Abstract
Telomerase is tightly regulated in humans relative to mice, owing to the differential regulation of TERT genes. To explore hTERT regulation in vivo, we engineered mice with a 160-kb transgenic bacterial artificial chromosome (BAC) spanning the hTERT locus with a Renilla luciferase (Rluc) cassette downstream of its promoter. Analysis of multiple founder lines revealed that the Rluc expression profile from the transgenic hTERT reporter locus reproduced that of the native hTERT gene in all tissues and organs examined, demonstrating that genetic sequence determined the species-specific developmental regulation of the hTERT gene and that mouse epigenetic and transcription machineries faithfully regulated hTERT transcription. Thus, these mice allowed detailed analyses of developmental hTERT regulation. Both the transgenic hTERT reporter and the endogenous mTERT locus were expressed in early embryonic stages, and their mRNA levels progressively decreased throughout embryonic and postnatal development. Whereas hTERT transcription was much lower than mTERT expression in most organs, it increased significantly during postnatal development of thymus, testis, and ovary. In testis, the Rluc mRNA was enriched in elongating spermatids of seminiferous tubules. In addition, the transcription of transgenic hTERT reporter, but surprisingly not the endogenous mTERT gene, was activated during Wnt1-induced mammary tumorigenesis, allowing the monitoring of tumor development via noninvasive bioluminescent imaging. Collectively, our results demonstrate that the hTERT transgenic reporter system recapitulates the developmental regulation of the hTERT gene in a chromosomal position-independent manner and serves as a legitimate model to explore telomerase regulation in the development of normal and neoplastic tissues in vivo.—Jia, W., Wang, S., Horner, J. W., Wang, N., Wang, H., Gunther, E. J., DePinho, R. A., Zhu, J. A BAC transgenic reporter recapitulates in vivo regulation of human telomerase reverse transcriptase in development and tumorigenesis.
Keywords: transgenic mouse, hTERT gene, mammary tumor, bioluminescent imaging
Telomeres serve as essential protective caps of linear chromosomal ends in all eukaryotic cells, and their maintenance enables limitless proliferative potential. The telomeric TTAGGG repeats are replenished by telomerase (1), a ribonucleoprotein complex that consists of a catalytic subunit (TERT; refs. 2–4), an RNA template (TERC; ref. 5), dyskerin (6), and other accessory proteins (7). Telomerase activity is controlled on a number of levels (8) and, in many normal and cancer cells, tracks closely with TERT gene expression, supporting the view that TERT transcriptional control is a major factor in the regulation of telomerase activity (9). In both humans and mice, the TERT gene is expressed during embryonic development but down-regulated in adult tissues (10, 11). In adult human tissues, telomerase is either below detection or expressed at extremely low levels (12), with the exception of some highly proliferative tissues, such as activated lymphocytes (13). Consequently, low telomerase activity in somatic cells results in telomere attrition on successive divisions, culminating in replicative senescence, as telomere reserves are depleted and uncap on critical shortening. TERT mRNA and telomerase activity are strongly up-regulated in >90% of cancers and have been proposed to be biomarkers of a broad spectrum of cancers (14). Indeed, limitless proliferation potential engendered by telomerase reactivation is regarded as a hallmark of cancer (15).
Because of its critical role in cell proliferation and survival, telomerase-directed telomere maintenance is important for the development of many tissues and organs (16–18). For example, telomerase is highly expressed in pluripotent stem cells (PSCs), including embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs) (19, 20), and some somatic stem cells (21). The hTERT gene undergoes repression as stem cells differentiate into adult somatic cells (22). In addition, telomerase was also shown to be crucial for germline cell development and spermatogenesis (17, 23). However, the regulation of telomerase, particularly on the level of TERT transcriptional control, during in vivo development remains poorly understood.
Transcriptional regulation of the hTERT promoter has been a subject of intense investigation. Available evidence indicates that hTERT expression is regulated by both positive and negative control mechanisms (24). Experiments utilizing transient transfection assays of reporter constructs have led to the identification of a core region that is seemingly sufficient for maximum hTERT promoter activity (25). Yet additional studies demonstrated that transiently transfected hTERT promoters were highly active and did not recapitulate the endogenous promoter in some cell types, suggesting that distal regulatory elements and/or chromatin environment of the native hTERT locus were required for proper regulation of hTERT transcription (9, 26).
Laboratory mice provide faithful models of human development and diseases, including aging and cancer, but cross-species differences in humans and mice can be exploited to provide vital insights into the genetic regulation of various biological processes. One important interspecies difference is telomerase regulation and telomere length control. Specifically, whereas human cells have relatively short telomeres (<15 kb), and telomere shortening/uncapping is a prominent mechanism of cellular senescence in humans, mouse cells have much longer telomeres (50–150 kb) and do not exhibit telomere-mediated replicative aging (27). Unlike in humans, telomerase expression in mice is less restricted, with many tissues expressing a moderate level of mTERT mRNA (10, 11, 28, 29). This difference may contribute to more ample telomere reserves in mouse cells and to some of the distinct characteristics of tumor development in mice vs. humans (30), providing opportunities for dissecting these differences and understanding human telomerase regulation.
Therefore, we generated transgenic mouse lines using a bacterial artificial chromosome (BAC) containing the hTERT locus. Our experiments revealed that that the human-specific hTERT expression was determined in a chromosomal position-independent manner by cis elements within the BAC construct. The regulation of hTERT expression was markedly different from that of the mTERT gene during normal mouse development and mammary tumorigenesis. The transgenic lines generated in this study are invaluable tools for studying telomerase regulation under physiological and pathological conditions.
MATERIALS AND METHODS
Generation of transgenic mice
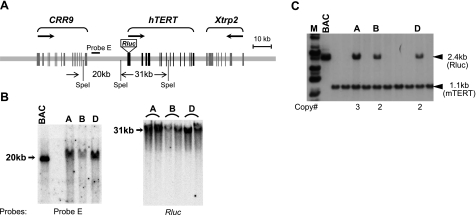
The BAC clone RPCI11-117B23, containing 158 kb of human genomic sequence encompassing 3 loci, CRR9, hTERT, and Xtrp2, was modified as shown in Fig. 1A. A reporter cassette containing a codon-optimized Renilla reniformis luciferase (Rluc) ORF and a poly(A) signal from phRL-TK (Promega, Madison, WI, USA) was inserted into the hTERT ATG codon as we previously described (31). BAC DNA (50 μg), linearized with homing endonuclease PI-Sce I, was microinjected into the pronuclei of fertilized eggs of FVB/NJ background. Five founder lines (A to E) were generated, as detected by Southern blot analysis of genomic DNAs from tail biopsies. Integrity of the transgenic BAC construct was determined by restriction enzyme digestion and pulsed-field gel electrophoresis, followed by Southern blot analysis using probes against various regions, including the CRR9, hTERT, and Xtrp2 loci and both ends of the BAC clone (representative Southern blots are shown in Fig. 1B, C).
