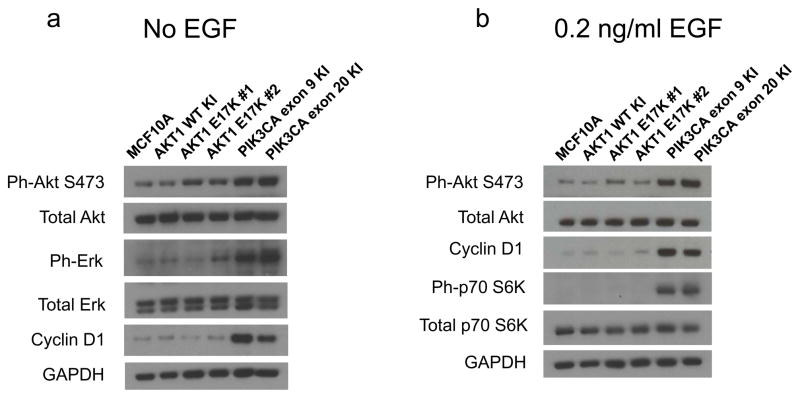
The AKT1 E17K mutation has been previously studied by overexpression in several cell types and has been shown to lead to increased phosphorylation and activation of Akt1 under conditions of serum starvation. This in turn activates downstream Akt1 phosphorylation targets (Carpten et al., 2007). We previously studied the effect of knocking in activating PIK3CA mutations on PI3K signaling in MCF10A cells (Gustin et al., 2009b). In this system, mutant PIK3CA expression led to increased phosphorylation of Akt at serine 473 in the absence of EGF. Interestingly, mutant PIK3CA also led to increased phosphorylation of Erk1 and Erk2 in the absence of EGF, indicating crosstalk between the PI3K and MAP Kinase pathways, which has been noted by others (Denley et al., 2008; Isakoff et al., 2005; Wennstrom & Downward, 1999). Additionally, we found that despite activating Akt under EGF-free conditions, mutant PIK3CA did not activate mTOR, as indicated by lack of phosphorylation of the mTOR target p70S6 Kinase (Gustin et al., 2009b). However, at physiologic EGF concentrations (0.2 ng/mL), mutant PIK3CA did lead to mTOR activation, whereas parental MCF10A cells fail to activate mTOR at this concentration of EGF.
To test whether AKT1 E17K results in a phenotype identical to oncogenic PIK3CA mutations, we subjected AKT1 E17K knock in cells to similar analyses. AKT1 E17K knock in clones show increased phosphorylation of Akt at serine 473 in the absence of EGF, although to a lesser extent than the PIK3CA mutant knock in cells (Figure 2a and Supplementary Information Figure S1). In contrast, Akt1 mutant cells do not show any appreciable increase in phosphorylation of Erk1 and Erk2 compared to PIK3CA knock in clones (Figure 2a). Moreover, despite increased constitutive activation of Akt1, these cells do not show downstream consequences of increased Akt signaling. For example, levels of cyclin D1 are not increased, in contrast with elevated cyclin D1 seen in PIK3CA mutant knock in clones. Cyclin D1 levels are regulated by glycogen synthase kinase beta (GSK3β), which is a direct phosphorylation target of Akt. Compared to PIK3CA mutant cells, AKT1 E17K cells showed a minimal increase in phospho-GSK3β serine 9 (Supplementary Information, Figure S1). In addition, mTOR activation is not increased in AKT1 E17K cells. Using physiologic EGF concentrations (0.2 ng/ml), PIK3CA mutant cells demonstrate p70S6 Kinase phosphorylation, a substrate of activated mTOR. However, AKT1 E17K mutant cells do not have increased p70S6 Kinase phosphorylation, similar to parental and wild type control cells (Figure 2b). In addition, with 0.2 ng/ml EGF there is only a slight increase in Akt S473 phosphorylation in the AKT1 E17K mutant clones relative to parental MCF10A and wild type knock in controls, but substantially less than that seen in mutant PIK3CA knock in clones. Consistent with this relative lack of signaling, Akt1 mutant cells show low levels of cyclin D1, which is comparable to controls in 0.2 ng/mL EGF, despite active cell proliferation. This is again in contrast to PIK3CA mutant cells where increased levels of cyclin D1 are readily apparent relative to controls.
Figure 2. AKT1 and PIK3CA mutants differentially affect PI3K and MAPK signaling.
Immunoblot analysis of parental MCF10A cells and their isogenic AKT1 and PIK3CA knock in derivatives under (a) EGF-free conditions and (b) physiologic 0.2 ng/mL EGF growth conditions.
