Abstract
Fyn, an Src kinase family member, acts as a negative regulator of NF-E2-related factor 2 (Nrf2). Under stressful conditions, Nrf2 translocates into the nucleus and binds to the antioxidant response element (ARE), activating defensive gene expression. Once Nrf2 completes activation, Fyn phosphorylates tyrosine 568 of Nrf2, resulting in the nuclear export and degradation of Nrf2. The present studies demonstrate that within 0.5 h of antioxidant treatment in human hepatoblastoma (HepG2) cells, Fyn exports out of the nucleus, allowing Nrf2 unimpeded movement to the ARE. Mutation of tyrosine 213 of Fyn stymied nuclear export, suggesting that tyrosine phosphorylation controls nuclear export. Mass spectrometry confirmed tyrosine 213 as the site of phosphorylation. ChIP and real-time PCR assays revealed that FynY213A mutant caused decreased binding of Nrf2 to the promoter of defensive gene NAD(P)H:quinone oxidoreductase 1 (NQO1) and decreased NQO1 expression by 5-fold (P<0.0001) compared to wild-type Fyn. In addition, a putative nuclear export signal (NES) was identified, and mutation of it also inhibited nuclear export of Fyn. Furthermore, FynY213A caused an increased susceptibility to cell death following treatment with etoposide in mouse hepatoma (Hepa-1) cells. The preinduction regulation of Nrf2 is controlled by the nuclear export of Fyn, allowing for activation of defensive gene expression.—Kaspar, J. W., Jaiswal, A. K. Tyrosine phosphorylation controls nuclear export of Fyn, allowing Nrf2 activation of cytoprotective gene expression.
Keywords: INrf2, Keap1, oxidative stress, cytoprotective gene induction, cellular protection, cell survival
Nf-e2-related factor 2 (Nrf2) is a member of the family of leucine zipper/cap'n'collar-containing nuclear factor proteins (1). Nrf2 binds to the antioxidant response element (ARE) and regulates expression and induction of many genes encoding chemopreventative proteins, including NAD(P)H:quinone oxidoreductases (NQO1 and NQO2), glutathione S-transferase Ya subunit, and heme oxygenase-1 (1). This induction involves a mechanism essential for cellular protection against oxidative and electrophilic stress and cellular survival (4). Nrf2 resides predominantly in the cytoplasm, where it interacts with actin-associated cytosolic protein, INrf2 (inhibitor of Nrf2), or Keap1 (Kelch-like ECH-associated protein 1) (2–4). The INrf2-Nrf2 complex serves as a cellular sensor of oxidative and electrophilic stress generated from endogenous reactions, exogenous chemicals, xenobiotics, drugs, and UV and ionizing radiation (1). The exposure to oxidative/electrophilic stress leads to dissociation of Nrf2 from INrf2. Nrf2 will then stabilize and translocate into the nucleus and activate the transcription of several defensive genes.
Several reports suggest that continuous accumulation of Nrf2 in the nucleus is detrimental. INrf2-null mice demonstrated persistent accumulation of Nrf2 in the nucleus that led to postnatal death from malnutrition, resulting from hyperkeratosis in the esophagus and forestomach (5). Reversed phenotype of INrf2 deficiency by breeding to Nrf2-null mice suggested that tightly regulated negative feedback might be essential for cell survival (6). Systemic analysis of the INrf2 genomic locus in human lung cancer patients and cell lines showed that deletion, insertion, and missense mutations in functionally important domains of INrf2 results in reduction of INrf2 affinity for Nrf2 and elevated expression of cytoprotective genes (7, 8). Taken together, uncontrolled activation of Nrf2 in cells increases the risk of adverse effects, including tumorigenesis. On the contrary, stress-induced activation of the Nrf2 pathway in normal cells is regulated tightly and confers cytoprotection against oxidative/electrophilic stress and carcinogens. Therefore, it is evident that cells contain multiple mechanisms that regulate cellular abundance of Nrf2.
The abundance of Nrf2 inside the nucleus is tightly controlled by positive and negative regulators that control nuclear import, binding to the ARE, nuclear export, and degradation of Nrf2 under normal and stressful conditions (9–13). Of these factors, Fyn kinase has been identified as a negative regulator of Nrf2 (14). On completion of induction of defensive genes, Fyn phosphorylates Nrf2 at tyrosine residue 568, which leads to a chromosomal region maintenance-1 (Crm-1) mediated nuclear export and degradation (16). Fyn is expressed ubiquitously across tissue types and is among 3 Src family members known to be responsive to oxidative stress (15–17). The biological functions of Fyn are diverse, including T-cell signaling, mitogenic signaling, and cell adhesion-mediated signaling (18).
Recently, we have shown that activated GSK-3β phosphorylated Fyn at threonine residues, leading to nuclear localization of Fyn (19). The nuclear localization of Fyn was a delayed response occurring between 4 and 5 h. In this report we investigated the early response of Fyn in reaction to oxidative stress and the mechanisms associated with it. We propose that Fyn will export out of the nucleus in response to oxidative stress, and the mechanism of export is associated with tyrosine phosphorylation. In this study we also examined whether the export is dependent on a nuclear export signal and the dominant negative effects a mutation of Fyn might have on downstream defensive genes and cell survival.
MATERIALS AND METHODS
Cell cultures
Human hepatoblastoma (HepG2) and mouse hepatoma (Hepa-1) cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). HepG2 cells were grown in minimum essential α-medium, and Hepa-1 was grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (40 U/ml), and streptomycin (40 μg/ml). The cells were grown in a monolayer in an incubator at 37°C in 95% air and 5% CO2.
