Abstract
In this study, the principles of surface sensing of translation (SUnSET) were used to develop a nonradioactive method for ex vivo and in vivo measurements of protein synthesis (PS). Compared with controls, we first demonstrate excellent agreement between SUnSET and a [3H]phenylalanine method when detecting synergist ablation-induced increases in skeletal muscle PS ex vivo. We then show that SUnSET can detect the same synergist ablation-induced increase in PS when used in vivo (IV-SUnSET). In addition, IV-SUnSET detected food deprivation-induced decreases in PS in the heart, kidney, and skeletal muscles, with similar changes being visualized with an immunohistochemical version of IV-SUnSET (IV-IHC-SUnSET). By combining IV-IHC-SUnSET with in vivo transfection, we demonstrate that constitutively active PKB induces a robust increase in skeletal muscle PS. Furthermore, transfection with Ras homolog enriched in brain (Rheb) revealed that a PKB-independent activation of mammalian target of rapamycin is also sufficient to induce an increase in skeletal muscle PS. Finally, IV-IHC-SUnSET exposed the existence of fiber type-dependent differences in skeletal muscle PS, with PS in type 2B and 2X fibers being significantly lower than that in type 2A fibers within the same muscle. Thus, our nonradioactive method allowed us to accurately visualize and quantify PS under various ex vivo and in vivo conditions and revealed novel insights into the regulation of PS in skeletal muscle.—Goodman, C. A., Mabrey, D. M., Frey, J. W., Miu, M. H., Schmidt, E. K., Pierre, P., Hornberger, T. A. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique.
Keywords: hypertrophy, atrophy, growth
Various cells and tissues undergo hypertrophy or atrophy in response to factors that include altered nutritional and hormonal status, physical activity/inactivity, disease states, aging, and pharmacological therapies. The extent of hypertrophy or atrophy is determined, in part, by changes in the relative rates of protein synthesis. For decades, the gold standard for measuring changes in protein synthesis has been the measurement of radioactively labeled tracer incorporation into proteins (for review, see ref. 1). Although very successful, these methodologies require the use of expensive and/or specialized techniques (e.g., scintillation counters, HPLC or gas chromatography–isotope ratio mass spectrometry, or others) and are complicated by issues such as extensive administration and record-keeping and the costs associated with the tracer and its disposal. Moreover, although useful for whole-body and whole-tissue studies, available techniques that use radioactive tracers lack the sensitivity needed to detect differences in protein synthesis between individual cells within the same tissue. Therefore, development of a highly sensitive nonradioactive technique that uses common laboratory methods, such as Western blot (WB) and immunohistochemistry (IHC), would be a major advancement in the methods available for measuring protein synthesis under various conditions.
Recently, a nonradioactive technique known as surface sensing of translation (SUnSET) was developed and validated against radioactive-based methods for measuring changes in protein synthesis in cell cultures (2). This technique involves the use of the antibiotic puromycin (a structural analog of tyrosyl-tRNA), and anti-puromycin antibodies to detect the amount of puromycin incorporation into nascent peptide chains (2, 3). It has been demonstrated that incorporation of puromycin into nascent peptide chains terminates their elongation, and when puromycin is used at low concentrations, the accumulation of puromycin-conjugated peptides accurately reflects the rate of protein synthesis (2, 4). Notably, this technology alleviates the problems associated with the use of radioactive materials for measuring changes in protein synthesis. Thus, the goal of this study was to establish a technique, based on the principles of SUnSET, for ex vivo and in vivo measurements of protein synthesis in whole tissues using WB and at the single-cell level using IHC.
To accomplish our goal, we focused primarily on skeletal muscle, with some experiments also performed on the heart and kidney. We focused mainly on skeletal muscle because there is currently an intense interest in identifying the molecular mechanisms that regulate skeletal muscle protein synthesis, and it is believed that identifying these mechanisms could lead to the development of therapies that can preserve muscle mass during conditions such as immobilization, bedrest, spaceflight, cancer cachexia, aging, and dystrophy (5). Therefore, we first tested the validity of our technique by using a variety of experimental models that are known to induce increases and decreases in skeletal muscle protein synthesis. Once validated, we then used our technique to examine the following for the first time at the single fiber/cell level: the in vivo effect of constitutively active PKB (CA-PKB) on skeletal muscle protein synthesis; the ability of a PKB-independent activation of the mammalian target of rapamycin (mTOR) to induce an increase in skeletal muscle protein synthesis in vivo; and the in vivo differences in protein synthesis rates among various fiber types within the same skeletal muscle.