Mutant Akt1 does not confer growth factor independence, colony formation, or abnormal acini morphology in Matrigel
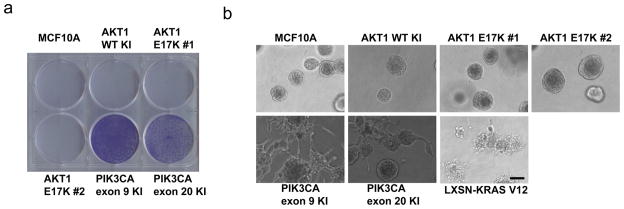
A hallmark of oncogenes is their ability to confer growth factor independence in culture and colony formation in soft agar when expressed in nontumorigenic cell lines. MCF10A cells are strictly dependent on exogenous EGF for growth. In contrast to isogenic MCF10A cells with targeted knock in of PIK3CA exon 9 and exon 20 hotspot mutations, AKT1 E17K knock in clones remained fully dependent on EGF for cell proliferation in culture. EGF dependence for growth was indistinguishable from parental MCF10A cells and wild type AKT1 knock in control cells across a range of EGF concentrations as assessed by cell proliferation assays and crystal violet staining (Figure 3a and data not shown). In addition, we previously found that knock in of activating oncogenic KRAS or PIK3CA mutations in MCF10A cells was not sufficient for formation of colonies in soft agar, despite the fact that overexpressed mutant K-ras did result in colony formation (Gustin et al., 2009b; Konishi et al., 2007a). Consistent with these data, colony formation in soft agar was not observed in either AKT1 E17K knock in clones, or any of the control cell lines (data not shown).
Figure 3. AKT1 mutation does not affect EGF independent growth or acinar morphogenesis.
(a) Equal numbers of parental MCF10A cells and derivatives with knock in of wild type or mutant AKT1, or exon 9 or exon 20 PIK3CA mutations were seeded in 6-well plates in EGF-free growth medium with 1% charcoal dextran treated FBS. Media was replaced daily. Plates were stained with crystal violet after 6 days. A representative experiment is shown. (b) MCF10A cells and knock in derivatives, as well as KrasV12 overexpressing derivatives, were seeded on growth factor-reduced Matrigel in medium containing 20 ng/mL EGF, (Bar = 100 μM).
Human mammary epithelial cells, including MCF10A, are known to form acini when cultured in three-dimensional Matrigel. In addition, previous studies have shown that over-expression of oncogenes in human breast epithelial cells can lead to abnormal acini formation when cultured under these conditions (Debnath et al., 2003b; Isakoff et al., 2005). We have previously described that mutant PIK3CA knock in cells do not form aberrant acini when grown in Matrigel (Gustin et al., 2009b). However, we have recently discovered that varying the levels of EGF used in these assays can alter the morphology of acini in mutant PIK3CA cells relative to control cell lines. Using EGF at a concentration of 20 ng/ml, mutant PIK3CA knock in cells displayed abnormal projections from acini after two weeks in culture (Figure 3b). In contrast, AKT1 mutant MCF10A derivatives and control cell lines formed typical acini, irrespective of the concentration of EGF used (20 ng/mL or 0.2 ng/mL). As a positive control cell line, MCF10A cells overexpressing K-rasV12 formed abnormal structures under both sets of growth conditions, as previously described (Konishi et al., 2007a).
Mutant Akt1 does not affect proliferative responses to estrogen and tamoxifen
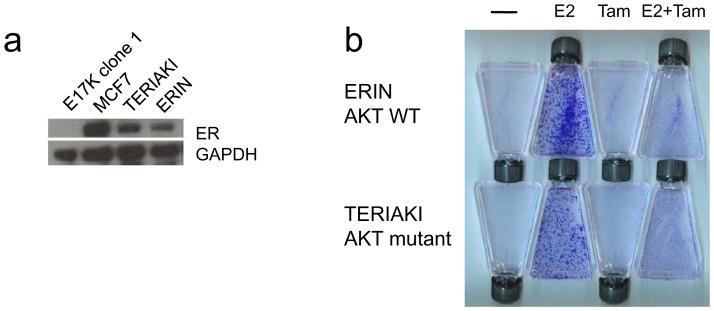
Although our work demonstrates a minimal phenotype for AKT1 E17K in nontumorigenic human breast epithelial cells, this could be due to the fact that most if not all mutations of AKT1 E17K have been found in ER-positive breast cancers (Carpten et al., 2007; Stemke-Hale et al., 2008), yet MCF10A are known to be ER-negative. However, human cancer cell line models are unavailable to study the potential interaction of this mutation and ER signaling, since no human cell lines with this mutation have been identified. Additionally, the majority of ER-positive human breast cancer cell lines currently available already contain known genetic alterations of the PI3K pathway including PIK3CA mutations or PTEN loss (COSMIC Sanger database). To address this issue, we took advantage of a physiologic model of ER signaling in nontumorigenic MCF10A cells developed in our laboratory (Abukhdeir et al., 2006). ERIN cells are MCF10A derivatives that express physiologic levels of ER and proliferate and activate transcription of ER target genes in response to estrogen. Therefore, we stably expressed the same ER construct in an AKT1 E17K knock in clone to generate TERIAKI cells (transgenic ER in AKT1 knock in). Western blot revealed high levels of ER expression in MCF7, ERIN, and TERIAKI cells (Figure 4a). To assess the growth response to ER signaling, we employed growth assays previously used with ERIN cells (Abukhdeir et al., 2006). ERIN and TERIAKI cells were seeded in phenol red-free medium with charcoal dextran stripped serum and no EGF, which led to cell cycle arrest of both ERIN and TERIAKI cells. 17 β-estradiol, 4-hydroxy-tamoxifen, or both were then added to cells, changing the medium daily until the cells were stained with crystal violet to assess cell proliferation and viability. Quantitative cell counting experiments were also performed to assess dose-response of estrogen-induced proliferation and growth-inhibition by tamoxifen (Supplementary Information, Figure S2). As demonstrated in Figures 4B and S2, both ERIN and TERIAKI cells were growth-stimulated by 17 β-estradiol, which was effectively blocked by the addition of 4-hydroxy-tamoxifen. Mutant AKT1 did not lead to dramatically increased cell proliferation in response to estrogen and did not alter the response to tamoxifen. These results suggest that despite the fact that AKT1 E17K mutations are found exclusively in breast cancers that are ER positive, this mutation does not appear to overtly affect ER signaling or the response to tamoxifen.