Plasmid construction
Mouse Fyn cDNA was amplified from the image clone obtained from ATCC using the following primers: forward, 5′-GCGCTCTAGAGAATTCGTCGAGACCATGGGCTGTGTG-3′, and reverse, 5′-CGCGGATCCGATATCCAGGTTTTCACCAGGTTGGTA-3′. The PCR-amplified DNA contained XbaI and BamHI restriction sites at the 5′ and 3′ end, respectively. The amplified DNA was digested with aforementioned enzymes and subcloned into the Flag vector digested with similar enzymes. The resultant plasmid was designated as Flag-Fyn. The Flag-Fyn was amplified with forward primer 5′-GCCACCATGGGCTGTGTGCAATGT-3′ and reverse primer 5′-CAGGTTTTCACCAGGTTGGTACTG-3′ and was also subcloned into pcDNA3.1/V5-His Topo Vector by TA cloning (Invitrogen, Carlsbad, CA, USA). This plasmid encodes the V5-tagged Fyn Wild Type Protein, designated Fyn-V5. Five tyrosine residues (Y132, Y213, Y336, Y417, and Y528) present in Fyn were mutated to alanine by using a site-directed mutagenesis kit (Invitrogen). Mutant Y132A was generated by PCR using mutant forward primer 5′-ACAACTGGAGAGACAGGTGCAATTCCCAGCAAT-3′ and reverse primer 5′-ACCTGTCTCTCCAGTTGTCAAGGAGCGGGC-3′. Mutant Y213A was generated by PCR using mutant forward primer 5′-TCCACCATTGTCAAGTTTGCGAATTTTATA-3′ and reverse primer 5′-TCCACCATTGTCAAGTTTGCGAATTTTATA-3′. Mutant Y336A was generated by PCR using mutant forward primer 5′-GTGTCTGAGGAGCCCATCGCAATCGTCACCGAG-3′ and reverse primer, 5′-GATGGGCTCCTCAGACACCACTGCATAGAG-3′. Mutant Y417A was generated by PCR using mutant forward primer 5′-TTGATAGAAGACAATGAGGCAACAGCAAGACAA-3′ and reverse primer 5′-CTCATTGTCTTCTATCAATCGGGCCAATCC-3′. Mutant Y528A was generated by PCR using mutant forward primer 5′-ACCGCGACAGAGCCCCAGGCACAACCTGGTGAA-3′ and reverse primer 5′-CTGGGGCTCTGTCGCGGTAAAGTAGTCTTC-3′. These plasmids will be designated Flag-FynY132A, Flag-FynY213A, Flag-FynY336A, Flag-FynY417A, and Flag-FynY528A. FynY213A-V5 was also cloned using the above corresponding primers and Fyn-V5 as template, using a site-directed mutagenesis kit. The putative nuclear export sequence mutant was generated in a 3-step cloning process. All three sets of primers correspond to each leucine residue being mutated to alanine in the NES. First set of primers used forward primer 5′-GATGGTGAAGGAAGAGCTGCAAAGTTGCCA-3′, reverse primer 5′-AGCTCTTCCTTCACCATCTTTTAAGAAGTC-3′, and Fyn-V5 template. Subsequent sets of primers used previous sets of cDNA to maintain all leucine mutations. Second set: forward, 5′-GAAGGAAGAGCTGCAAAGGCACCAAACCTT-3′, and reverse, 5′-CTTTGCAGCTCTTCCTTCACCATCTTTTAA-3′. Third set: forward, 5′-GCTGCAAAGGCACCAAACGCAGTGGACATG-3′, and reverse, 5′-GTTTGGTGCCTTTGCAGCTCTTCCTTCACC. This plasmid will be designated as Fyn-Mutant-Nes-V5. The construction of the pCMX-Flag-Crm1, HA-Ub, and reporter plasmid human NQO1 pGL2-hARE-Luc were described previously (15, 20). All of the plasmids were confirmed by sequencing.
In vitro transcription/translation
In vitro transcription/translation of the plasmids encoding tyrosine mutations and NES mutations were performed using the TNT-coupled rabbit reticulocyte lysate system (Promega Corp., Madison, WI, USA) as described previously (20). All of the in vitro transcribed/translated proteins gave the expected size bands.
Subcellular fractionation and Western blotting
Subcellular fractionation and Western blotting were described previously (13). Antibodies used in this study were as follows: anti-Fyn (1:1000) and anti-Src(pY416) (1:1000), purchased from Cell Signaling (Danvers, MA, USA); anti-V5 HRP (1:5000) and anti-Flag HRP, purchased from Invitrogen; and anti-phosphotyrosine (1:1000) and anti-actin (1:5000), purchased from Sigma-Aldrich Corp. (St. Louis, MO, USA). For immunoprecipitations, anti-Fyn (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was used. To confirm the purity of nuclear-cytoplasmic fractionation, the membranes were reprobed with cytoplasm-specific anti-lactate dehydrogenase (LDH; Chemicon, Billerica, MA, USA) and nuclear-specific anti-lamin B antibodies (Santa Cruz). In related experiments, the cells were treated with 100 μM tert-butylhydroquinone (t-BHQ; Sigma); 20 ng/ml leptomycin B (LMB), 30 μg/ml cycloheximide (CHX), 2 μM MG132, 50 μm genistein (Calbiochem, La Jolla, CA, USA); 10 μg/ml cadmium chloride (CdCl) (Sigma), or DMSO as a vehicle for different time intervals.
Immunoprecipitation
Immunoprecipitations were performed based on methods previously described (13).
Transient transfection and luciferase assay
Methods for transient transfection and luciferase assay were described previously (13). For luciferase assays in this study, 1.0-μg samples of Fyn or FynY213A plasmids were transfected with other previously mentioned plasmids.
Real-time PCR
The real-time PCR analysis and primer/probe sequence information was previously described (13). Fyn primer and probe amplicon Mm00433373_m1 (Applied Biosystems, Foster City, CA, USA) was also used in this study.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed using kits from Active Motif (Carlsbad, CA, USA) as per protocol. Briefly, HepG2 cells were transfected with the indicated plasmid and treated with DMSO or 100 μM t-BHQ for 0.5 or 4 h and then fixed in 1% formaldehyde for 15 min. Cells were lysed, and nuclei were pelleted by centrifugation. Nuclei were resuspended and sheared using a sonicator (Misonix Inc., Farmingdale, NY, USA) with 5 pulses of 20 s at 25% of maximum output. Sheared chromatin was immunoprecipitated with 2 μg of anti-Nrf2 or control IgG antibody. The cross-links were reversed overnight at 65°C and deproteinated with 20 μg/ml proteinase K. PCR was performed with a primer pair spanning the human NQO1 gene ARE. The NQO1 ARE spanning primers and PCR procedures were described previously (13).
In-gel digestion
Coomassie-stained Fyn bands were excised, cut into ∼1- × 1-mm pieces and dehydrated with methanol for 5 min. The gel pieces were then washed as follows: 1 × 5 min with 30% methanol/70% water, 2 × 10 min with water, and 3 × 10 min with 100 mM ammonium bicarbonate (NH4HCO3)/30% acetonitrile. Gel pieces were dried in a SpeedVac (Thermo Scientific, Waltham, MA, USA). Protein disulfide bonds were reduced with 10 mM tris(hydroxypropyl)phosphine (TCEP) in 100 mM NH4HCO3 for 60 min at 56°C, followed by alkylation with 55 mM iodoacetamide in 100 mM NH4HCO3 for 45 min at room temperature in the dark. The gel pieces were washed with 100 mM NH4HCO3 for 15 min and dehydrated with acetonitrile, followed by complete drying in a SpeedVac. Gel pieces were rehydrated in trypsin solution (15 ng/μl trypsin in 50 mM NH4HCO3) on ice for 45 min. Excess trypsin solution was discarded, replaced with 50 mM NH4HCO3, and incubated overnight at 37°C. Digestion buffer was collected and saved. Peptides were extracted once with 50 mM NH4HCO3, once with acetonitrile, and twice with 5% formic acid in 50% acetonitrile; each extraction was performed by incubating at 37°C for 15 min with vortexing. All supernatants were combined, dried in a SpeedVac, and stored at −20°C before LC-MS/MS analysis.
LC-MS/MS analysis and protein identification
Reversed-phase separation of peptides was performed using a Surveyor liquid chromatography system (Thermo Scientific); solvent A: 0.1% formic acid in water; solvent B: 0.1% formic acid in acetonitrile. Peptides were loaded onto an online desalting peptide trap (Michrom Bioresources, Auburn, CA, USA) using an autosampler. A 40 min gradient from 2–40% B was then used to elute the peptides. All MS analyses were performed using an LCQ Deca mass spectrometer equipped with a nanospray ionization source (Thermo Scientific). Peptides were introduced into the mass spectrometer via a 75-μm i.d./15-μm tip i.d. C18-packed PicoFrit column (New Objective, Woburn, MA, USA). The spray voltage was 2.0 kV, and the heated capillary temperature was 200°C. MS/MS data were acquired using a top-3 data-dependent acquisition method with dynamic exclusion enabled. MS/MS spectra were searched against a mouse protein database (downloaded on December 11, 2007, from the National Center for Biotechnology Information; 88,212 sequences; http://www.ncbi.nlm.nlh.gov) using Bioworks with the SEQUEST algorithm. Peptides passing the following Xcorr vs. charge-state filter were accepted as confident identifications: +2, ≥2.5; +3, ≥3.0; +1, peptides were ignored. All mass spectrometry was done by the University of Maryland Proteomics Core Facility (Baltimore, MD, USA).