Figure 1.
Generation of transgenic hTERT reporter mice. A) Schematic illustration of the BAC reporter. BAC clone RPCI11–117B23 was used, in which an Rluc cassette was inserted at the hTERT ATG codon. Exons are designated as vertical lines. Arrows indicate transcriptional directions of the CRR9, hTERT, and Xtrp2 genes. B) Analysis of transgenic reporter locus. Top panel: diagram of the BAC reporter. Bottom panel: characterization of transgenic founders. Genomic DNAs were digested with SpeI and subjected to pulsed-field gel electrophoresis, followed by Southern blotting hybridized to probe E (47) and Rluc ORF. C) Identification of transgenic founder lines. Tail genomic DNAs were digested with PvuII and analyzed by Southern blot analysis using a mixture of Rluc and mTERT promoter probes. Numbers at bottom are copy numbers calculated based on the relative intensities of Rluc and mTERT fragments. BAC, BAC construct; M, molecular mass marker.
Gene expression analysis measured by qRT-PCR
Total RNAs were extracted from mouse tissues, human tissues (Penn State Cancer Institute Tissue Bank), mouse embryonic fibroblasts (MEFs), mouse iPSCs (clone III-9; ref. 20), human normal foreskin fibroblasts (NHF), and human embryonic stem cell (hESC) H1 line using Tripure isolation reagent (Roche Diagnostics, Mannheim, Germany). Total RNAs (0.8 μg) were reverse transcribed with a primer mix of oligo dT and random hexamers (8:1 ratio) using the SuperScript System (Invitrogen, Carlsbad, CA, USA). Quantitative PCR (Taqman) analyses were performed in triplicates using FastStart Master Mix (Roche), and data were normalized to 18S ribosomal RNA. Primer and probe sequences are provided in Table 1.
Table 1.
Primer and probe sequences for qRT-PCR
| RNA | Sequence, 5′ to 3′ |
|---|---|
| mTERT mRNA | |
| Probe | TTCAGGCCTACAGGTTCCATGCATGT |
| Forward | TTCTAGACTTGCAGGTGAACAGCC |
| Reverse | TTCCTAACACGCTGGTCAAAGGGA |
| mTERC RNA | |
| Probe | TTTTCTCGCTGACTTCCAGC |
| Forward | GCTGTGGGTTCTGGTCTTTT |
| Reverse | CTGCAGGTCTGGACTTTCCT |
| hTERT mRNA | |
| Probe | TGGATTTGCAGGTGAACAGCCTCCA |
| Forward | GAACATGCGTCGCAAACTCTTTGG |
| Reverse | TGCAGCAGGAGGATCTTGTAGATG |
| Rluc mRNA | |
| Probe | CGACCAACTTCTGCAGCTTAAGTTCG |
| Forward | GCTTGATTCTTCTGACACAACAG |
| Reverse | AGTGGACACCCAGTGCCT |
| 18S rRNA | |
| Probe | AGCAATAACAGGTCTGTGATG |
| Forward | TAGAGGGACAAGTGGCGTTC |
| Reverse | CGCTGAGCCAGTCAGTGT |
Mammary tumor induction
All transgenic mouse experiments were performed in FVB/NJ inbred background. To induce the expression of transgenic Wnt1 oncogene, triple-transgenic female mice (MTB/TWNT/hTERT) were fed with mouse chow containing 2 g/kg doxycycline (Dox; Bio-Serv, Frenchtown, NJ, USA). To accelerate mammary tumor development, a 1-time dose of 2.5 mg N-methyl-N-nitrosourea (MNU), an alkylating agent, was administered intraperitoneally into each triple-transgenic female 1 wk after starting the Dox diet. In most cases, palpable mammary tumors are developed within 5 wk (32).
Luciferase imaging
For in vivo bioluminescent imaging (BLI), mice were transiently anesthetized by intraperitoneally administering 0.1 ml of a solution containing 10 mg/ml ketamine and 2 mg/ml xylazine. Whole-mouse BLI of Rluc and Firefly luciferase (Fluc) was acquired using the Xenogen IVIS 50 Imaging System (Xenogen Corp., Alameda, CA, USA), following tail-vein injection of 50 μg coelenteranzine/100 μl (Prolume, La Jolla, CA, USA) and intraperitoneal (i.p.) injection of 150 μg d-luciferin/100 μl (Prolume), respectively. Because the signal of Rluc decays much faster than that of Fluc, sequential BLI was performed first for Rluc on injection of coelenteranzine, followed by Fluc BLI on subsequent injection of d-luciferin.
RNA in situ hybridization
To generate riboprobes, a 468-bp BamHI-StuI mTERT cDNA fragment and a 459-bp EcoRV-NotI Rluc fragment from phRL-TK (Promega) were cloned into pBluescript SK(−), generating pSK-mTERT and pSK-Rluc, respectively. Antisense and sense riboprobes were synthesized from linearized plasmids using T3 or T7 RNA polymerases and digoxigenin (DIG) RNA labeling kit (Roche). Whole-mount in situ hybridization was performed as described previously by Nissim et al. (33). Briefly, mouse embryos [embryonic day (E)9.5 and E10.5] were fixed in 4% paraformaldehyde, permeabilized with 5 μg/ml proteinase K, and postfixed in 4% paraformaldehyde. Following hybridization with riboprobes, embryos were incubated overnight at 4°C with alkaline phosphatase (AP)-conjugated anti-DIG Fab fragments (1:2500; Roche). Color detection was performed with BM purple AP substrate (Roche). RNA in situ hybridization of fresh cryosections was performed as described previously (34) using the same riboprobes.