Figure 4. AKT1 mutation does not alter the proliferative response to estrogen and tamoxifen.
(a) Establishment of ER expressing MCF10A AKT1 E17K mutant cells. Immunoblot analysis of ER and GAPDH expression in MCF7, ERIN (parental MCF10A cells transfected with ER), TERIAKI (AKT1 E17K knock in MCF10A cells transfected with ER), and untransfected AKT1 E17K knock in cells. (b) Proliferative response to estrogen and tamoxifen. ERIN and TERIAKI cells were seeded in EGF-free, phenol red-free growth medium supplemented with vehicle, 17 β-estradiol (E2), tamoxifen (Tam), or both. Medium was replaced daily. Flasks were stained with crystal violet after six days in culture.
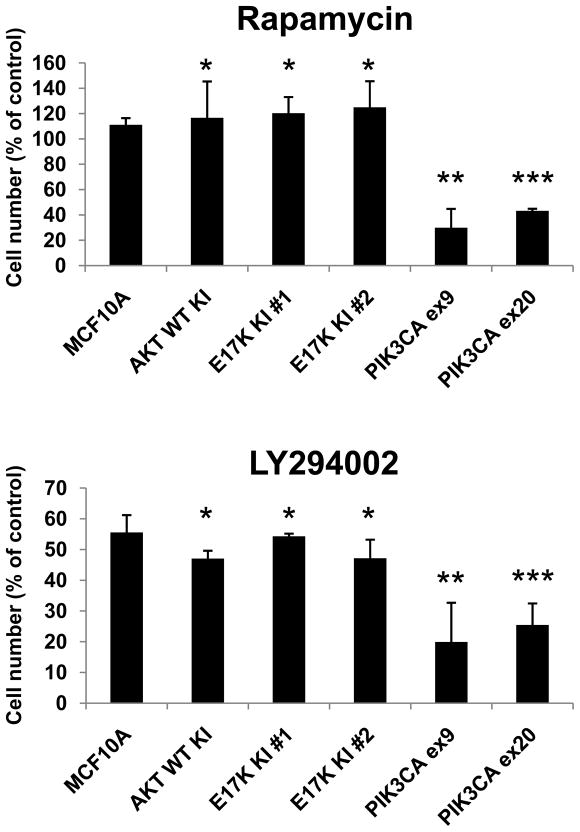
Oncogenic PIK3CA, but not AKT1 E17K, confers sensitivity to PI3K and mTOR inhibition
There is considerable interest in the potential ability of common oncogenic mutations to predict for response or resistance to both conventional chemotherapy and targeted agents. In particular one might expect activating mutations in the PI3K pathway to predict for sensitivity to mTOR inhibitors such as rapamycin. To assess whether AKT1 E17K led to increased sensitivity to pathway inhibitors, we analyzed our AKT1 knock in clones in cell proliferation assays when exposed to the mTOR inhibitor rapamycin or the PI3K inhibitor LY294002. As shown previously, PIK3CA mutant knock in clones were more sensitive to growth inhibition by rapamycin than parental MCF10A cells at physiologic EGF levels [Figure 5a and (Gustin et al., 2009b)]. However, AKT1 mutant clones were relatively resistant to rapamycin, similar to control cells. These results were consistent with the fact that AKT1 E17K clones and control cells do not activate mTOR under these growth conditions (Figure 2b). In contrast, control cells and AKT1 E17K clones did demonstrate partial growth inhibition with the PI3K inhibitor LY294002, though predictably, PIK3CA knock in cells were more sensitive than either control cells or AKT1 mutant cells [Figure 5b and (Gustin et al., 2009b)]. Interestingly, it could theoretically be hypothesized that mutant Akt1 might confer resistance to LY294002 under these conditions, because it is downstream of PIK3CA, but this did not occur. However, these results are difficult to interpret because LY294002 also targets other relevant kinases that are not bypassed by activated Akt1, as has been described (Workman, 2004). It remains to be determined whether AKT1 E17K would confer resistance to PIK3CA-specific inhibitors that are currently under development. We also tested the sensitivity of our isogenic cell line panel to the chemotherapeutic agents adriamycin and paclitaxel. Similar to the previous assays, there was no obvious difference in sensitivity to any of these agents (data not shown).