Pulse-chase assay
Hepa-1 cells were plated in 6-well plates and allowed to adhere overnight. Cells were then transfected with 500 ng of Fyn-V5/well for 24 h. Methods for pulse-chase assays have been described previously (13).
MTT cell survival assay
Hepa-1 cells were plated at a density of 5000 cells/well in 96-well plates, transfected with Fyn and FynY213A, treated with etoposide (10 μM) for 30 h, and further treated with 100μM DMSO or t-BHQ for an additional 24 h. Cells were incubated with fresh MTT solution (200 μl/well; stock 5 mg/ml in PBS) for 2 h, and absorbance at 490 nm was measured. Each experiment was repeated 3 times. Each data point represents a mean ± sd normalized to the value of the corresponding control cells.
Statistical analysis
Data from luciferase assays, MTT assays, real-time PCR, and immunoblotting band intensities were analyzed using a 2-tailed Student's t test. Data are expressed as means ± sd of 3 independent experiments. Values of P < 0.05 were considered significant; levels of significance are indicated in the figures.
RESULTS
Antioxidant and xenobiotic treatment causes nuclear export of Fyn
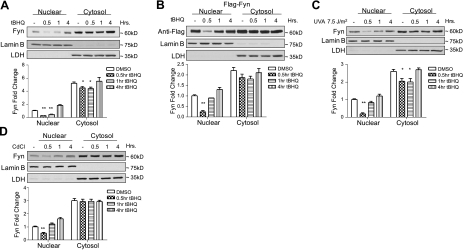
To investigate the early response of Fyn in reaction to oxidative stress, the subcellular localization of Fyn was followed by immunoblotting. Western blot analysis shows that endogenous and overexpressed Fyn exported out of the nucleus within 0.5 h of treatment (Fig. 1A, B; top panels; also see quantitative densitometry graphs, bottom panels). To determine whether the nuclear export of Fyn is inducible by other factors other than antioxidants, UVA radiation (Fig. 1C) and cadmium chloride (Fig. 1D), both of which have been shown to induce Nrf2 (21, 22), were used. In both instances, Fyn exported out of the nucleus within 0.5 h. These results suggest that in the presence of oxidative stress, Fyn exports out of the nucleus within 0.5 h to allow Nrf2 to bind to the ARE. This finding is similar to earlier reports that Fyn exported from the nucleus within 1 h following hydrogen peroxide treatment (19). Once Fyn exported out of the nucleus, a corresponding accumulation of Fyn in the cytoplasm was not seen. This finding suggests that Fyn may be degraded on nuclear export.
Figure 1.
Subcellular localization of endogenous Fyn and Flag-Fyn. HepG2 cells were treated with chemicals for varying time points and harvested, and nuclear and cytosolic extracts were prepared. Lysates were immunoblotted (top panels). Densitometry measurements of bands were quantitated and shown in graphs (bottom panels). below Anti-LDH and anti-lamin B were probed in all blots. A) Cells were treated with vehicle control (DMSO) or 100 μM t-BHQ. Endogenous Fyn was probed. B) Flag-Fyn (1 μg) was transiently transfected, cells were treated with DMSO or 100 μM t-BHQ, and anti-Flag was probed. C) Cells in 100-mm dishes with PBS were exposed to UVA for 10 s; PBS was replaced with medium, and cells were placed back in incubator for indicated time points. Endogenous Fyn was probed. D) Cells were treated with 10 μg/ml of cadmium chloride (CdCl) for indicated time points. Endogenous Fyn was probed. Comparison between vehicle control and t-BHQ treated time points shows a significant difference. *P < 0.05, **P < 0.005; 2-tailed Student's t test.
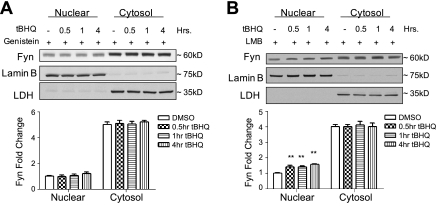
Crm-1 inhibitor and tyrosine kinase inhibitor block Fyn nuclear export
To study the means by which Fyn exports out of the nucleus, preliminary experiments were performed using inhibitors. Genistein, a tyrosine kinase inhibitor, given concurrently with t-BHQ prevented the transient decrease in nuclear Fyn protein levels (compare Fig. 2A to Fig. 1A). These results suggest that the nuclear export might be due to tyrosine phosphorylation. LMB, a specific inhibitor of proteins containing nuclear export signals (23), was also given simultaneously with t-BHQ (Fig. 2B). LMB blocked the antioxidant-induced nuclear export of endogenous Fyn. These results demonstrate that Fyn might be interacting with Crm-1 and that Fyn might contain a nuclear export sequence.
Figure 2.
Subcellular localization of endogenous Fyn with nuclear export inhibitor and tyrosine kinase inhibitor. A) HepG2 cells were pretreated with 100 μM genistein for 2 h; cells were then treated with either DMSO or 100 μM t-BHQ along with genistein for indicated time points. Lysate was immunoblotted with anti-Fyn. B) HepG2 cells were pretreated with 20 ng/ml of LMB for 2 h; cells were then treated with either 100 μM DMSO or t-BHQ along with LMB for indicated time points. Lysate was immunoblotted with anti-Fyn. Comparison between DMSO and t-BHQ treated time points shows a significant difference. **P < 0.005; 2-tailed Student's t test.
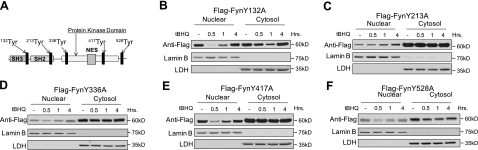
Tyrosine mutation causes nuclear accumulation of Fyn
Analysis of the mouse Fyn amino acid sequence identified 5 different putative tyrosine phosphorylation sites, shown with the different domains within the Fyn protein (Fig. 3A). Site-directed mutagenesis mutations were performed on the 5 aforementioned tyrosine residues. Subcellular localization followed by immunoblotting was performed to investigate whether any of the tyrosine residues were implicated in the nuclear export of Fyn. Fyn mutants Y132A, Y336A, Y417A, and Y528A all showed an antioxidant-mediated nuclear export at 0.5 h when exposed to t-BHQ (Fig. 3B, D–F). Interestingly, Fyn mutant Y213A was the only mutant to show nuclear accumulation in the presence of t-BHQ (Fig. 3C). These results convey that phosphorylation at tyrosine residue 213 might be required for the nuclear export of Fyn.
Figure 3.
Subcellular localization of mutant forms of Fyn. A) Schematic diagram of mouse Fyn gene showing corresponding domains, a putative nuclear export signal, and tyrosine phosphorylation sites. B–F) HepG2 cells were transfected with 1 μg of FynY132A (B), FynY213A (C), FynY336A (D), FynY417A (E), or FynY528A (F) mutant plasmid. Cells were then treated with either DMSO or 100 μM t-BHQ for indicated time points. Cells were harvested, and nuclear and cytosolic extracts were prepared. Lysates were immunoblotted with anti-Flag, anti-LDH, and anti-lamin B.