RESULTS
Generation of transgenic hTERT reporter mice
To study the in vivo regulation of the hTERT gene, we set out to generate transgenic mouse lines using a BAC clone containing the hTERT locus. This BAC also contains neighboring genes, CRR9 and Xtrp2. The CRR9 gene is expressed abundantly and uniformly in all tissues and cells examined (35), whereas the Xtrp2 gene is expressed exclusively in kidney (36). Given the sizable genomic region spanning the hTERT gene, we hypothesized that this BAC clone contained the cis regulatory elements required for hTERT regulation. To facilitate analysis of the hTERT gene expression, an Rluc expression cassette was inserted into, and thus disabled, the hTERT initiation codon, designated reporter 117B23-tR (Fig. 1A). Linearized 117B23-tR was injected into fertilized FVB/NJ oocytes, producing 5 founders. On the basis of restriction enzyme digestion, pulsed-field gel electrophoresis, and Southern blot analyses, 3 founder lines (A, B, and D) were shown to contain no detectable rearrangements of the transgenic BAC (Fig. 1B). The integrity of the transgenic BAC construct in these lines was verified by conventional restriction enzyme digestion and Southern blot analysis using probes within the 5′ intergenic region of hTERT, as well as its introns and exons. The transgene copy number in each line was determined by comparing the intensities of bands derived from the transgenic BAC with the endogenous mTERT locus on Southern blots. A, B, and D lines were found to contain 3, 2, and 2 copies of the transgenic BAC reporter, respectively (Fig. 1C). Although A and D lines were chosen to be used in most of the subsequent experiments, available data showed that all three lines had very similar Rluc expression patterns.
Expression profile of the transgenic hTERT promoter in adult tissues
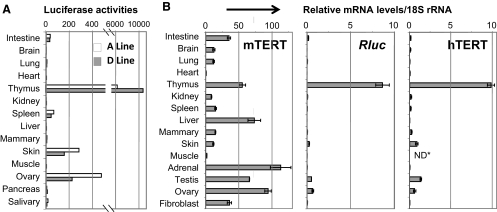
The promoter activity of transgenic hTERT reporters, as reflected by Rluc activity, was determined in various organs and tissues of adult A-line and D-line mice (Fig. 2A). Among the tissues examined, thymus contained the highest level of Rluc activity, whereas skin and ovary expressed modest levels of luciferase activity. To compare the tissue expression patterns of the transgenic hTERT and endogenous mTERT promoters, the mRNA levels of Rluc and mTERT were determined by qRT-PCR analyses. As shown in Fig. 2B, while mTERT mRNA was readily detected in the majority of mouse tissues examined (left panel), Rluc expression was very restricted (middle panel), with a level highest in thymus and detectable in skin, testis, and ovary. This expression profile closely mirrored the endogenous hTERT expression in human tissues (Fig. 2B, right panel). Thus, these results demonstrated that regulation of the transgenic BAC reporter recapitulated that of the native hTERT gene in humans.
Figure 2.
Expression of transgenic hTERT reporters. A) Luciferase expression in adult tissues. Rluc activities in 2-mo-old A-line and D-line mice were normalized to the amounts of total proteins. B) mRNA expression in adult tissues. Total RNAs were extracted from tissues of multiple 2- to 4-mo-old D-line mice and normal human tissues. mRNA expressions were determined by qRT-PCR in triplicates, normalized to 18S rRNA. mTERT and Rluc mRNA levels in left and middle panels are expressed as percentage of those in iPSC clone III-9. Right panel shows relative levels of hTERT mRNA. *ND, not determined.
Developmental silencing of the transgenic hTERT promoter
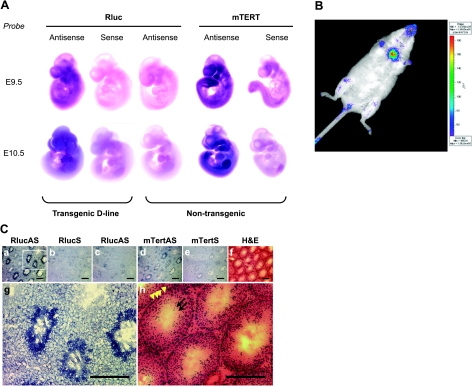
To determine the regulation of the transgenic hTERT promoter in the context of normal development, expression of the Rluc reporter in embryos was determined by whole-mount RNA in situ hybridization using digoxigenin-labeled riboprobes. As shown in Fig. 3A, the Rluc message was uniformly expressed in the majority of tissues in E9.5 transgenic embryos. In E10.5 embryos, where both forelimb and hind limb buds became noticeable, the Rluc expression began to concentrate in the head/neck area, as well as in forelimb buds. These expression signals, as detected by the Rluc antisense riboprobe, were specific because the sense riboprobe derived from the same DNA sequence did not hybridize to transgenic embryos. As expected, the same antisense riboprobe did not produce significant signals in nontransgenic embryos. The mTERT antisense riboprobe also detected uniform signal in many tissues of E9.5 embryos, with significantly stronger signals in the head and tail. In E10.5 embryos, this signal became more localized to the neck region/branchial bars, both forelimb and hind limb buds, and the tail tips. This embryonic mTERT expression pattern aligns well with that reported by Martin-Rivera et al. (11). In addition, we also conducted an in vivo BLI analysis on adult animals to monitor the transgenic hTERT promoter activity. As shown in Fig. 3B, the BLI signal of Rluc in a transgenic animal was concentrated at the anatomic position of the thymus, as well as the legs, consistent with transgene reporter expression in bone marrow cells. In contrast, the rest of the body showed low or no BLI signal, indicating the restricted expression in adult tissues (Fig. 3B).
Figure 3.
In vivo expression of the transgenic hTERT reporter. A) Whole-mount in situ RNA hybridization of Rluc and mTERT mRNAs in developing embryos. E9.5 and E10.5 transgenic (D-line) and wild-type embryos were hybridized to antisense and sense riboprobes of Rluc and mTERT, as indicated. B) Whole-body BLI detection of Rluc expression in a D-line adult animal. C) Reporter expression in testis by RNA in situ hybridization. All panels except c are consecutive cryosections of a testis from a D-line male; panel c is from a nontransgenic wild-type mouse. a–e) RNA in situ hybridization using antisense (a, c, d) and sense (b, e) riboprobes of Rluc (a–c) and mTERT (d, e). g, h) Magnified images of brackets in a (g) and f (h). Dotted lines outline one seminiferous tubule. Arrowheads indicate basal layer of spermatogonial stem cells; arrows demarcate elongating spermatids. Weak Rluc signals around feet, rectal area, and oral nasal area likely resulted from autofluorescence of dried urine. Scale bars = 200 μm.