MATERIALS AND METHODS
Animals
Female FVB/N mice, 8–10 wk old, were used for all conditions. Mice were housed under a 12-h light/dark cycle with ad libitum access to food and water unless otherwise stated. Before all surgical procedures, mice were anesthetized with an intraperitoneal injection of ketamine (100 mg/kg) and xylazine (10 mg/kg). After tissue extraction, the mice were euthanized by cervical dislocation. All methods were approved by the Institutional Animal Care and Use Committee of the University of Wisconsin-Madison.
Experimental models
Synergist ablation (SA) of the plantaris (PLT) muscle
SA surgical procedures involved the bilateral removal of the soleus (SOL) and distal half of the gastrocnemius muscle. Mice in control groups received sham surgery, for which an incision was made on the lower leg and then closed. After the surgical procedures, the incision was closed with Vetbond surgical glue (Henry Schein, Melville, NY, USA). Mice were allowed to recover for 7 d and then the PLT muscles were subjected to various measurements described below.
Food deprivation
Food was withdrawn from mice for 48 h, but they still had ad libitum access to water (control mice were maintained on the ad libitum diet). After 48 h, tissues were subjected to nonradioactive in vivo measurements of protein synthesis as described below.
In vivo transfection of green fluorescent protein (GFP), CA-PKB, and wild-type Ras homolog enriched in brain (Rheb)
The pEGFP-C3 (GFP) plasmid was purchased from Clontech (Mountain View, CA, USA). CA-PKB plasmid [pCMV6 myristoylated hemagglutinin (HA)-PKB] (6) was kindly provided by Dr. Jie Chen (University of Illinois, Urbana, IL, USA). The wild-type Rheb plasmid (pCDNA3 HA-Rheb; ref. 7) was a generous gift from Dr. Kun-Liang Guan (University of California–San Diego, La Jolla, CA, USA). All plasmid DNA was grown in DH5α Escherichia coli, purified with an EndoFree plasmid kit (Qiagen, Valencia, CA, USA), and resuspended in sterile PBS. Plasmid DNA (30 μg) was transfected into mouse tibialis anterior (TA) muscles by electroporation as described previously (8). In brief, mice were anesthetized, and a small incision was made through the skin covering the TA muscle. A 27-gauge needle was used to inject plasmid DNA solution (2.5 μg/μl GFP or HA-Rheb or CA-PKB) into the proximal (6 μl) and distal (6 μl) ends of the muscle belly. After the injections, electric pulses were applied through 2 stainless steel pin electrodes (1-cm gap, Harvard Apparatus, Holliston, MA, USA) laid on top of the proximal and distal myotendinous junctions. Eight 20-ms square-wave electric pulses at a frequency of 1 Hz were delivered with an ECM 830 electroporation unit (BTX-Harvard Apparatus, Holliston, MA, USA) with a field strength of 180 V/cm. After the electroporation procedure, the incision was closed with Vetbond surgical glue. At 3 or 4 d after transfection, muscles were subjected to nonradioactive in vivo measurements of protein synthesis as described below.
Radioactive measurement of protein synthesis
PLT muscles were preincubated in an organ culture bath with medium composed of Krebs-Henseleit buffer plus 1× modified essential medium amino acid solution (HyClone, Logan, UT, USA) and 25 mM glucose for 15 min as described previously (9). After the preincubation period, fresh medium supplemented with 2.5 mM unlabeled phenylalanine and 20 μCi of [3H]phenylalanine (Amersham Biosciences, Little Chalfont, UK) was added. After 30 min, muscles were removed from the medium, and the rate of protein synthesis based on [3H]phenylalanine incorporation into nascent proteins was determined as described previously (9).
Nonradioactive measurements of protein synthesis with SUnSET
Cell culture
C2C12 myoblasts were cultured on a 6-well dish as described previously (10). On confluence, cells were switched to serum-free DMEM (HyClone) for 16 h. The medium was then replaced with fresh serum-free DMEM with or without 100 nM insulin (Humalog; Lilly, Indianapolis, IN, USA). After 60 min, 1 μM puromycin was added to all wells, and the cells were incubated for an additional 30 min. Cells were then collected and subjected to WB analysis or RNA measurements as described below.