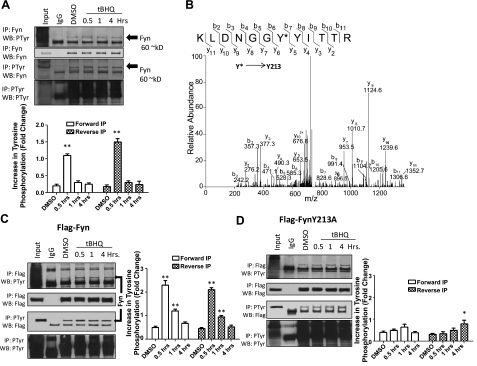
Immunoprecipitation and mass spectrometry of Fyn show tyrosine phosphorylation
Immunoprecipitation followed by immunoblotting was used to investigate tyrosine phosphorylation of endogenous Fyn, Flag-Fyn, and Flag-FynY213A. HepG2 cells were treated with t-BHQ, and immunoprecipitation was performed with anti-Fyn antibodies using only the nuclear fraction of each sample. Western blots were probed with anti-phosphotyrosine antibodies and then reprobed with anti-Fyn antibodies. Anti-Fyn antibodies immunoprecipitated phosphotyrosine proteins at basal levels as well as treated time points. However, antioxidant treatment immunoprecipitated more phosphotyrosine (Fig. 4A, top panels) at 0.5 h, suggesting that more tyrosine phosphorylation is likely taking place. The reverse immunoprecipitation confirmed an increase in interaction between endogenous Fyn and phosphotyrosine at 0.5 h (Fig. 4A, bottom panels).
Figure 4.
Mass spectrometry and immunoprecipitation of Fyn, Flag-Fyn, and Flag-FynY213A. A) HepG2 cells were treated with 100 μM DMSO or t-BHQ for indicated time points. Cells were then collected, and nuclear and cytosolic fractions were separated. Top panels: 1 mg nuclear lysate was immunoprecipitated with anti-Fyn antibody and Western blotted with anti-phosphotyrosine and anti-Fyn. Bottom panels: 1 mg nuclear lysate was immunoprecipitated with anti-phosphotyrosine antibody and Western blotted with anti-Fyn antibody. B) Mass spectroscopy of Fyn. Cells were treated with t-BHQ for 0.5 h. Cells were collected, and nuclear and cytosolic fractions were separated. Nuclear lysate was immunoprecipitated with anti-Fyn antibodies. C, D) HepG2 cells were transfected with 1 μg of Flag-Fyn (C) or Flag-FynY213A (D) and treated with 100 μM DMSO or t-BHQ for indicated time points. Cells were harvested, and nuclear and cytosolic fractions were separated. Top panels: 1 mg nuclear lysate was immunoprecipitated with anti-Flag antibody and Western blotted with anti-phosphotyrosine and anti-Flag. Bottom panels: 1 mg nuclear lysate was immunoprecipitated with anti-phosphotyrosine antibody and Western blotted with anti-Flag antibody.
Mass spectroscopy was used to confirm phosphorylation of Fyn Y213. Tandem mass spectrometry was used to confirm the phosphorylation of Fyn on Tyr213. Hepa-1 cells were treated with t-BHQ for 0.5 h, nuclear and cytosol fractions were separated, and the nuclear fraction was subjected to immunoprecipitation with anti-Fyn antibodies. Immunoprecipitated Fyn was identified by LC-MS/MS with 43% sequence coverage. A singly phosphorylated Fyn peptide (207KLDNGGYYITTR218) was identified with phosphorylation occurring at Tyr213 Xcorr = 3.83 (Fig. 4B).
In an analogous setting to the endogenous Fyn experiment, HepG2 cells were transfected with Flag-Fyn and treated with t-BHQ. As expected, immunoprecipitation with anti-Flag antibodies behaved similarly to the endogenous Fyn experiment. Anti-Flag immunoprecipitated phosphotyrosine at all time points, with an increase at the 0.5 h treatment (Fig. 4C, top panels). In the reverse experiment, immunoprecipitation with phosphotyrosine also immunoprecipitated an increase in Flag-tagged Fyn protein at the 0.5 h treatment. (Fig. 4C, bottom panels).
We then determined whether FynY213A mutant would be able to interact with any tyrosine phosphorylation. Anti-Flag antibodies did in fact immunoprecipitate phosphotyrosine protein in HepG2 cells transfected with FynY213A at all time points (Fig. 4D, top panels). Interestingly, FynY213A did not show an increase at the 0.5 h treatment time point, indicating less tyrosine phosphorylation when compared to endogenous Fyn and Flag-Fyn. In the reverse experiment, phosphotyrosine antibodies immunoprecipitated Flag tagged Fyn protein at all time points, with a decrease at 0.5 h (Fig. 4D, bottom panels) compared to the endogenous and Flag-Fyn experiments. Taken together, these results show that endogenous Fyn and Flag-Fyn show more tyrosine phosphorylation at 0.5 h than the FynY213A mutant, suggesting that the FynY213 residue is likely phosphorylated. Mass spectroscopy confirmed tyrosine residue 213 as the site of phosphorylation.
Fyn nuclear export is dependent on NES and Crm1
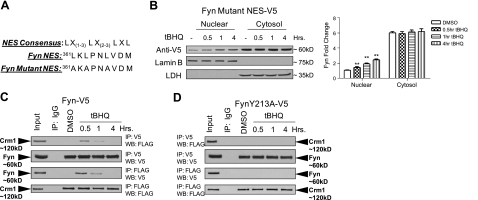
To further test our previous experiments where LMB blocked nuclear export of Fyn, we sought to investigate the possibility that Fyn might possess nuclear export signals that facilitate its nuclear export. Analysis of the Fyn amino acid sequence revealed a tentative NES, shown in Fig. 5A. When compared to a NES consensus sequence, Fyn NES exhibited a similar short leucine-rich residue motif, suggesting a NES might be present. However, the Fyn NES does not contain a fourth leucine residue as is present in other NES motifs, suggesting that the NES might be nonfunctional. Nonetheless, we mutated the leucine residues in the putative NES and investigated whether it was functional. Interestingly, t-BHQ did not induce the nuclear export of Fyn-Mutant-NES (Fig. 5B). These results indicate that Fyn translocation out of the nucleus requires the NES. Next, we examined whether Fyn-V5 and FynY213A-V5 could interact with Crm1, a nuclear exporter protein. Anti-V5 antibodies immunoprecipitated Flag-tagged Crm1 at 0.5 and 1 h (Fig. 5C, top panels). In the reverse experiment, anti-Flag antibodies immunoprecipitated Fyn-V5 at 0.5 and 1 h (Fig. 5C, bottom panels). In a similar experimental setting, cells were cotransfected with FynY213A-V5 and Flag-Crm1. Anti-V5 antibodies could not immunoprecipitate Flag-Crm1 (Fig. 5D, top panels), and anti-Flag antibodies could not immunoprecipitate Fyn-V5 (Fig. 5D, bottom panels). Together, these results show that Fyn-V5, and not FynY213A-V5, could interact with Crm1, suggesting that Fyn nuclear export is dependent on a NES interaction with Crm1.
Figure 5.