To understand the kinetics of hTERT repression during development, RNAs were harvested from tissues and organs at different developmental stages, starting from embryos of E14.5, a time when most internal organs are well formed, to adult stage. Levels of mRNA expression from the transgenic hTERT promoter and endogenous mTERT locus were determined by qRT-PCR (Fig. 4). To directly compare the Rluc and mTERT expression levels, data were shown as percentages of those in iPSC clone III-9 derived from MEFs of D-line transgenic mice following retroviral transduction of the four reprogramming factors, c-Myc, Oct4, Sox2, and Klf4 (20). We have previously shown that transcription of both the reporter and endogenous mTERT gene was reactivated in these iPSCs (20).
Figure 4.
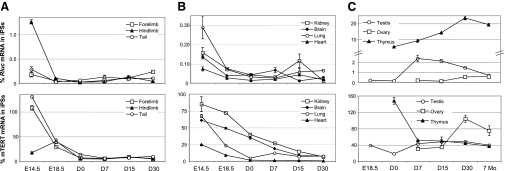
Developmental regulation of the transgenic hTERT and endogenous mTERT promoters. Total RNAs were isolated from tissues and organs at different developmental stages. Levels of Rluc and mTERT mRNAs were measured by qRT-PCR, normalized to 18S rRNA. Data are shown as percentages of those in iPSC clone III-9. A) Limb and tail tips. B) Kidney, brain, lung, and heart. C) Testis, ovary, and thymus.
As shown in Fig. 4, the Rluc mRNA levels in almost all tissues of E14.5 embryos were <1% of that in iPSCs, except for thymus, which was too small to be unambiguously identified at this stage. Starting from E18 to adult stage, the Rluc expression further decreased to <0.1% of that in iPSCs in most tissues and organs, including forelimb, hind limb, tail, kidney, brain, lung, and heart. Similarly, the mTERT expression, which was at a level ranging from 20 to 120% of that in iPSCs in various tissues of E14.5 embryos, also decreased significantly during fetal development, albeit at slower rates. Yet, most adult tissues still expressed mTERT mRNA at a level of ∼10% of that in iPSCs, with the exception of the heart, where the mTERT mRNA was barely detectable (Figs. 2B and 4B).
Tissue-specific expression of the transgenic hTERT reporter
Thymus, testis, and ovary are the three organs that expressed significant amounts of the transgenic hTERT reporter in adult mice. Expression of Rluc in thymus was readily detectable in E18 embryos and continued to increase throughout prenatal and postnatal development, whereas that of the endogenous mTERT gene decreased in neonatal mice and maintained at a significant level thereafter (Fig. 4C). Rluc expression in testis increased in neonatal mice and peaked in the second week after birth, coinciding with the start of spermatogenesis, when gonocytes resume mitosis to give rise to spermatogonia (37). On the other hand, Rluc expression in ovary only became detectable 4 wk after birth, which likely correlated with puberty on ovary stimulation.
Testes are composed of seminiferous tubules with many types of cells, including male germline cells at different stages of differentiation, from spermatogonia to mature sperms (38). To determine the specific differentiation stages at which the transgenic hTERT promoter is active, RNA in situ hybridization was performed in testes. As shown in Fig. 3Ca, g, with the antisense Rluc riboprobe, relatively weak hybridization signals were detected in all seminiferous tubules in the cryosections of a transgenic testis, whereas very intense signals were found in a subset of tubules. In contrast, the sense Rluc riboprobe did not hybridize to a consecutive cryosection of the same testis (Fig. 3Cb), and the antisense probe detected no signals from a nontransgenic testis (Fig. 3Cc), indicating that the antisense riboprobe hybridized specifically to the Rluc mRNA. An antisense, but not the sense, riboprobe of mTERT revealed a similar hybridization pattern as the Rluc probe (Figs. 3Cd, e). On the basis of the positions of hybridization signals and that only a subset of seminiferous tubules contained TERT-expressing cells, these cells were most likely elongating spermatids. Because of synchronous development of clonal units of germ cells, the sequential order of spermatogenesis in the seminiferous epithelium appears as “waves” along the length of the tubules. As a result, each seminiferous tubular section was expected to contain only spermatids at a subset of differentiation steps. This result was consistent with an earlier report that higher levels of telomerase activity, detected by in situ telomeric repeat amplification protocol (TRAP) signals, were detected in elongating spermatids at a subset of developmental steps (39).
Activation of transgenic hTERT reporter in Wnt1-initiated breast tumors
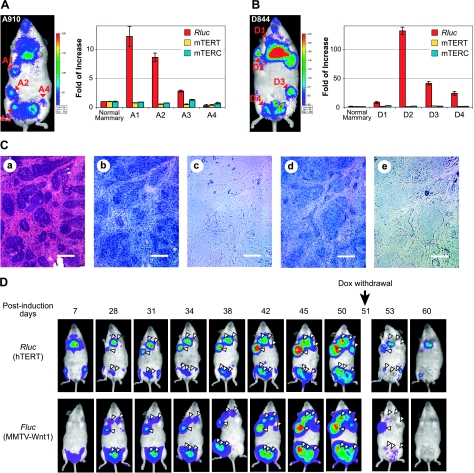
While telomerase and hTERT expression are tightly regulated in normal human somatic tissues, a large body of data showed that telomerase expression was elevated during the development of the majority of tumors. In fact, limitless proliferation resulted from telomerase activation and telomere maintenance has been regarded as one of the hallmarks of cancer (15). To determine the regulation of transgenic hTERT promoter during tumor development, we employed a Dox-dependent, Wnt1-initiated mammary tumor model (40). In this model, the Wnt1 oncogene is controlled by a Dox-regulated promoter (TetO-Wnt1 responder transgene in the TWNT line), activated via mammary-specific expression of the reverse tetracycline transactivator (rtTA) driven by the MMTV-LTR promoter in the MTB transactivator line (41). Mammary adenocarcinomas developed consistently in TWNT/MTB female mice, with a relatively long latency, following Dox treatment (40). Transgenic reporter A-line and D-line mice were crossed to TWNT/MTB mice, generating triple-transgenic female mice in 1/16 of the offspring. To reduce the latency of mammary tumor development, triple-transgenic females were treated with alkylating agent MNU 1 wk after Dox treatment, leading to the simultaneous development of multiple tumors in each female mouse (Fig. 5). Multiple BLI signals of Rluc, in addition to the one for thymus, were detected at all positions of palpable breast tumors in triple-transgenic TWNT/MTB/A or TWNT/MTB/D females (Fig. 5A, B). The levels of Rluc mRNA from most of the individually dissected tumors were significantly elevated, compared to those in normal mammary tissues. However, the degree of activation was variable among different tumors, even within the same animal, suggesting that additional tumor-specific factors contributed to hTERT transcriptional activation. The tumor-specific hTERT-promoter activation was especially strong in D-line mice. For example, the Rluc mRNA level in tumor D2 was increased by >100-fold. Overall, levels of the Rluc expression, and thus the hTERT promoter activity, were consistent with intensities of the in vivo BLI signals in all mammary tumors analyzed. On the other hand, levels of the endogenous expression of mTERT mRNA and mTERC RNA, encoding the two essential subunits of mouse telomerase, did not alter in any of the tumors examined. Therefore, transcription of the transgenic hTERT reporter, but not that of the mTERT or mTERC genes, was activated during the development of Wnt1-initiated mammary tumors. This result further highlighted the differential regulation of human and mouse telomerase genes.