Figure 5. PIK3CA mutants, but not AKT1 mutants, display increased sensitivity to rapamycin and the PI3K inhibitor LY294002.
Cells were seeded in triplicate wells on day 0 in medium containing 0.2 ng/ml EGF. Drugs or vehicle (DMSO) were added on days 1 and 4. Cells were harvested and counted on day 6. Values are shown as percentage of vehicle control cell number and represent averages of three independent experiments with standard deviations. * p=NS compared to MCF10A. ** p < 0.02 compared to MCF10A. ***p < 0.005 compared to MCF10A.
Discussion
There has recently been great emphasis on somatic hotspot mutations in cancer as these invariant mutations represent ideal candidates for targeted therapies. The discovery of the AKT1 E17K mutation in human cancers, albeit at low frequency, is of particular interest due to Akt’s known importance in acting as a central mediator of signal transduction pathways that are altered in human malignancies. Although many human cancer cell lines exist with somatic mutations that represent the spectra and types found in primary human tissue samples, currently, there are no available cell lines with a naturally occurring AKT1 E17K mutation, though Mills and colleagues recently reported two human melanoma cell lines with AKT3 E17K mutations (Davies et al., 2008). To further study the effects of AKT1 E17K when expressed in a physiologic manner, we have created an isogenic set of nontumorigenic human breast epithelial cell lines with and without the AKT1 E17K mutation. The use of nontumorigenic cell lines has several advantages over human cancer cell lines. First, the MCF10A cell line is genetically stable as assessed by chromosomal instability analysis with FISH (Yoon et al., 2002), microsatellite and karyotype analyses (unpublished data), sequencing of p53 (Abukhdeir et al., 2006; Konishi et al., 2007a), and copy number variation analysis (manuscript submitted). This provides stronger support that gene targeted knock in clones derived from MCF10A are truly isogenic. Second, the ability to study the effects of a single oncogenic mutation can be easily observed using the various assays we have employed in past and present studies. Indeed, our previous work with mutant PIK3CA knock in clones emphasizes this fact. Third, knock in of AKT1 E17K into breast cancer cell lines would likely not yield any definitive data, as most breast cancer cell lines would be expected to already have activation of the PI3K pathway, as evidenced by the high frequency of PIK3CA mutations found in these cell lines (Bachman et al., 2004). Thus, for these reasons, MCF10A cells enable us to definitively address the phenotypic consequences of introducing oncogenic mutations via gene targeting.
Somewhat surprisingly, knock in of mutant AKT1 failed to transform cells in vitro, in contrast to observations with overexpressed mutant AKT1 (Carpten et al., 2007). This finding is similar to what we and others have observed with knock in of mutant KRAS and may reflect differences between knock in and transgenic overexpression as well as differences between cell types in their susceptibility to transformation (Arena et al., 2007b; Konishi et al., 2007a). Although only biochemical evidence of Akt activation was seen as evidenced by a slight increase in Akt phosphorylation under conditions of no EGF, there are limitations to the present study. Notably, it should be pointed out that our results are derived from a single cell line, MCF10A. Therefore the effects of AKT1 E17K with MCF10A cells, which are ER negative, may be masking a true phenotype since the AKT1 E17K mutation has only been seen in ER positive breast cancers. Arguing against this is the fact that knock in of mutant PIK3CA has dramatic effects in vitro, despite the fact that the majority of PIK3CA oncogenic mutations are also found in ER positive breast tumors. In addition, physiologic expression of ER in AKT1 E17K cells did not demonstrate any overt difference compared to ER expressing MCF10A cells with wild type AKT1. Thus, it may be that AKT1 E17K requires additional genetic events in other pathways to realize its full oncogenic potential. In support of this view is the fact that mutant PIK3CA knock in cells were shown to unexpectedly activate the MAP kinase pathway, a phenotype not observed with AKT1 E17K knock in cell lines. Thus, mutant PIK3CA activates multiple pathways simultaneously likely accounting for its transforming properties in vitro. Engineering additional oncogenic events within the AKT1 E17K knock in cell lines will allow us to address this readily testable hypothesis.