Ex vivo SUnSET (EV-SUnSET)
PLT muscles were preincubated for 15 min in the organ culture bath as described above. After the preincubation period, fresh medium supplemented with 1 μM puromycin (Calbiochem; EMD Chemicals, Gibbstown, NJ, USA) was added. After 30 min, the muscles were removed from the medium, frozen in liquid N2, and stored at −80°C. The extent of puromycin incorporation into nascent peptides was then assessed by WB analysis as described below.
In vivo SUnSET (IV-SUnSET) and in vivo IHC SUnSET (IV-IHC-SUnSET)
For all in vivo measurements of protein synthesis, mice were anesthetized as described above and then given an intraperitoneal injection of 0.040 μmol/g puromycin dissolved in 100 μl of PBS. At exactly 30 min after injection, tissues were extracted and either frozen in liquid N2 for WB analysis (IV-SUnSET) or prepared for IHC analysis (IV-IHC-SUnSET) as detailed below.
WB analysis
Frozen tissues were homogenized with a Polytron homogenizer (Kinematica AG, Lucerne, Switzerland) for 20 s in ice-cold WB buffer (40 mM Tris, pH 7.5; 1 mM EDTA; 5 mM EGTA; 0.5% Triton X-100; 25 mM β-glycerophosphate; 25 mM NaF; 1 mM Na3VO4; 10 μg/ml leupeptin; and 1 mM PMSF), and the whole homogenate was used for further analysis. For cell culture experiments, myoblasts were lysed in the ice-cold WB blot buffer and centrifuged at 500 g for 5 min, and the supernatant was used for further analysis. Sample protein concentrations were determined with a DC protein assay kit (Bio-Rad Laboratories, Hercules, CA, USA), and equivalent amounts of protein (55 μg) from each sample were dissolved in Laemmli buffer and subjected to electrophoretic separation on 10% SDS-PAGE acrylamide gels as described previously (8). Electrophoresis was terminated when the dye front was ∼1.5 cm from the bottom of the gel. Proteins were transferred to a PVDF membrane in transfer buffer (242 mM Tris, 58 mM glycine, and 20% v/v methanol). Membranes were blocked with 5% powdered milk in TBST (Tris-buffered saline with 0.1% Tween 20) for 1 h followed by an overnight incubation at 4°C with mouse IgG2a monoclonal anti-puromycin antibody (clone 12D10, 1:5000) (2) dissolved in TBST containing 1% BSA. Membranes were washed for 30 min in TBST and then incubated for 1 h at room temperature in 5% milk-TBST containing horseradish peroxidase conjugated anti-mouse IgG Fc 2a antibody (1:50,000; Jackson ImmunoResearch Laboratories Inc., West Grove, PA, USA). After 30 min of washing in TBST, the blots were developed on film using regular enhanced chemiluminescence (ECL) reagent (Pierce; Thermo Fisher Scientific, Rockford, IL, USA) or ECL plus reagent (Amersham, Piscataway, NJ, USA). Once the appropriate image was captured, the membranes were stained with Coomassie Blue to verify equal loading in all lanes. Densitometric measurements were performed by determining the density of each whole lane (incorporating the entire molecular weight range of puromycin-labeled peptides) using the public domain ImageJ software (U.S. National Institutes of Health, Bethesda, MD, USA; http://rsb.info.nih.gov/nih-image/).