Figure 5.
hTERT activation in Wnt1-induced mammary tumors. A, B) Expression of transgenic hTERT reporter in mammary tumors. MTB (MMTV-rtTA)/TWNT (TetO–Wnt1)/hTERT triple-transgenic female mice A910 (60 d old) and D844 (40 d old) were put on a diet containing 2 mg/ml Dox and subjected to MNU treatment 7 d later. A910 (A) and D844 (B) mice were imaged at 36 and 46 d after MNU treatment, respectively, and tumors were dissected on the same days. Left panels: whole-body BLI of Rluc expression. Right panels: expression of Rluc and mTERT mRNAs, as well as mTERC in mammary tumors. Total RNAs were extracted from tumor biopsies and nearby normal mammary tissues. RNA levels were determined by qRT-PCR, normalized to 18S rRNA. Data are shown as fold of increase as compared to normal mammary tissue. C) RNA in situ hybridization of D2 tumor shown in B. H&E staining, riboprobes used (a), Rluc antisense (b), Rluc sense (c), mTERT antisense (d), and mTERT sense (e). Scale bars = 100 μm. D) Noninvasive imaging of mammary tumor development. MTB/TWNT/D-line triple-transgenic female mouse (81 d old) was treated with MNU after 1 wk of consuming a Dox-containing diet, followed by Dox withdrawal on d 51 postinduction. For each imaging session, the mouse was treated with 50 μg coelenteranzine in 100 μl via tail-vein injection, and Rluc images were captured, followed by IP injection of 150 μg d-luciferin and imaging of Fluc expression from the Wnt1 transgene. Triangles indicate positions of mammary tumors. Shift of the Fluc image on d 38 was from a slight movement of the mouse position during imaging. Weak Rluc signals around feet, rectal area, and oral nasal area likely resulted from autofluorescence of dried urine.
To identify the cells in which the transgenic hTERT promoter was activated, RNA in situ hybridization was performed using cryosections of tumor D2 (Fig. 5C). Although the signal intensities were variable across each tumor section, the Rluc expression was detected in the majority of tumor cells by an antisense, but not a sense, Rluc riboprobe. Similarly, the mTERT expression was also detected, albeit at a lower level, in a consecutive cryosection of tumor D2, by an antisense mTERT riboprobe. Thus, transcription from both hTERT and mTERT occurred in the majority of breast tumor cells, but not in stromal cells within the tumors.
Noninvasive imaging of hTERT reporter during mammary tumor development
Because telomerase activation occurs in many types of cancer, we hypothesized that the BLI could be used for noninvasive imaging of hTERT transcription and for monitoring in vivo tumor development in mouse models. To test this hypothesis, TWNT/MTB/D-line triple-transgenic female mice were supplied a Dox-containing diet, followed by MNU treatment 1 wk later. BLI was carried out throughout the entire process of tumor development, for a period of 8 to 10 wk. The TetO-Wnt1 responder transgene in the TWNT line contained a bicistronic mRNA that encoded Fluc translated from an internal ribosome entry site downstream of the Wnt1 coding region. Hence, expression of the tumor-initiating Wnt1 oncogene could be monitored by Fluc activity (40) and used as an independent assessment for tumor progression. Figure 5D shows the results from a representative time course study on mammary tumor development in a triple-transgenic mouse. The Rluc BLI signals were initially detected only in thymus and bone marrows of thighbones (first panel at upper left). Starting at 4 wks after induction, Rluc BLI signals from individual tumors began to be noticeable, which correlated with the corresponding Fluc BLI signals. In the following days, the intensities of these tumor signals increased progressively as the sizes of tumors got larger. At 50 d after Dox induction, the triple-transgenic female mouse bore ≥7 tumors, as detected by BLI of both Rluc and Fluc, and some of these tumors were >1 cm in diameter. At this point, the Dox diet was replaced by a normal diet, since the movement of the mouse became restricted due to the large tumor burden. Within 3 d of Dox withdrawal, the sizes of tumors were reduced dramatically and so were the BLI signals of Rluc and Fluc. One week later, no palpable tumors were present, and the tumor BLI signals were significantly down-regulated. Thus, this experiment provided a proof of principle that the transgenic hTERT BAC reporter locus could serve as a BLI system for noninvasive imaging of in vivo tumor development and therapeutic responses in mouse models of cancer.
DISCUSSION
Telomerase and its regulation play important roles in cancer and aging. However, differences in the regulation of telomerase activity in humans and laboratory mice have hampered the effective use of mouse models to study human telomerase RT gene regulation. Using a large BAC construct that contains the hTERT gene and its neighboring loci, we revealed that the regulation of hTERT transcription in transgenic mice could mirror that of the native hTERT gene in human tissues. Our results support the view that cis-acting regulatory elements, rather than human-specific trans-acting factors, determine the tight regulation of hTERT gene and that mouse cells express the regulatory factors required for this regulation. Using hepatocytes from an aneuploid mouse strain with a human chromosome 21, Wilson et al. (42) revealed that, on a chromosomal scale, interspecies differences in transcriptional regulation were primarily directed by human genetic sequence and that species differences in transcription factors and epigenetic machineries played secondary roles. Our study extended this conclusion and further demonstrated that, in virtually all tissues, genetic sequences were responsible for directing hTERT transcription. Thus, understanding the mechanisms of human telomerase regulation will require the identification of human-specific cis elements of the hTERT gene.
It has been well documented that telomerase expression is much more restricted in humans than in mice. In fact, the highest level of hTERT expression was found in PSCs, including ESCs and iPSCs, consistent with the limitless proliferative potentials of these cell types. In most adult somatic cells, hTERT is expressed at a level <0.1% of that in iPSCs. As a result, differentiation of PSCs into somatic cells often leads to hTERT repression by as much as 10,000-fold (22). On the contrary, all adult mouse tissues that have been examined, with the exception of heart and skeletal muscle, expressed a moderate level of mTERT mRNA, ≥10% of that in iPSCs. Some organs/tissues expressed >50% of mTERT mRNA of that in iPSCs. Thus, the regulation of transgenic hTERT promoter in the transgenic mice is in line with all previously published results on human telomerase regulation and these transgenic mice are valid models for studying the in vivo regulation of human telomerase.