Given the lack of naturally occurring AKT1 E17K cell lines, we feel that these knock in clones will be a valuable resource for understanding the biological and clinical implications of mutant AKT1, as well as other PI3K pathway mutations and alterations found in human cancers. Although the prognostic implications of PI3K pathway mutations in breast cancer are controversial and still emerging, there has been a suggestion from the available data that AKT1 mutant cancers may have a more favorable prognosis (Stemke-Hale et al., 2008). Certainly as a class effect, AKT1 mutant cancers largely belong to the more-favorable hormone receptor-positive subset of breast cancer (Carpten et al., 2007; Stemke-Hale et al., 2008). A previous study of primary human breast cancers found that cancers with AKT1 mutations or PTEN mutations/loss, displayed high levels of in vivo Akt, mTOR, and p70S6 Kinase phosphorylation, as compared to tumors with PIK3CA mutations (Stemke-Hale et al., 2008). This result was surprising given the current body of knowledge of the PI3K pathway, and in light of in vitro data from traditional oncogene overexpression experiments as well as our own PIK3CA knock in cells. In contrast, our data suggest that the Akt1 mutant is a weaker activator of the PI3K pathway than mutant PIK3CA. However, our data do support the notion that not all PI3K pathway mutations are equivalent, a concept that has recently emerged in breast cancers as Perez-Tenorio et al. and Stemke-Hale et al. have demonstrated PIK3CA mutations with concordant PTEN loss (Perez-Tenorio et al., 2007; Stemke-Hale et al., 2008). During review of this manuscript, Fine et al. and Vasudevan et al. presented studies suggesting that differences in P-REX2a and PDK-1 activity may account for the variability seen in PI3K pathway signaling depending on the genetic lesions present within cancer cells (Fine et al., 2009; Vasudevan et al., 2009). In our isogenic system, PIK3CA mutations appear to generate a stronger proliferative signal than the AKT1 E17K mutation, conferring growth factor independence and altering the threshold for mTOR activation. In addition, mutant PIK3CA stimulates mTOR-independent growth even under EGF-free conditions, suggesting that mutant PIK3CA provides a stronger oncogenic signal, either via quantitative increases in Akt1 phosphorylation, activation of other Akt isoforms, crosstalk with the MAP Kinase pathway, and/or other Akt1-independent mechanisms.
The results presented here also have potential clinical implications involving pathway inhibitors that are being rapidly studied in clinical trials. In keeping with the quantitative and qualitative differences in signaling, PIK3CA mutations but not AKT1 E17K, increased sensitivity to the PI3K inhibitor LY294002 and the mTOR inhibitor rapamycin under appropriate growth conditions. It will be of interest to determine in larger clinical studies whether these mutations predict for differential sensitivity to various PI3K, MAP Kinase and mTOR inhibitors currently in clinical trials.
Materials and methods
Cell lines
The nontransformed human breast epithelial cell line MCF10A and its AKT1 knock in derivatives were maintained in DMEM/F12 (1:1) supplemented with 5% horse serum (HS; Invitrogen), 20ng/mL EGF, 10 μg/mL insulin, 0.5 μg/mL hydrocortisone, and 0.1 μg/mL cholera toxin, 100 U/mL penicillin and 100 μg/mL streptomycin(Invitrogen) (hereafter denoted as “supplemented DMEM/F12”). All supplements were purchased from Sigma-Aldrich unless otherwise noted. MCF10A cells with targeted knock in of the PIK3CA exon 9 E545K and exon 20 H1047R mutations were generated in our laboratory as described and were maintained in supplemented DMEM/F12 without EGF (Gustin et al., 2009b). TERIAKI cells were generated from MCF10A AKT1 E17K knock in clone #1 by stable transfection with pIRESneo-ERα as described (Abukhdeir et al., 2006). ERα-expressing MCF10A derivative ERIN9 and TERIAKI cells were maintained in phenol red-free DMEM/F12 (1:1) supplemented with 2% charcoal dextran-stripped fetal bovine serum (CD; Hyclone), 20ng/mL EGF, 10 μg/mL insulin, 0.5 μg/mL hydrocortisone, and 0.1 μg/mL cholera toxin, 100 U/mL penicillin, 100 μg/mL streptomycin(Invitrogen), and 120 μg/mL G418 (Invitrogen). All cells were cultured at 37°C at 5% CO2.