IHC analysis
IV-IHC-SUnSET in food deprivation and in vivo transfected samples
TA muscles were extracted at exactly 30 min after the injection of puromycin (described above), placed at resting length in ice-cold PBS containing 4% paraformaldehyde, and rocked at 4°C for 30 min. Samples were then submerged in optimal cutting temperature (OCT) compound (Tissue-Tek; Sakura, Torrance, CA, USA) and frozen in liquid N2-chilled isopentane. Cross sections (10 μm thick) were taken from the midbelly and fixed in −20°C acetone for 10 min. Sections were warmed to room temperature for 5 min and rehydrated with cool steam vapors and then incubated in PBS for 15 min, followed by a 20-min incubation in solution A (PBS containing 0.5% BSA and 0.5% Triton X-100). Samples were then incubated for 1 h in solution A containing anti-mouse IgG Fab (1:10; Jackson ImmunoResearch). After three 5-min washes with PBS, samples were incubated for 1 h with solution A containing primary antibodies [mouse anti-puromycin (clone 12D10, 1:1000) and, when appropriate, rat anti-HA (clone 3F10, 1:200; Roche Diagnostics, Mannheim, Germany)]. Samples were washed 3 times for 5 min each with PBS and then incubated for 1 h at room temperature with solution A containing secondary antibodies [DyLight 594-conjugated anti-mouse IgG Fc 2a (1:500; Jackson ImmunoResearch) and, when appropriate, FITC-conjugated anti-rat IgG F(ab′)2 (1:100; Santa Cruz Biotechnology, Santa Cruz, CA, USA)]. Samples were then washed 3 times for 10 min each with PBS and were covered with mounting medium (Vectashield, Vector Laboratories, Burlingame, CA, USA) and a coverslip. Grayscale monochrome images of the DyLight 594 and FITC signals were captured with a Nikon DS-QiMc camera (Nikon, Tokyo, Japan), which has a >33-fold working linear range (r=0.9997, data not shown). The camera was mounted on a Nikon 80i epifluorescence microscope, and images were captured through tetramethylrhodamine isothiocyanate (TRITC) and FITC cubes, respectively. When appropriate, monochrome images were merged with Nikon NIS-Elements D software to identify transfected and nontransfected fibers. Fiber cross-sectional area measurements on the resulting images were performed as described previously (8). For protein synthesis measurements, the intensity of the TRITC signal was measured in 60 to 120 randomly selected fibers per section with Nikon NIS-Elements D software. For the food-deprivation experiments, the TRITC signal in individual fibers from both ad libitum-fed and food-deprived sections was expressed relative to the mean TRITC signal intensity of fibers in ad libitum-fed sections (Note that differences in incubation conditions and exposure time were avoided by capturing images of food-deprived and ad libitum-fed samples that were mounted immediately adjacent to one another on the same slide; see Fig. 4.) Relative rates of protein synthesis in transfected muscles were calculated by expressing the TRITC signal of both individual nontransfected and transfected fibers to that of the mean TRITC signal in the nontransfected fibers from the same section. All analyses were performed on images that had signal intensities within the linear range of the camera, and the analyses were performed by investigators masked to the sample identification.
Figure 4.
Food deprivation induces a decrease in protein synthesis as determined with IV-SUnSET and IV-IHC-SUnSET. Mice were fed ad libitum (control) or deprived of food for 48 h (fasted), and the in vivo rates of protein synthesis in the heart (A, B), kidney (C, D), and TA skeletal muscle (E, F) were measured with IV-SUnSET as described in Materials and Methods. A, C, E) Representative images of WB analysis for puromycin (Puro) followed by Coomassie Blue (CB) staining to verify equal loading of proteins. B, D, F) Quantification of the puromycin-labeled peptides, expressed as a percentage of the values obtained in the control group. RLU, relative light units. A–F) All values are means ± se (for heart, n=8/group; for kidney and TA, n=4–5/group). G, H) In vivo rates of protein synthesis in the TA muscle were determined with an IV-IHC-SUnSET as described in Materials and Methods. G) Representative image of cross sections from TA muscles that were subjected to control or food-deprivation conditions, mounted adjacent to one another on a slide, and then subjected to IHC for puromycin. H) Puromycin staining intensity in individual fibers from both control and food-deprivation sections was expressed relative to the mean puromycin signal intensity of fibers in control sections and then plotted on a histogram. Average ± se staining intensity is indicated next to group labels in legend; n = 300 fibers/group from 5 independent pairs of muscles. *P < 0.05 vs. control.