By far, thymus expresses the highest levels of hTERT mRNA in humans and Rluc reporter activity in transgenic mice. It has been previously reported that human lymphocytes contained a detectable level of telomerase activity, which increased significantly on activation of peripheral T cells (43). In our hTERT transgenic mice, both CD4+ and CD8+ T cells, as well as the double-negative CD4−/CD8− cells isolated from thymus contained very high levels of luciferase and Rluc mRNA (unpublished results). The high level of telomerase expression might be required for the functions of these lymphocytes in humans, as an extremely large number of rapid cell divisions occurred during immune responses. It remains to be determined whether the high level of hTERT transcription in thymus results from the lack of repression or the presence of T-cell-specific enhancers. Previously, Ritz et al. (44) generated a transgenic mouse reporter line using an 8-kb hTERT promoter fragment, which displayed the highest expression in testis, but not thymus. Thus, it is possible that distal regulatory elements, such as tissue-specific enhancers, absent in this 8-kb fragment, is required for hTERT expression in adult tissues.
Several transgenic mouse lines containing the hTERT locus or its promoter fragments have been published (29, 44). Among these previous studies, the hTERT expression in the transgenic line reported by Horikawa et al. (29) was the closest to the endogenous hTERT expression in humans. These researchers generated a transgenic line using a smaller BAC containing a 54-kb human genomic fragment, with an 11-kb upstream sequence, all of the hTERT exons and introns, and a 1.2-kb downstream region. In this line, testis expressed the highest level of hTERT mRNA, ∼20-fold of that in fibroblasts, similar to the testis Rluc mRNA expression in both A and D lines of our current study. Although the thymus also expressed hTERT mRNA in their transgenic mice, its expression was determined to be much lower than that in our transgenic lines. In addition, the transgenic hTERT mRNA was also detected in lung and brain, suggesting that hTERT silencing was incomplete in these organs. On the other hand, the hTERT promoter in transgenic reporter lines published by Ritz et al. (44) was repressed in most somatic tissues, similar to that in many human somatic tissues. Yet unlike its endogenous gene, the transgenic promoter was not detected in skin, ovary, and thymus (44). Thus, these previous data, together with our current study, strongly indicate that transcriptional regulation of the hTERT gene requires distal regulatory elements.
Of all of the organs and tissues, telomerase expression is arguably most important in testis and ovary, where germline cells develop, because resetting telomere lengths is essential for precisely transmitting genetic information to the next generation. In mice, spermatogenesis in seminiferous tubule is divided into 3 sequential phases: mitotic proliferation of spermatogonial cells, meiosis of spermatocytes, and spermiogenic differentiation of haploid spermatids. Following meiosis, the secondary spermatocytes differentiate into round spermatids (steps 1–7), which develop further into elongating spermatids (steps 8–12), elongated spermatids (steps 13–16), and finally mature sperms (45). Indeed, elongated spermatids have longer telomeres than earlier spermatogenic cells (39). Our RNA in situ hybridization data showed that both transgenic Rluc and endogenous mTERT mRNAs were detected throughout the seminiferous epithelium, but the strongest expression was observed in a subset of elongating spermatids. This result was in accordance with that reported by Tanemura et al. (39), who showed that in situ TRAP signals were detected in elongating spermatids (steps 9–11). Thus, activation of TERT expression and telomerase activity, and consequently telomere elongation, all occurred in differentiating spermatids, indicating that activation of TERT transcription in these cells is critical for restoration of telomere length in male germline cells. Interestingly, at these stages of spermatogenesis, the chromatin is undergoing a dramatic genome-wide change involving the replacement of most canonical histones by protamines and extensive DNA methylation (46). In mature sperms, the vast majority of genomic DNA is tightly packaged by protamines. Because the hTERT gene is embedded in a condensed chromatin domain in somatic cells (47), it is, therefore, tempting to speculate that nucleosomes may be decondensed in elongating spermatids in preparation for protamine replacement, resulting to dramatically increased TERT transcription, a process reminiscent of the nuclear reprogramming during induction of pluripotency (20).
Telomerase activation also occurs in the majority of cancer cells. It has been reported that the quantitative level of hTERT mRNA might have prognostic significance in human breast cancers (48). Indeed, we found that hTERT transcription was dramatically increased in Wnt1-initiated mammary tumorigenesis. However, the activation was not uniform, as multiple tumors within the same animal had different levels of Rluc mRNA. Furthermore, in both transgenic mouse models and human mammary tumors, the status of hTERT activation did not directly correlate with the progression of mammary tumors. Since transgenic Wnt1 expression was the tumor-initiating event that occurred in all tumors, the variable levels of hTERT promoter activation among different tumors likely resulted from subsequent tumor-specific events during tumor development, rather than directly from Wnt1 activation. Surprisingly, we also found that, while hTERT transcription was elevated in many tumors, the endogenous levels of mTERT mRNA and mTERC RNA did not alter significantly, suggesting that mouse cells contained trans-acting factors responsible for hTERT activation, but cis elements mediating this activation were not conserved in mouse genomic sequences. In human cells, such trans factors would be critical for tumorigenesis as telomeres are short and limiting. Overall, the result was consistent with the notion that mouse cells have longer telomeres that were not limiting for tumorigenesis.
Noninvasive and real-time monitoring of biochemical reactions in live animals, or molecular imaging, is playing an increasingly important role in defining molecular events in the field of cancer biology (49). The strong activation of transgenic hTERT promoter in mammary tumor cells, represented by Rluc expression, suggests that the transgenic reporter lines may be useful for noninvasive monitoring of tumor development in other mouse models by BLI. Because telomerase was shown to be activated in >90% of cancers (14), it is likely that these transgenic lines can be used to image other cancers in mice. As compared to previously reported transgenic mouse lines (32, 50), the transgenic hTERT reporters generated and utilized in this study have several features important for tumor imaging. First, hTERT activation is a tumor-specific pathological event during the development of most human cancers and may occur at distinct stages of tumorigenesis. Thus, it may be feasible to use these transgenic lines to report the in vivo transcriptional activities of hTERT promoter that are associated with the pathological states of tumors. In contrast, most published reporter expressions were driven by tissue-specific promoters and linked to specific tumor-initiating transgenes. Second, our transgenic reporter utilized Rluc to monitor hTERT transcription. Hence, these mice can be used in conjunction with other transgenic Fluc reporters, linked to tumor-inducing oncogenes, for quantitative measurement of hTERT activation and further analysis of the pathological states of tumors. Third, thymus expresses a high level of Rluc activity and thus can serve as an internal positive control for Rluc imaging. In summary, our experiments demonstrated that the transgenic hTERT reporters faithfully mimicked the regulation of telomerase in human tissues. These transgenic reporters can potentially be used to monitor tumor progression in a broad spectrum of cancer models and may provide a useful tool for preclinical drug evaluation.