Fyn NES mutation and Crm1 interaction. A) A putative nuclear export signal was identified in Fyn as shown, consensus NES is also shown. Leucines in Fyn were mutated to alanine in Fyn Mutant NES. B) Cells were transfected with 1 μg of Fyn-Mutant-NES and then treated with either DMSO or 100 μM t-BHQ for indicated time points. Lysates were immunoblotted (left panel). Densitometry measurements of bands were quantitated and graphed (right panel). C, D) HepG2 cells were cotransfected with 1 μg of Fyn-V5 (C) or FynY213A-V5 (D) and 1 μg of Flag-Crm1 and then treated with either DMSO or 100 μM t-BHQ for indicated time points; cells were harvested. Top panels: 1 mg lysate was immunoprecipitated with anti-V5 antibody and Western blotted with anti-Flag and anti-V5. Bottom panels: 1 mg lysate was immunoprecipitated with anti-Flag antibody and Western blotted with anti-V5 and anti-Flag antibodies.
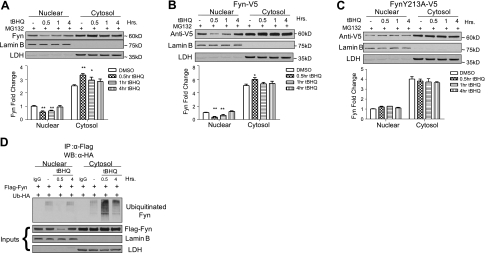
Proteasome inhibitor causes cytosolic accumulation of Fyn
To elucidate the fate of Fyn after nuclear export has taken place, experiments were performed using proteasome inhibitor MG132 and t-BHQ. After nuclear export of endogenous Fyn at 0.5 h, accumulation of Fyn in the cytosol appeared (Fig. 6A). In a similar experiment, Flag-Fyn was transfected into HepG2 cells and pretreated with proteasome inhibitor MG132 followed by t-BHQ. Flag-Fyn behaved analogously to endogenous Fyn, revealing accumulation in the cytosol at 0.5 h (Fig. 6B). Next, Flag-FynY213A was transfected into HepG2 cells and pretreated with MG132, followed by t-BHQ treatment. As expected, FynY213A failed to transport out of the nucleus and obviously did not indicate any cytosolic accumulation (Fig. 6C). The effect of MG132 seemed to mediate cytosolic accumulation of Fyn after nuclear export, suggesting that Fyn is degraded after nuclear export. To confirm that Fyn is ubiquitinated and degraded after nuclear export, an ubiquitination assay was performed. HepG2 cells were transfected with Flag-Fyn and HA-Ub plasmids. Cells were then immunoprecipitated with anti-Flag antibodies and immunoblotted with anti-HA antibodies. Cells treated with DMSO showed little ubiquitinated Fyn in the cytosol and almost undetectable levels in the nucleus (Fig. 6D). Fyn ubiquitination was highest in the cytosol after 30 min of t-BHQ treatment following nuclear export. Ubiquitination returned to near basal levels at 4 h after antioxidant treatment. Taken together, these results demonstrate that Fyn is ubiquitinated and degraded following nuclear export.
Figure 6.
Proteasome inhibitor causes cytosolic accumulation of Fyn. HepG2 cells were pretreated with 2 μM MG132 for 16 h. HepG2 cells were then treated with 100 μM t-BHQ and MG132 for indicated time points. Cells were harvested, and nuclear and cytosolic extracts were prepared. Lysates were immunoblotted (top panels). Densitometry measurements of bands were quantitated and graphed (bottom panels). A) Endogenous Fyn, anti-LDH, and anti-lamin B were probed. B) Fyn-V5 (1 μg) was transiently transfected; anti-V5, anti-LDH, and anti-lamin B were probed. C) FynY213-V5 mutant (1 μg) was transiently transfected; anti-V5, anti-LDH, and anti-lamin B were probed. D) HepG2 cells were transfected with Flag-Fyn and HA-Ub plasmids and pretreated with 2 μM MG132 for 16 h. Cells were then treated with 100 μM t-BHQ and MG132 for indicated time points. Protein was aliquoted from samples and used for inputs. Rest of the sample (1 mg) was immunoprecipitated with anti-Flag antibodies and immunoblotted with anti-HA antibodies. Comparison between DMSO and t-BHQ treated time points shows a significant difference. *P < 0.05, **P < 0.005; 2-tailed Student's t test.
FynY213A mutant represses ARE-mediated NQO1 activity resulting in cell death
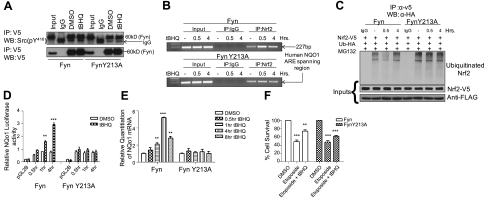
Before determining the downstream effects a mutation in Fyn would have on Nrf2 downstream genes, the activity of FynY213A was assessed. HepG2 cells were transfected with Fyn-V5 and FynY213A-V5 constructs and treated with DMSO and t-BHQ. Immunoprecipitation experiments were performed with anti-V5 antibody, followed by Western blotting using anti-Src(pY416) that recognizes Fyn tyrosine phosphorylation (Fig. 7A). The results confirmed that both Fyn and FynY213A had tyrosine kinase activity and autophosphorylated.
Figure 7.
Dominant negative effects of FynY213A mutant. A) HepG2 cells were transfected with 1 μg of Fyn-V5 and FynY213A-V5 and treated with DMSO or 100 μM t-BHQ. Lysate (1 mg) was immunoprecipitated with anti-V5 antibody and Western blotted with anti-Src(pY416). B) ChIP assay. HepG2 cells were treated with 100 μM t-BHQ for 0.5 and 4 h. Extracts were prepared as described in Materials and Methods. C) Hepa-1 cells were transfected with Flag-Fyn or Flag-FynY213A as well as Nrf2-V5 and Ub-HA, and pretreated with 2 μM MG132 for 16 h. Cells were then treated with DMSO or t-BHQ. Cell lysate (1 mg) was immunoprecipitated with anti-V5 and Western blotted with anti-HA. D) Hepa-1 cells were treated with either DMSO or 100 μM t-BHQ for indicated time points. Cells were lysed, and relative luciferase activity was measured. E) Hepa-1 cells were transfected with either Flag-Fyn or Flag-FynY213A. Cells were treated with 100 μm DMSO or t-BHQ. RNA was extracted, and quantitation of mRNA was measured. F) Cell survival assay. Absorbance was measured; data were normalized to the value of the corresponding control cells. Comparison between DMSO and t-BHQ within each transfection concentration shows a significant difference. All experiments were repeated 3 times; values are means ± sd. *P < 0.05, **P < 0.005, ***P < 0.0001; 2-tailed Student's t test.
To investigate the hypothesis that FynY213A mutant would inhibit Nrf2 binding to the ARE, we performed a ChIP assay in HepG2 cells using Nrf2 antibodies and PCR primers covering the human NQO1 region in the NQO1 promoter. The results demonstrated that in the presence of FynY213A and t-BHQ, binding of Nrf2 to the ARE was decreased at 0.5 h compared to Fyn (Fig. 7B, compare top and bottom panels). Since FynY213A cannot export out of the nucleus, Nrf2 cannot bind effectively to the ARE.
To examine the differences between the negative regulation of Fyn and FynY213A mutant on Nrf2, ubiquitination assays were performed. Hepa-1 cells were transfected with either Flag-Fyn or Flag-FynY213A and Nrf2-V5 and Ub-HA, and pretreated with MG132 and then treated with either DMSO or t-BHQ. When Fyn is overexpressed, ubiquitinated Nrf2 is lowest at 0.5 h because Fyn is exporting out of the nucleus and cannot phosphorylate Nrf2, allowing it to up-regulate cytoprotective genes (Fig. 7C). However, when FynY213A is overexpressed, Nrf2 ubiquitination at 0.5 h remains constant compared to DMSO and 4 h t-BHQ treatment. Increased Nrf2 ubiquitination at 0.5 h is due to the phosphorylation of Nrf2 by FynY213A and subsequent nuclear export and degradation of Nrf2.