IV-IHC-SUnSET in different skeletal muscle fiber types
At exactly 30 min after the injection of puromycin (described above), the TA, PLT, or SOL muscles were extracted and immediately submerged in OCT at resting length and frozen in liquid N2-chilled isopentane. The samples were then triple stained for puromycin and various myosin heavy chain (MHC) isoforms using slight modifications of the procedures described above. Specifically, the primary antibody solutions contained mouse IgG2a monoclonal anti-puromycin and mouse IgG1 monoclonal anti-type 2a MHC (clone SC-71, 1:100), and one of the following: mouse IgG2b monoclonal anti-type 1 MHC (clone BA-D5, 1:100), mouse IgM monoclonal anti-type 2b MHC (clone BF-F3, 1:10), or mouse IgM monoclonal anti-type 2x MHC (clone 6H1, 1:2). All MHC antibodies were obtained from the Developmental Studies Hybridoma Bank at the University of Iowa (Ames, IA, USA). The secondary antibody solutions contained DyLight 594-conjugated anti-mouse IgG Fc 2a (1:500), FITC-conjugated anti-mouse IgG Fc 1 (1:100; Jackson ImmunoResearch), and, depending on the other anti-MHC primary antibody applied, Alexa 350-conjugated anti-mouse IgG Fc 2b (1:500; Invitrogen, Carlsbad, CA, USA) or aminomethylcoumarin acetate (AMCA)-conjugated anti-mouse IgM (1:150; Jackson ImmunoResearch). Grayscale monochrome images of the DyLight 594, FITC, and Alexa 350 or AMCA signals were captured through TRITC, FITC, and DAPI cubes, respectively, on the microscope described above. The monochrome images were merged with Nikon NIS-Elements D software to identify the fibers expressing the specific types of MHC. Thirty type 2A and either type 1, 2X, or 2B fibers were randomly selected from each section, and the intensity of the TRITC signal in individual type 1, 2A, 2X, or 2B fibers was expressed relative to the mean TRITC signal in type 2A fibers from the same section. All analyses were performed on images that had signal intensities within the linear range of the camera, and the analyses were performed by investigators masked to the sample identification.
Free puromycin concentration assay
A 250-μl aliquot of sample homogenate made for WB analysis (described above) was precipitated with 28 μl of 100% trichloroacetic acid, incubated on ice for 30 min, and then centrifuged for 5 min at 4200g. The supernatant was then neutralized with 30 μl of 5.25 M NaOH and 15 μl of buffer containing 1 M Tris, 3 M NaCl, and 1% Tween 20 (pH 7.0). Sample pH was adjusted to 9.0, and the sample was filtered through a >3-kDa exclusion filter (Amicon Ultra-0.5 ml; Millipore, Carrigtwohill, Ireland) for 60 min at 14,000 g. A range of standards (0.025–40.0 pmol in 100 μl) was also made by adding free puromycin to the supernatant made from samples taken from animals that were not given puromycin injections but were treated exactly as just described above. A 100-μl aliquot of the sample or standard, per well, was added to a 96-well amine-binding maleic anhydride-activated plate (Pierce; Thermo Fisher Scientific) and rocked overnight at 4°C. The plate was washed 4 times for 1 min each with PBS with 1% Tween 20 (PBST) and then blocked with 1% BSA-PBST for 1 h at room temperature. Next, 100 μl of anti-puromycin antibody (clone 12D10, 1:38,400) in 1% BSA-PBST was added to each well and rocked for 2 h at room temperature. After four 1-min washes with PBST, 100 μl of horseradish peroxidase-conjugated anti-mouse IgG Fc 2a in 1% BSA-PBST (1:10,000; Jackson ImmunoResearch) was added to each well and rocked for 1 h at room temperature. The wells were washed 4 times for 1 min each with PBST, and finally a 100-μl mixture of o-phenylenediamine dihydrochloride (horseradish peroxidase substrate; Pierce; Thermo Fisher Scientific) and stable peroxide substrate buffer (Pierce; Thermo Fisher Scientific) was added to each well and rocked. After 10–15 min, the reaction was stopped by addition of 100 μl of 5 N sulfuric acid to each well, and the absorbance at 492 nm was measured on a plate reader (FLUOstar Optima; BMG Labtech, Durham, NC, USA). With use of the values from the standard curve, the free puromycin content was calculated and normalized to the protein content within the original 250-μl aliquot of sample.
Total and ribosomal RNA
Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions. PLT muscle RNA was resuspended in 4 μl of nuclease-free, diethyl pyrocarbonate-treated water (Genemate, Kaysville, UT, USA) per milligram of muscle, and RNA from each well of C2C12 myoblasts was resuspended in 35 μl. The RNA concentration was determined by spectrophotometric analysis at 260 nm. RNA samples were then run on a 1% agarose gel and viewed under UV light. Densitometric measurements of the 28S and 18S rRNA were performed with ImageJ software as described above.