Acknowledgments
The authors thank Longgui Chen for technical assistance, Aimin Liu for RNA in situ hybridization, and Kang Li for the preparation of tissue cryosections. The authors also thank the Macromolecular and Molecular Genetics Core Facilities at Penn State College of Medicine for the excellent services.
The work was supported in part by U.S. National Institutes of Health grants R21CA106470 and R01GM071725 (J.Z.), a 973 grant (2009CB941002) from the Chinese National Programs for Fundamental Research and Development (H.W.), and a Ph.D. student scholarship from the Chinese Ministry of Education (W.J.). None of the authors have professional or financial affiliations that can be perceived to bias the presentation of this work.
REFERENCES
- 1. Morin G. B. (1989) The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 59, 521–529 [DOI] [PubMed] [Google Scholar]
- 2. Lingner J., Hughes T. R., Shevchenko A., Mann M., Lundblad V., Cech T. R. (1997) Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276, 561–567 [DOI] [PubMed] [Google Scholar]
- 3. Meyerson M., Counter C. M., Eaton E. N., Ellisen L. W., Steiner P., Caddle S. D., Ziaugra L., Beijersbergen R. L., Davidoff M. J., Liu Q., Bacchetti S., Haber D. A., Weinberg R. A. (1997) hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 90, 785–795 [DOI] [PubMed] [Google Scholar]
- 4. Harrington L., McPhail T., Mar V., Zhou W., Oulton R., Bass M. B., Arruda I., Robinson M. O. (1997) A mammalian telomerase-associated protein. Science 275, 973–977 [DOI] [PubMed] [Google Scholar]
- 5. Shippen-Lentz D., Blackburn E. H. (1990) Functional evidence for an RNA template in telomerase. Science 247, 546–552 [DOI] [PubMed] [Google Scholar]
- 6. Cohen S. B., Graham M. E., Lovrecz G. O., Bache N., Robinson P. J., Reddel R. R. (2007) Protein composition of catalytically active human telomerase from immortal cells. Science 315, 1850–1853 [DOI] [PubMed] [Google Scholar]
- 7. Venteicher A. S., Abreu E. B., Meng Z., McCann K. E., Terns R. M., Veenstra T. D., Terns M. P., Artandi S. E. (2009) A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323, 644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aisner D. L., Wright W. E., Shay J. W. (2002) Telomerase regulation: not just flipping the switch. Curr. Opin. Genet. Dev. 12, 80–85 [DOI] [PubMed] [Google Scholar]
- 9. Wang S., Zhu J. (2003) Evidence for a relief of repression mechanism for activation of the human telomerase reverse transcriptase promoter. J. Biol. Chem. 278, 18842–18850 [DOI] [PubMed] [Google Scholar]
- 10. Greenberg R. A., Allsopp R. C., Chin L., Morin G. B., DePinho R. A. (1998) Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene 16, 1723–1730 [DOI] [PubMed] [Google Scholar]
- 11. Martin-Rivera L., Herrera E., Albar J. P., Blasco M. A. (1998) Expression of mouse telomerase catalytic subunit in embryos and adult tissues. Proc. Natl. Acad. Sci. U. S. A. 95, 10471–10476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wright W. E., Piatyszek M. A., Rainey W. E., Byrd W., Shay J. W. (1996) Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 18, 173–179 [DOI] [PubMed] [Google Scholar]
- 13. Broccoli D., Young J. W., de Lange T. (1995) Telomerase activity in normal and malignant hematopoietic cells. Proc. Natl. Acad. Sci. U. S. A. 92, 9082–9086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim N. W., Piatyszek M. A., Prowse K. R., Harley C. B., West M. D., Ho P. L., Coviello G. M., Wright W. E., Weinrich S. L., Shay J. W. (1994) Specific association of human telomerase activity with immortal cells and cancer [see comments]. Science 266, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 15. Hanahan D., Weinberg R. A. (2000) The hallmarks of cancer. Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 16. Blasco M. A., Lee H. W., Hande M. P., Samper E., Lansdorp P. M., DePinho R. A., Greider C. W. (1997) Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91, 25–34 [DOI] [PubMed] [Google Scholar]
- 17. Lee H. W., Blasco M. A., Gottlieb G. J., Horner J. W., 2nd, Greider C. W., DePinho R. A. (1998) Essential role of mouse telomerase in highly proliferative organs. Nature 392, 569–574 [DOI] [PubMed] [Google Scholar]
- 18. Rudolph K. L., Chang S., Lee H. W., Blasco M., Gottlieb G. J., Greider C., DePinho R. A. (1999) Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell 96, 701–712 [DOI] [PubMed] [Google Scholar]
- 19. Marion R. M., Strati K., Li H., Tejera A., Schoeftner S., Ortega S., Serrano M., Blasco M. A. (2009) Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell 4, 141–154 [DOI] [PubMed] [Google Scholar]
- 20. Mathew R., Jia W., Sharma A., Zhao Y., Clarke L. E., Cheng X., Wang H., Salli U., Vrana K. E., Robertson G. P., Zhu J., Wang S. (2010) Robust activation of the human but not mouse telomerase gene during the induction of pluripotency. FASEB J. 24, 2702–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sharpless N. E., DePinho R. A. (2004) Telomeres, stem cells, senescence, and cancer. J. Clin. Invest. 113, 160–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang S., Hu C., Zhu J. (2007) Transcriptional silencing of a novel hTERT reporter locus during in vitro differentiation of mouse embryonic stem cells. Molec. Biol. Cell 18, 669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Coussens M., Yamazaki Y., Moisyadi S., Suganuma R., Yanagimachi R., Allsopp R. (2006) Regulation and effects of modulation of telomerase reverse transcriptase expression in primordial germ cells during development. Biol. Reprod. 75, 785–791 [DOI] [PubMed] [Google Scholar]