Luciferase and real-time PCR assays were used in the determination of whether Fyn activity differs from FynY213A activity on t-BHQ-induced expression of ARE-mediated cytoprotective gene expression. Hepa-1 cells were cotransfected with either Fyn or FynY213A and human NQO1 pGL2-hARE-Luc. Luciferase activity of NQO1 exhibited a substantial increase in activity at 1 and 4 h with t-BHQ treatment in the presence of Fyn (Fig. 7D). However, luciferase activity of NQO1 in the presence of FynY213A revealed a decrease in activity at 1 and 4 h compared to Fyn. Similarly, NQO1 mRNA increased 2-fold at 1 h and 5-fold at 4 h when transfected with Fyn after antioxidant treatment (Fig. 7E). When Hepa-1 cells were transfected with FynY213A, NQO1 mRNA levels remained low compared to Fyn. Since FynY213A represses downstream cytoprotective genes, we examined whether FynY213A can cause cell death. When cells were transfected with Fyn and FynY213A and treated with the chemotherapeutic drug etoposide, cell survival decreased to levels near 50% (Fig. 7F). However, when t-BHQ was given concurrently with etoposide, cell survival increased in cells transfected with Fyn, but only marginal increases in cell survival were found in cells transfected with FynY213A. These results suggest that FynY213A blocks Nrf2 activation, causing more susceptibility to cellular death.
Taken together, these experiments elucidate that Nrf2 is ineffective in the presence of FynY213A and t-BHQ because the mutant can avoid nuclear export and at the same time constantly negatively regulate Nrf2 in the nucleus, interfering with defensive gene up-regulation. Nrf2 can activate defensive genes in the presence of Fyn because Fyn is able to export out of the nucleus when treated with t-BHQ. This mutation in Fyn ultimately leads to decreases in cellular survival.
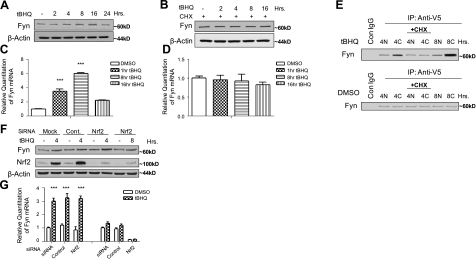
Antioxidant induction of Fyn protein expression and Fyn mRNA
Western blot analysis was used to investigate the antioxidant induction of Fyn. The t-BHQ treatment of Hepa-1 cells shows that endogenous Fyn is inducible with antioxidant treatment, reaching maximum protein levels at 16 h (Fig. 8A). Cycloheximide, a protein synthesis inhibitor, was used to investigate whether the increase in protein levels seen at 16 h was from new synthesis. As expected, cells pretreated with cycloheximide and then treated with t-BHQ showed no increase in protein expression, suggesting that the increase in protein levels is from new synthesis (Fig. 8B). To confirm the antioxidant induction of Fyn, mRNA was also measured using real-time PCR. Treatment of cells with t-BHQ for the indicated time points showed almost a 4-fold induction of Fyn mRNA at 1 h (Fig. 8C). Fyn mRNA levels increased to 6-fold at 8 h and decreased to around 2-fold at 16 h over DMSO control. In a similar experiment, preincubation with the transcription inhibitor actinomycin D blocked the t-BHQ-mediated induced expression of Fyn mRNA (Fig. 8D). Since t-BHQ is inducing Fyn protein and mRNA levels, we studied the effects of t-BHQ on the newly synthesized Fyn using pulse-chase experiments. Hepa-1 cells were metabolically labeled with 35S methionine for 1 h and chased with unlabeled methionine for 4 h and then treated with t-BHQ for indicated time points. Cytosolic and nuclear fractions were also separated. Labeled cytosolic Fyn-V5 showed an increase in expression at 4 h, while nuclear levels at 4 h remained low (Fig. 8E). Cytosolic levels at 8 h were the highest, suggesting that most of the newly synthesized Fyn is accumulating in the cytosol between 4 and 8 h. Cycloheximide was used at 4 h as a negative control. We then investigated whether Nrf2 regulates the induction of Fyn on t-BHQ treatment. Nrf2 siRNA was utilized to see whether knocking down Nrf2 would also decrease the antioxidant-induced Fyn induction. Western blot experiments revealed a t-BHQ induction of Fyn at 4 and 8 h even in the presence of Nrf2 siRNA (Fig. 8F). Real-time PCR experiments showed similar results to the Nrf2 siRNA Western blot. Fyn mRNA was increased with t-BHQ treatment in the presence of Nrf2 siRNA (Fig. 8G).
Figure 8.
Effect of t-BHQ on Fyn protein and gene expression. A, B) Hepa-1 cells were treated with 100 μM DMSO or t-BHQ (A) or pretreated with 30 μg/ml CHX and then treated with 100 μM DMSO or t-BHQ (B) for indicated time points. Cells were harvested; lysates were immunoblotted with anti-Fyn and anti-actin antibodies. C, D) Hepa-1 cells were treated with DMSO or 100 μM t-BHQ (C) and/or 20 ng/ml of actinomycin D (ActD; D) for varying time points. Cells were collected and RNA was extracted, and quantitation of mRNA was measured. Experiments were repeated 3 times; values are means ± sd. E) Pulse-chase assay, Hepa-1 cells transfected with Fyn-V5 were metabolically labeled with [S35] methionine (pulse). After 1 h, the medium was replaced with complete medium containing sufficient cold methionine, and the cells were harvested at 4 h (chase). t-BHQ (100 μM) and/or 30 μg/ml of cycloheximide was added to the medium at different time points. Nuclear (N) or cytosolic (C) cell lysate (500 μg) was immunoprecipitated with 2 μg of anti-V5 antibody. F, G) Hepa-1 cells were transfected with 100 nM control or Nrf2 siRNA. After 24 h, cells were stimulated with 100μM t-BHQ for 4 h. Fyn and Nrf2 expression was examined by Western blot (F) and real time PCR analysis (G). Comparison between DMSO and t-BHQ treatment time points shows a different significance. ***P < 0.0001; 2-tailed Student's t test.
Taken together, these results indicate that after the nuclear export of Fyn has taken place, t-BHQ induces Fyn transcription and subsequent increases in protein levels. Levels of protein, labeled protein, and mRNA are increasing, presumably to replenish the Fyn that was lost due to degradation. Results also demonstrated that Nrf2 is not responsible for the regulation of Fyn, suggesting that another transcription factor might be activated in the presence of t-BHQ that causes increased expression of Fyn mRNA and protein.
DISCUSSION
The regulation of Nrf2, especially its abundance in the nucleus, is important for controlling expression of cytoprotective genes in response to oxidative stress (4). Since persistent increases in cytoprotective gene expression threaten cell survival (24), Nrf2 is exported out of the nucleus and degraded. The nuclear export of Nrf2 is activated after Fyn accumulates inside the nucleus and phosphorylates tyrosine 586 of Nrf2 (14). Fyn phosphorylation of Nrf2 and subsequent nuclear export are delayed responses to oxidative stress (14, 19). In the current study, we investigated the early response of Fyn to oxidative stress.