Targeted knock in of the AKT1 E17K mutation
Targeted knock in the AKT1 E17K mutation was conducted with an adeno-associated viral vector as described (Gustin et al., 2009b; Lauring et al., 2008). The targeting cassette contains a short synthetic intron, an internal ribosome entry site and a promoterless neomycin resistance gene followed by a polyadenylation signal, all flanked by loxP sites. 5′- and 3′-homology arms were constructed by high-fidelity PCR using genomic DNA (gDNA) fromMCF10A as template for the homology arms. The targeting vector was transduced into cells and antibiotic selection was done with 120 μg/mL of G418 in multiple 96-well plates. Neomycin-resistant colonies were expanded, replica-plated and pooled and PCR-based screening was performed to identify cells that had undergone homologous integration of the targeting vectors followed by further PCR screening of individual colonies from positive pools (Konishi et al., 2007c). Targeted cells were infected with an adenovirus encoding Cre recombinase to remove the selection cassette followed by single-cell dilution and screening by PCR for successful Cre recombination. Primer sequences for PCR are available on request.
gDNA and RNA extraction, cDNA synthesis, PCR, and direct sequencing
Sequencing of tissue samples was performed as previously described (Sjoblom et al., 2006). Briefly, frozen breast cancer tissues and adjacent normal controls were subjected to laser capture microdissection and genomic DNA was extracted using QIAamp DNA Blood Kits (Qiagen). For cell lines, gDNA and total RNA were prepared from cells using QIAamp DNA Blood kits and RNeasy kits (Qiagen), respectively. cDNA was synthesized with First-Strand cDNA Synthesis kits (GE Biosciences). PCR amplification was done using GeneAmp 9700 (Applied Biosystems) and Platinum Taq polymerase (Invitrogen). PCR primers to amplify cDNA were designed so that forward and reverse primers were located indistinct exons. Automated direct sequencing of PCR products was carried out by the Johns Hopkins DNA Synthesis and Sequencing Facility. Primer sequences for PCR and direct sequencing are available on request.
Immunoblotting
Whole-cell protein extracts prepared in Laemmli sample buffer were resolved by SDS-PAGE using NuPage gels (Invitrogen), transferred to Invitrolon polyvinylidene difluoride membranes (Invitrogen), and probed with primary and horseradish peroxidase–conjugated secondary antibodies. The primary antibodies used in this study are anti-p44/p42 mitogen-activated protein kinase (MAPK) rabbit antibody, anti–phospho-p44/p42 MAPK (Thr202/Tyr204) mouse antibody, anti-AKT rabbit antibody, anti–phospho-AKT (Ser473) rabbit antibody, anti-cyclin D1, anti-p70 S6K, anti-phospho-p70 S6K, anti-phospho-GSK3β (Ser9) (all from Cell Signaling Technology), anti-estrogen receptor alpha mouse monoclonal antibody (Beckman Coulter), and anti–glyceraldehyde-3-phosphatedehydrogenase (GAPDH) mouse monoclonal antibody (6C5; Abcam). Blots were exposed to Kodak XAR film using chemiluminescence for detection (Perkin Elmer).
Growth assays
MCF10A cells and their derivative clones were plated at low confluency in assay medium consisting of supplemented DMEM/F12 with 1% CD serum without EGF for 7 days, changing media daily until the assay endpoint. ERIN9 and TERIAKI cells were grown in phenol red-free supplemented DMEM/F12 with 2% CD-FBS without EGF, to which 10 nM 17-β-estradiol, ethanol, 1 μM 4-OH-tamoxifen, or combinations of these drugs were added, for 14 days. Media were replaced daily until the time of harvest. Cell counts were obtained using a Vi-Cell automated counter (Beckman Coulter). To visualize differences in cell proliferation, cells were stained with crystal violet.
Colony formation assay in semisolid medium and acinar morphogenesis assay
For colony formation assays, 3 × 104 exponentially growing cells were cast in 3 mL of top layer medium composed of supplemented DMEM/F12 and 0.4% UltraPure agarose (Invitrogen) and poured on top of a 2 mL bottom layer containing 0.6% agarose in six-well tissue culture plates. Supplemented DMEM/F12 was added to the wells once a week. Two independent experiments were done in triplicate. Morphogenesis assays were carried out as described previously (Debnath et al., 2003a), except that EGF was supplemented to the medium at a final concentration of either 0.2 ng/mL or 20 ng/mL. Plates were viewed after 2 weeks of incubation with an Eclipse TE2000-S microscope using a CFI Plan Fluor DL 4x/0.17 lens (Nikon, Melville, NY). Images were acquired using a SPOT RT KE monochrome cooled CCD camera and SPOT version4.0.1 software (Diagnostic Instruments, Sterling Heights, MI).
Drug sensitivity assays
Cells were seeded in triplicate in supplemented DMEM/F12 medium containing 1% CD serum at 2.2×104 cells per well in 6-well plates. The following day fresh medium was added containing rapamycin 1 nM, LY294002 10 μM, or an equivalent volume of DMSO (always less than 1:5000 dilution). Fresh medium and drug was added on day 4 and cells were counted on day 6 with a Z1 Cell and Particle Counter (Beckman Coulter).
Statistical analysis
Student’s t tests were calculated with Excel version 2007 software (Microsoft, Redmond, WA) using an unpaired 2-tailed analysis. A P value less than .05 was considered statistically significant.