- 24. Zhu J., Zhao Y., Wang S. (2010) Chromatin and epigenetic regulation of the telomerase reverse transcriptase gene. Protein Cell 1, 22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu K. J., Grandori C., Amacker M., Simon-Vermot N., Polack A., Lingner J., Dalla-Favera R. (1999) Direct activation of TERT transcription by c-MYC. Nat. Genet. 21, 220–224 [DOI] [PubMed] [Google Scholar]
- 26. Ducrest A. L., Amacker M., Mathieu Y. D., Cuthbert A. P., Trott D. A., Newbold R. F., Nabholz M., Lingner J. (2001) Regulation of human telomerase activity: repression by normal chromosome 3 abolishes nuclear telomerase reverse transcriptase transcripts but does not affect c-Myc activity. Cancer Res. 61, 7594–7602 [PubMed] [Google Scholar]
- 27. Forsyth N. R., Elder F. F., Shay J. W., Wright W. E. (2005) Lagomorphs (rabbits, pikas and hares) do not use telomere-directed replicative aging in vitro. Mech. Ageing Dev. 126, 685–691 [DOI] [PubMed] [Google Scholar]
- 28. Prowse K. R., Greider C. W. (1995) Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc. Natl. Acad. Sci. U. S. A. 92, 4818–4822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Horikawa I., Chiang Y. J., Patterson T., Feigenbaum L., Leem S. H., Michishita E., Larionov V., Hodes R. J., Barrett J. C. (2005) Differential cis-regulation of human versus mouse TERT gene expression in vivo: identification of a human-specific repressive element. Proc. Natl. Acad. Sci. U. S. A. 102, 18437–18442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. DePinho R. A. (2000) The age of cancer. Nature 408, 248–254 [DOI] [PubMed] [Google Scholar]
- 31. Wang S., Zhao Y., Leiby M., Zhu J. (2009) A new positive/negative selection scheme for precise BAC recombineering. Mol. Biotechnol. 42, 110–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gestl S. A., Leonard T. L., Biddle J. L., Debies M. T., Gunther E. J. (2007) Dormant Wnt-initiated mammary cancer can participate in reconstituting functional mammary glands. Mol. Cell. Biol. 27, 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nissim S., Allard P., Bandyopadhyay A., Harfe B. D., Tabin C. J. (2007) Characterization of a novel ectodermal signaling center regulating Tbx2 and Shh in the vertebrate limb. Dev. Biol. 304, 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schaeren-Wiemers N., Gerfin-Moser A. (1993) A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry 100, 431–440 [DOI] [PubMed] [Google Scholar]
- 35. Wang S., Robertson G. P., Zhu J. (2004) A novel human homologue of Drosophila polycomblike gene is up-regulated in multiple cancers. Gene 343, 69–78 [DOI] [PubMed] [Google Scholar]
- 36. Nash S. R., Giros B., Kingsmore S. F., Kim K. M., el-Mestikawy S., Dong Q., Fumagalli F., Seldin M. F., Caron M. G. (1998) Cloning, gene structure and genomic localization of an orphan transporter from mouse kidney with six alternatively-spliced isoforms. Recept. Channels 6, 113–128 [PubMed] [Google Scholar]
- 37. De Rooij D. G. (1998) Stem cells in the testis. Int. J. Exp. Pathol. 79, 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hess R. A. (1998) Spermatogenesis overview. In Encyclopedia of Reproduction, Vol. 4 (Knobil E. ed) pp. 539–545, Academic Press, San Diego, CA, USA [Google Scholar]
- 39. Tanemura K., Ogura A., Cheong C., Gotoh H., Matsumoto K., Sato E., Hayashi Y., Lee H. W., Kondo T. (2005) Dynamic rearrangement of telomeres during spermatogenesis in mice. Dev. Biol. 281, 196–207 [DOI] [PubMed] [Google Scholar]
- 40. Gunther E. J., Moody S. E., Belka G. K., Hahn K. T., Innocent N., Dugan K. D., Cardiff R. D., Chodosh L. A. (2003) Impact of p53 loss on reversal and recurrence of conditional Wnt-induced tumorigenesis. Genes Dev. 17, 488–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gunther E. J., Belka G. K., Wertheim G. B., Wang J., Hartman J. L., Boxer R. B., Chodosh L. A. (2002) A novel doxycycline-inducible system for the transgenic analysis of mammary gland biology. FASEB J. 16, 283–292 [DOI] [PubMed] [Google Scholar]
- 42. Wilson M. D., Barbosa-Morais N. L., Schmidt D., Conboy C. M., Vanes L., Tybulewicz V. L., Fisher E. M., Tavare S., Odom D. T. (2008) Species-specific transcription in mice carrying human chromosome 21. Science 322, 434–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weng N. P. (2008) Telomere and adaptive immunity. Mech. Ageing Dev. 129, 60–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ritz J. M., Kuhle O., Riethdorf S., Sipos B., Deppert W., Englert C., Gunes C. (2005) A novel transgenic mouse model reveals humanlike regulation of an 8-kbp human TERT gene promoter fragment in normal and tumor tissues. Cancer Res. 65, 1187–1196 [DOI] [PubMed] [Google Scholar]
- 45. Russell L. D., Ettlin R. A., Sinha Hikim A. P., Clegg E. D. (1990) Histological and Histopathological Evaluation of the Testis, Cache River Press, Clearwater, FL, USA [Google Scholar]
- 46. Hammoud S. S., Nix D. A., Zhang H., Purwar J., Carrell D. T., Cairns B. R. (2009) Distinctive chromatin in human sperm packages genes for embryo development. Nature 460, 473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang S., Zhu J. (2004) The hTERT gene is embedded in a nuclease-resistant chromatin domain. J. Biol. Chem. 279, 55401–55410 [DOI] [PubMed] [Google Scholar]
- 48. Bieche I., Nogues C., Paradis V., Olivi M., Bedossa P., Lidereau R., Vidaud M. (2000) Quantitation of hTERT gene expression in sporadic breast tumors with a real-time reverse transcription-polymerase chain reaction assay. Clin. Cancer Res. 6, 452–459 [PubMed] [Google Scholar]
- 49. Massoud T. F., Gambhir S. S. (2003) Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 17, 545–580 [DOI] [PubMed] [Google Scholar]
- 50. Vooijs M., Jonkers J., Lyons S., Berns A. (2002) Noninvasive imaging of spontaneous retinoblastoma pathway-dependent tumors in mice. Cancer Res. 62, 1862–1867 [PubMed] [Google Scholar]