Statistical methods
All data analysis was performed using GraphPad Prism 5.0 (GraphPad Software Inc., La Jolla, CA, USA). Student's 2-tailed, unpaired t tests were used for all 2-group comparisons. The statistical significance level was set at P < 0.05.
RESULTS
Effect of synergist ablation on protein synthesis as measured with radioactive-based and nonradioactive SUnSET-based techniques
To determine whether our EV-SUnSET technique could detect an increase in skeletal muscle protein synthesis, we subjected PLT muscles to bilateral SA surgery. In this surgical procedure, the SOL and distal half of the gastrocnemius muscles are removed, leaving the PLT as the sole plantar flexor muscle. Consequently, the PLT muscle is placed under chronic mechanical overload, and after 7 d, there was a 2-fold increase in the muscle weight/body weight ratio (Fig. 1A) and a 1.6-fold increase in total protein content (Fig. 1B). Furthermore, SA induced a 1.9-fold increase in total RNA per milligram of muscle (Fig. 1C) and a 2.2-fold increase in 28S and 18S rRNA per milligram of muscle (Fig. 1D). SA also induced a 3.6-fold increase in the amount of puromycin-labeled peptides (i.e., protein synthesis) when measured with EV-SUnSET (Fig. 1E, F). To verify that the difference in the amount of puromycin-labeled peptides accurately reflected differences in rates of protein synthesis, ex vivo measurements of protein synthesis were performed on the contralateral muscle with a conventional radioactive-based method (9, 11). This method indicated that SA induced a 3.4-fold increase in the rate of protein synthesis (Fig. 1G), which was not significantly different from that determined with EV-SUnSET (P=0.65). Furthermore, a strong correlation (r=0.9, P=0.015) between the two methodologies was observed when protein synthesis rates in the contralateral muscles of the same animal were compared.
Figure 1.
SA induces hypertrophy, increased rRNA, and increased protein synthesis as measured ex vivo with radioactive- and SUnSET-based techniques. PLT muscles were subjected to sham (control) or SA surgical procedures and after 7 d were assessed for changes in parameters. A) Muscle weight (MW)/body weight (BW) ratio (n=5/group). B) Total protein content (n=5/group). C) Total RNA/MW (n=5/group). D) Relative 28S and 18S rRNA/MW (n=5/group). E–G) Ex vivo rates of protein synthesis (n=3/group). E, F) SUnSET measurements of protein synthesis were performed by incubating muscles in an organ culture bath with medium containing puromycin as described in Materials and Methods. E) Representative image of WB analysis for puromycin followed by Coomassie Blue staining to verify equal loading of proteins. Note that the specificity of the anti-puromycin blot was demonstrated by a sample that was not incubated with puromycin (Puro, far left lane). F) Quantification of the puromycin-labeled peptides, expressed as a percentage of the values obtained in the control group. RLU, relative light units. G) Radioactive measurements of protein synthesis rate were performed by incubating muscles in an organ culture bath with medium containing a flooding dose of [3H]phenylalanine as described in Materials and Methods. All values are means + se. *P < 0.05 vs. control.
It has been proposed that SA promotes an increase in protein synthesis via both enhanced translational capacity (rRNA) and translational efficiency (rate/rRNA) (12, 13). Consistent with this hypothesis, we found that the SA-induced increase in protein synthesis (3.6-fold) greatly exceeded the increase in 28S and 18S rRNA concentration (2.2-fold; Fig. 1D). To determine whether SUnSET could detect acute changes in protein synthesis in the absence of changes in rRNA, we measured the amount of puromycin-labeled peptides in C2C12 myoblasts after 90 min of insulin stimulation. Compared with controls, insulin induced a significant increase in the amount of puromycin-labeled peptides (protein synthesis) while having no effect on total RNA concentration (16.8±1.5 vs. 16.0±0.8 μg/well total RNA, respectively; P=0.68) or the concentration of the 28S or 18S rRNA (Fig. 2). These results show that SUnSET can also detect relatively acute changes in protein synthesis in the absence of changes in rRNA.
Figure 2.
SUnSET measurements of enhanced protein synthesis in the absence of changes in rRNA. Serum-starved C2C12 myoblasts were stimulated with or without 100 nM insulin (INS) for 90 min, and rates of protein synthesis were measured with the SUnSET technique described in Materials and Methods. A) Representative image of WB analysis for puromycin followed by Coomassie Blue staining to verify equal loading of proteins. B) Quantification of the puromycin-labeled peptides (n=8 wells/group from 3 independent experiments). RLU, relative light units. C) Relative concentration of 28S and 18S rRNA per cell culture well (n=6 wells/group from 3 independent experiments). All values are expressed as a percentage of the values obtained in control samples and are presented as means + se. *P < 0.05 vs. control.