Supplementary Material
Acknowledgments
J.L. receives support from the Flight Attendant Medical Research Institute FAMRI), the V Foundation, and the Maryland Cigarette Restitution Fund, Breast Cancer SPORE grant #PO CA088843-08, and the AVON Foundation. D.C. receives support from National Institutes of Health Grant T32 CA09071–27 and a Young Investigator Award from the American Society of Clinical Oncology Foundation. J. P. Gustin is a recipient of a Department of Defense Breast Cancer Research Program Predoctoral Fellowship Award W81XWH-06–1-0325. J. P. Garay is a recipient of a Research Supplement to Promote Diversity in Health-Related Research. H.K. is a recipient of a Young Clinical Scientist Award from FAMRI. A.M.A is supported by a Susan G. Komen Foundation Postdoctoral Fellowship Award. G.W. is a recipient of a Medical Scientist Training Program Fellowship. M.M. is recipient of a Department of Defense Breast Cancer Predoctoral Training Award (BC083057). B.H.P. acknowledges support from the Avon Foundation, Susan G. Komen for the Cure, the Mary Kay Ash Charitable Foundation, the Stewart Trust Fund, National Institutes of Health Grants CA109274 and CA88843, FAMRI, and the Breast Cancer Research Foundation.
Footnotes
Conflict of interest statement
B.H.P. has received research funding from GlaxoSmithKline in the past, though none of the studies presented here were supported by GlaxoSmithKline. B.H.P. is also a consultant for GlaxoSmithKline and Horizon Discovery LTD. Under separate licensing agreements between Genzyme Corporation and The Johns Hopkins University, B.V. is entitled to a share of royalties received by the University on sales of products, though none described in this work. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies.
References
- Abubaker J, Bavi PP, Al-Harbi S, Siraj AK, Al-Dayel F, Uddin S, Al-Kuraya K. Leukemia. 2007;21:2368–70. doi: 10.1038/sj.leu.2404873. [DOI] [PubMed] [Google Scholar]
- Abukhdeir AM, Blair BG, Brenner K, Karakas B, Konishi H, Lim J, Sahasranaman V, Huang Y, Keen J, Davidson N, Vitolo MI, Bachman KE, Park BH. Breast Cancer Res Treat. 2006;99:23–33. doi: 10.1007/s10549-006-9177-0. [DOI] [PubMed] [Google Scholar]
- Arena S, Isella C, Martini M, de Marco A, Medico E, Bardelli A. Cancer Res. 2007a;67:8468–76. doi: 10.1158/0008-5472.CAN-07-1126. [DOI] [PubMed] [Google Scholar]
- Arena S, Isella C, Martini M, de Marco A, Medico E, Bardelli A. Cancer Res. 2007b;67:8468–8476. doi: 10.1158/0008-5472.CAN-07-1126. [DOI] [PubMed] [Google Scholar]
- Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair BG, Lin C, Peters BA, Velculescu VE, Park BH. Cancer Biol Ther. 2004;3:772–5. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- Bleeker FE, Felicioni L, Buttitta F, Lamba S, Cardone L, Rodolfo M, Scarpa A, Leenstra S, Frattini M, Barbareschi M, Grammastro MD, Sciarrotta MG, Zanon C, Marchetti A, Bardelli A. Oncogene. 2008 doi: 10.1038/onc.2008.170. [DOI] [PubMed] [Google Scholar]
- Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, Uhlik M, Lin A, Du J, Qian YW, Zeckner DJ, Tucker-Kellogg G, Touchman J, Patel K, Mousses S, Bittner M, Schevitz R, Lai MH, Blanchard KL, Thomas JE. Nature. 2007;448:439–44. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- Cohen Y, Shalmon B, Korach J, Barshack I, Fridman E, Rechavi G. Gynecol Oncol. 2009 doi: 10.1016/j.ygyno.2009.09.038. [DOI] [PubMed] [Google Scholar]
- Davies MA, Stemke-Hale K, Tellez C, Calderone TL, Deng W, Prieto VG, Lazar AJ, Gershenwald JE, Mills GB. Br J Cancer. 2008;99:1265–8. doi: 10.1038/sj.bjc.6604637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Methods. 2003a;30:256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Debnath J, Walker SJ, Brugge JS. J Cell Biol. 2003b;163:315–26. doi: 10.1083/jcb.200304159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denley A, Gymnopoulos M, Hart JR, Jiang H, Zhao L, Vogt PK. Methods Enzymol. 2008;438:291–305. doi: 10.1016/S0076-6879(07)38020-8. [DOI] [PubMed] [Google Scholar]