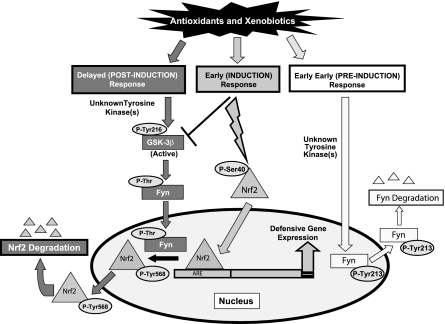
In this report, studies demonstrated that Fyn was exported out of the nucleus soon after exposure to oxidative stress (Fig. 9). The nuclear export of Fyn allowed Nrf2 to bind to the ARE and activate NQO1 gene expression. The antioxidant-induced nuclear export of Fyn appears to be an integral part of the ARE/Nrf2-mediated activation of cytoprotective genes. Treatment with LMB, an inhibitor of Crm-1 mediated nuclear export, stymied nuclear export of Fyn, resulting in nuclear accumulation of the protein. These findings are similar to what was reported with Lyn, another member of the Src family of kinases (25). The mechanism of Fyn nuclear export was due to phosphorylation of tyrosine 213 after oxidative stress stimulation. Once Fyn was phosphorylated, the newly identified nuclear export signal interacted with Crm-1, which allowed for nuclear export and subsequent degradation. Interestingly, the short leucine-rich residue motif present in Fyn lacked a fourth leucine residue, which is present in other NES motifs among different proteins. However, several other functional NES motifs have been characterized in proteins, such as p53 and IκB-α, that lack 4 leucine residues, indicating that 4 leucine residues are not required for an NES to be functional (26, 27).
Figure 9.
Model depicting the role of tyrosine phosphorylation-mediated nuclear export of Fyn and regulation of Nrf2.
The regulation of Src family kinase activity is controlled through phosphorylation of 2 tyrosine residues (28). Autophosphorylation of tyrosine 417 in Fyn increases kinase activity, and phosphorylation of tyrosine 528 in Fyn inhibits kinase activity. Phosphorylation at tyrosine 528 inhibits kinase activity through an intramolecular Src homology 2 (SH2)-phosphotyrosine interaction. This interaction stabilizes a closed or inactive conformation of the kinase (17). In this study, we found phosphorylation at tyrosine 213 in Fyn, which controls nuclear export. In addition, we also found phosphorylation at tyrosine 417 (Xcorr=3.05, data not shown), revealing an active kinase in response to antioxidant treatment. This kinase activity was later confirmed with anti-Src(pY416) antibodies. The kinase responsible for phosphorylation of tyrosine 213 has not been identified. However, since Fyn kinase activity is active during its nuclear export, it might be possible that Fyn autophosphorylates tyrosine 417 and 213, controlling its own nuclear export. If autophosphorylation is not responsible for the export of Fyn, we believe that the tyrosine kinase that phosphorylates Fyn has to be stress responsive and is rapidly activated in response to antioxidants. At this time, Fyn autophosphorylation at FynY213A remains unknown.
Interestingly, on Fyn nuclear export, subsequent accumulation in the cytosol was absent. When cells were treated with proteasome inhibitors, Fyn accumulation could be seen in the cytosol, alluding to the conclusion that antioxidants as well as other inducers of Nrf2 might induce proteasome-dependent degradation following nuclear export. Previous findings have shown that constitutive fibroblast growth factor receptor-2 (FGFR2) activation also induces Fyn ubiquitination and proteasomal degradation via a Casitas B-lineage lymphoma (Cbl)-mediated pathway (29). Although, it is unclear whether the Cbl ubiquitin ligase is responsible for the ubiquitination of Fyn in response to antioxidants, it appears that several mechanisms can induce Fyn proteasomal degradation.
Our previous reports have shown that GSK-3B phosphorylated Fyn at threonine residues, leading to nuclear localization of Fyn occurring around 4 h after oxidant treatment (19). The current study shows that Fyn mRNA and protein levels increased between 4 and 16 h of antioxidant treatment. Pulse-chase assays also revealed an increase in Fyn-labeled protein around 8 h in the cytosol only. Taken together, these results suggest that Fyn reentering the nucleus after nuclear export has taken place is not from de novo synthesis. However, increases in Fyn transcription is probably due to replenishing and replacing the Fyn that was lost to degradation after nuclear export.
Previous studies have reported that Nrf2 controls the regulation of some of its other negative regulators (30). This finding led to the obvious question, whether Nrf2 is responsible for the up-regulation of Fyn mRNA and protein in response to antioxidant treatment. Experiments using Nrf2 siRNA showed that Nrf2 does not regulate the transcription and induction of Fyn. Previous reports have suggested that in the presence of oxidative stress, transcription factors Sp1 and Egr1 might be responsible for up-regulation of Fyn mRNA and protein levels (31).
The chemotherapeutic agent etoposide kills cells by increasing topoisomerase II-induced DNA fragmentation, which triggers programmed cell death (32). When HepG2 cells were treated with etoposide, cell survival decreased significantly in cells transfected with Fyn and FynY213A. When t-BHQ was given concurrently with etoposide to cells transfected with Fyn, cellular survival was increased due to Nrf2 activation. This finding is similar to other findings that reported activation of Nrf2 helps survival of cancer cells during treatment with chemotherapeutic agents (33). When etoposide and t-BHQ treatments were given simultaneously to cells transfected with FynY213A, a t-BHQ-induced activation of Nrf2 did not fully rescue the cells, resulting in only a marginal increase in cell survival. Since FynY213A blocked Nrf2 activation, cells became sensitive to etoposide, which resulted in an increase in cellular death. These results illuminate the post-translational modifications of Fyn that can be targeted for potential treatments to different forms of cancer where Nrf2 is overexpressed.
In summation, we have investigated the nuclear export of Fyn and the regulation by which Fyn is exported in the early response to oxidative stress. We demonstrate that Fyn exports out of the nucleus within 30 min after antioxidant treatment, allowing nuclear import of Nrf2 and activation of cytoprotective gene expression without any inadvertent phosphorylation/degradation of Nrf2. The nuclear export of Fyn is controlled by tyrosine phosphorylation of residue 213, and a nuclear export signal interaction with Crm-1 leading to Fyn degradation after nuclear export. FynY213A cannot export out of the nucleus and interferes with the ability of Nrf2 to bind to the ARE and activate defensive gene expression. This interference is caused probably by the ability of the mutant to phosphorylate Nrf2, causing nuclear export of Nrf2 and subsequent ubiquitination/degradation before it can activate defensive genes. Because of this interference, cell survival is impaired. After Nrf2's task of cytoprotective gene up-regulation is complete, Fyn is imported back into the nucleus to phosphorylate Nrf2, causing Nrf2 export and degradation. This study dissects the mechanisms involved in the early response of Fyn kinase to oxidative stress.
Acknowledgments
This investigation was supported by U.S. National Institutes of Health grant RO1 GM047466. The authors thank Dr. Suresh K. Niture (University of Maryland School of Medicine, Baltimore, MD, USA) for helpful suggestions.