- Di Nicolantonio F, Arena S, Gallicchio M, Zecchin D, Martini M, Flonta SE, Stella GM, Lamba S, Cancelliere C, Russo M, Geuna M, Appendino G, Fantozzi R, Medico E, Bardelli A. Proc Natl Acad Sci U S A. 2008;105:20864–9. doi: 10.1073/pnas.0808757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine B, Hodakoski C, Koujak S, Su T, Saal LH, Maurer M, Hopkins B, Keniry M, Sulis ML, Mense S, Hibshoosh H, Parsons R. Science. 2009;325:1261–5. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin JP, Karakas B, Vitolo MI, Lauring J, Abukhdeir AM, Konishi H, Garay JP, Rosen M, Denmeade S, Tamaki A, Mohseni M, Wang G, Bachman KE, Park BH. Proc Natl Acad Sci U S A. 2009a doi: 10.1073/pnas.0813351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin JP, Karakas B, Weiss MB, Abukhdeir AM, Lauring J, Garay JP, Cosgrove D, Tamaki A, Konishi H, Konishi Y, Mohseni M, Wang G, Rosen DM, Denmeade SR, Higgins MJ, Vitolo MI, Bachman KE, Park BH. Proc Natl Acad Sci U S A. 2009b;106:2835–40. doi: 10.1073/pnas.0813351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes MP, Wang H, Espinal-Witter R, Douglas W, Solomon GJ, Baker SJ, Ellenson LH. Clin Cancer Res. 2006;12:5932–5. doi: 10.1158/1078-0432.CCR-06-1375. [DOI] [PubMed] [Google Scholar]
- Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, Cantley LC, Brugge JS. Cancer Res. 2005;65:10992–1000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- Kim MS, Jeong EG, Yoo NJ, Lee SH. Br J Cancer. 2008;98:1533–5. doi: 10.1038/sj.bjc.6604212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi H, Karakas B, Abukhdeir AM, Lauring J, Gustin JP, Garay JP, Konishi Y, Gallmeier E, Bachman KE, Park BH. Cancer Res. 2007a;67:8460–7. doi: 10.1158/0008-5472.CAN-07-0108. [DOI] [PubMed] [Google Scholar]
- Konishi H, Karakas B, Abukhdeir AM, Lauring J, Gustin JP, Garay JP, Konishi Y, Gallmeier E, Bachman KE, Park BH. Cancer Res. 2007b;67:8460–7. doi: 10.1158/0008-5472.CAN-07-0108. [DOI] [PubMed] [Google Scholar]
- Konishi H, Lauring J, Garay JP, Karakas B, Abukhdeir AM, Gustin JP, Konishi Y, Park BH. Nat Protoc. 2007c;2:2865–74. doi: 10.1038/nprot.2007.409. [DOI] [PubMed] [Google Scholar]
- Lauring J, Abukhdeir AM, Konishi H, Garay JP, Gustin JP, Wang Q, Arceci RJ, Matsui W, Park BH. Blood. 2008;111:856–64. doi: 10.1182/blood-2007-05-088674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Stokoe D, Taketani Y, McCormick F. Cancer Res. 2005;65:10669–73. doi: 10.1158/0008-5472.CAN-05-2620. [DOI] [PubMed] [Google Scholar]
- Perez-Tenorio G, Alkhori L, Olsson B, Waltersson MA, Nordenskjold B, Rutqvist LE, Skoog L, Stal O. Clin Cancer Res. 2007;13:3577–84. doi: 10.1158/1078-0432.CCR-06-1609. [DOI] [PubMed] [Google Scholar]
- Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmstrom PO, Mansukhani M, Enoksson J, Hibshoosh H, Borg A, Parsons R. Cancer Res. 2005;65:2554–9. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- Shoji K, Oda K, Nakagawa S, Hosokawa S, Nagae G, Uehara Y, Sone K, Miyamoto Y, Hiraike H, Hiraike-Wada O, Nei T, Kawana K, Kuramoto H, Aburatani H, Yano T, Taketani Y. Br J Cancer. 2009;101:145–8. doi: 10.1038/sj.bjc.6605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo W-L, Davies M, Carey M, Hu Z, Guan Y, Sahin A, Symmans WF, Pusztai L, Nolden LK, Horlings H, Berns K, Hung M-C, van de Vijver MJ, Valero V, Gray JW, Bernards R, Mills GB, Hennessy BT. Cancer Res. 2008;68:6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, Hennessy BT, Tseng H, Pochanard P, Kim SY, Dunn IF, Schinzel AC, Sandy P, Hoersch S, Sheng Q, Gupta PB, Boehm JS, Reiling JH, Silver S, Lu Y, Stemke-Hale K, Dutta B, Joy C, Sahin AA, Gonzalez-Angulo AM, Lluch A, Rameh LE, Jacks T, Root DE, Lander ES, Mills GB, Hahn WC, Sellers WR, Garraway LA. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennstrom S, Downward J. Mol Cell Biol. 1999;19:4279–88. doi: 10.1128/mcb.19.6.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman P. Biochemical Society Transactions. 2004;32:393–396. doi: 10.1042/bst0320393. [DOI] [PubMed] [Google Scholar]
- Yoon DS, Wersto RP, Zhou W, Chrest FJ, Garrett ES, Kwon TK, Gabrielson E. Am J Pathol. 2002;161:391–7. doi: 10.1016/S0002-9440(10)64194-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.