REFERENCES
- 1. Kaspar J. W., Niture S. K., Jaiswal A. K. (2009) Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic. Biol. Med. 47, 1304–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dhakshinamoorthy S., Jaiswal A. K. (2001) Functional characterization and role of INrf2 in antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. Oncogene 20, 3906–3917 [DOI] [PubMed] [Google Scholar]
- 3. Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J. D., Yamamoto M. (1999) Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13, 76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kang M. I., Kobayashi A., Wakabayashi N., Kim S. G., Yamamoto M. (2004) Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc. Natl. Acad. Sci. U. S. A. 101, 2046–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wakabayashi N., Itoh K., Wakabayashi J., Motohashi H., Noda S., Takahashi S., Imakado S., Kotsuji T., Otsuka F., Roop D. R., Harada T., Engel J. D., Yamamoto M. (2003) Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 35, 238–245 [DOI] [PubMed] [Google Scholar]
- 6. Kwak M. K., Wakabayashi N., Itoh K., Motohashi H., Yamamoto M., Kensler T. W. (2003) Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J. Biol. Chem. 278, 8135–8145 [DOI] [PubMed] [Google Scholar]
- 7. Padmanabhan B., Tong K. I., Ohta T., Nakamura Y., Scharlock M., Ohtsuji M., Kang M. I., Kobayashi A., Yokoyama S., Yamamoto M. (2006) Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell 21, 689–700 [DOI] [PubMed] [Google Scholar]
- 8. Singh A., Misra V., Thimmulappa R. K., Lee H., Ames S., Hoque M. O., Herman J. G., Baylin S. B., Sidransky D., Gabrielson E., Brock M. V., Biswal S. (2006) Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 3, e420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bloom D. A., Jaiswal A. K. (2003) Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. J. Biol. Chem. 278, 44675–44682 [DOI] [PubMed] [Google Scholar]
- 10. Huang H. C., Nguyen T., Pickett C. B. (2002) Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 277, 42769–42774 [DOI] [PubMed] [Google Scholar]
- 11. Jain A. K., Bloom D. A., Jaiswal A. K. (2005) Nuclear import and export signals in control of Nrf2. J. Biol. Chem. 280, 29158–29168 [DOI] [PubMed] [Google Scholar]
- 12. Kannan S., Jaiswal A. K. (2006) Low and high dose UVB regulation of transcription factor NF-E2-related factor 2. Cancer Res. 66, 8421–8429 [DOI] [PubMed] [Google Scholar]
- 13. Kaspar J. W., Jaiswal A. K. (2010) Antioxidant induced phosphorylation of tyrosine486 leads to rapid nuclear export of Bach1 that allows Nrf2 to bind to ARE and activate defensive genes expression. J. Biol. Chem. 285, 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jain A. K., Jaiswal A. K. (2006) Phosphorylation of tyrosine 568 controls nuclear export of Nrf2. J. Biol. Chem. 281, 12132–12142 [DOI] [PubMed] [Google Scholar]
- 15. Abe J., Berk B. C. (1999) Fyn and JAK2 mediate Ras activation by reactive oxygen species. J. Biol. Chem. 274, 21003–21010 [DOI] [PubMed] [Google Scholar]
- 16. Courtneidge S.A., Fumagalli S., Koegl M., Superti-Furga G., Twamley-Stein G. M. (1993) The Src family of protein tyrosine kinases: regulation and functions. Dev. Suppl. 57–64 [PubMed] [Google Scholar]
- 17. Sanguinetti A. R., Cao H., Corley Mastick C. (2003) Fyn is required for oxidative- and hyperosmotic-stress-induced tyrosine phosphorylation of caveolin-1. Biochem. J. 376, 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Resh M. D. (1998) Fyn, a Src family tyrosine kinase. Int. J. Biochem. Cell Biol. 30, 1159–1162 [DOI] [PubMed] [Google Scholar]
- 19. Jain A. K., Jaiswal A. K. (2007) GSK-3beta acts upstream of Fyn kinase in regulation of nuclear export and degradation of NF-E2 related factor 2. J. Biol. Chem. 282, 16502–16510 [DOI] [PubMed] [Google Scholar]
- 20. Niture S. K., Jaiswal A. K. (2009) Prothymosin-alpha mediates nuclear import of the INrf2/Cul3 Rbx1 complex to degrade nuclear Nrf2. J. Biol. Chem. 284, 13856–13868 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21. Hirota A., Kawachi Y., Itoh K., Nakamura Y., Xu X., Banno T., Takahashi T., Yamamoto M., Otsuka F. (2005) Ultraviolet A irradiation induces NF-E2-related factor 2 activation in dermal fibroblasts: protective role in UVA-induced apoptosis. J. Invest. Dermatol. 124, 825–832 [DOI] [PubMed] [Google Scholar]
- 22. Stewart D., Killeen E., Naquin R., Alam S., Alam J. (2003) Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. J. Biol. Chem. 278, 2396–3402 [DOI] [PubMed] [Google Scholar]
- 23. Fukuda M., Asano S., Nakamura T., Adachi M., Yoshida M., Yanagida M., Nishida E. (1997) CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390, 308–311 [DOI] [PubMed] [Google Scholar]
- 24. Strachan G. D., Ostrow L. A., Jordan-Sciutto K. L. (2005) Expression of the fetal Alz-50 clone 1 protein induces apoptotic cell death. Biochem. Biophys. Res. Commun. 336, 490–495 [DOI] [PubMed] [Google Scholar]
- 25. Kikuko I., Nakayama Y., Togashi Y., Obata Y., Kuga T., Kasahara K., Fukumoto Y., Yamaguchi N. (2008) Nuclear localization of Lyn tyrosine kinase mediated by inhibition of kinase activity. Exp. Cell. Res. 314, 3392–3404 [DOI] [PubMed] [Google Scholar]
- 26. Stommel J. M., Marchenko N. D., Jimenez G. S., Moll U. M., Hope T. J., Wahl G. M. (1999) A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 18, 1660–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arenzana-Seisdedos F., Turpin P., Rodriguez M., Thomas D., Hay R. T., Virelizier J. L., Dargemont C. (1997) Nuclear localization of I kappa B alpha promotes active transport of NF-kappa B from the nucleus to the cytoplasm. J. Cell Sci. 110, 369–378 [DOI] [PubMed] [Google Scholar]
- 28. Mayer B. J. (1997) Signal transduction: clamping down on Src activity. Curr. Biol. 7, R295–298 [DOI] [PubMed] [Google Scholar]
- 29. Kaabeche K., Lemonnier J., Le Mée S., Caverzasio J., Marie P. J. (2004) Cbl-mediated degradation of Lyn and Fyn induced by constitutive fibroblast growth factor receptor-2 activation supports osteoblast differentiation. J. Biol. Chem. 279, 36259–36267 [DOI] [PubMed] [Google Scholar]
- 30. Kaspar J. W., Jaiswal A. K. (2010) An auto-regulatory between Cul3 and Rbx1 controls their cellular abundance. J. Biol. Chem. 285, 21349–21358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gao Y., Howard A., Ban K., Chandra J. (2009) Oxidative stress promotes transcriptional up-regulation of Fyn in BCR-ABL1-expressing cells. J. Biol. Chem. 284, 7114–7125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bromberg K. D., Burgin A. B., Osheroff N. (2003) A two-model for etoposide action against human topoisomerase IIalpha. J. Biol. Chem. 278, 7406–7412 [DOI] [PubMed] [Google Scholar]
- 33. Wang X. J., Sun Z., Villeneuve N. F., Zhang S., Zhao F., Li Y., Chen W., Yi X., Zheng W., Wondrak G. T., Wong P. K., Zhang D. D. (2008) Nrf2 enhances resistance to cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 29, 1235–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]