The next series of experiments determined whether SUnSET could detect an increase in protein synthesis when used in vivo (IV-SUnSET). To accomplish this goal, 7 d after SA surgery, mice were anesthetized and given an intraperitoneal injection of puromycin 30 min before muscle extraction. Similar to the ex vivo measurements, our IV-SUnSET measurements revealed that SA induced a 2.9-fold increase in the amount of puromycin-labeled peptides (Fig. 3A, B). Under in vivo conditions, it is important to consider that differences in the delivery/uptake of free puromycin may produce differences in the amount of puromycin-labeled proteins that are not a result of differences in the rate of protein synthesis. To address this concern, we developed an assay for measuring the free puromycin concentration in the same samples that were subjected to the WB analysis and found that SA had no effect on the free puromycin concentration (Fig. 3C). This observation indicates that the SA-induced increase in puromycin-labeled peptides was due to an increase in the rate of protein synthesis and, therefore, provides further evidence that our IV-SUnSET technique can be used to detect increases in protein synthesis in vivo.
Figure 3.
SA induces protein synthesis in vivo as measured with IV-SUnSET. PLT muscles were subjected to sham (control) or SA surgical procedures, and after 7 d in vivo rates of protein synthesis were determined with the IV-SUnSET technique described in Materials and Methods. A) Representative image of WB analysis for puromycin-labeled peptides followed by Coomassie Blue staining to verify equal loading of proteins. B) Quantification of the puromycin-labeled peptides, expressed as a percentage of the values obtained in the control group (n=3/group). RLU, relative light units. C) Measurement of the free puromycin concentration (n=6/group). Values are means + se. *P < 0.05 vs. control.
Food deprivation induces a decrease in protein synthesis in vivo as measured with IV-SUnSET and IV-IHC-SUnSET
To determine whether our IV-SUnSET technique could detect in vivo decreases in protein synthesis, we subjected mice to 48 h of food deprivation, which has previously been shown to attenuate the rate of protein synthesis in a variety of different tissues (14). Hence, we performed analyses on the TA, SOL, or extensor digitorum longus skeletal muscles, as well as heart and kidney. The results indicated that food deprivation induced a significant decrease in the amount of puromycin-labeled peptides (protein synthesis) in all of the tissues analyzed (Fig. 4A–F and Supplemental Fig. S1E, F). Furthermore, we determined that food deprivation did not significantly affect the free puromycin concentration in the kidney or TA muscle (Supplemental Fig. S1A, B). Although not statistically significant (P=0.12), a trend for food deprivation to decrease the free puromycin content in the heart was seen (Supplemental Fig. S1C). Therefore, to correct for the potentially lower free puromycin concentration in the food-deprived hearts, we normalized the relative light unit values obtained from the WB analysis for puromycin-labeled peptides to the free puromycin content in the same samples (Supplemental Fig. S1D). This normalization showed essentially the same decrease in protein synthesis as the uncorrected values (64 vs. 71%, respectively, P=0.49). Combined, these results demonstrate that our IV-SUnSET technique can detect an in vivo decrease in protein synthesis in a variety of different tissues.
Next, we investigated whether the IV-IHC-SUnSET technique could detect the food deprivation-induced decrease in protein synthesis. As shown in Supplemental Fig. S2, our IV-IHC-SUnSET staining technique was devoid of a nonspecific background signal. Furthermore, the puromycin-labeled peptides were detected primarily within individual muscles fibers, whereas the interstitial space was devoid of a signal. We also determined that puromycin-labeled peptides were highly colocalized with the presence of the ribosomal S6 protein. Combined, these results indicate that our staining technique does not detect free puromycin within the interstitial space and that a large proportion of the puromycin-labeled peptides are found in regions that are enriched with ribosomes. Furthermore, our IV-IHC-SUnSET technique revealed that food deprivation induced a 53% decrease in protein synthesis (Fig. 4H), an effect that was not significantly different from that observed with IV-SUnSET (65%; P=0.33). Thus, the IV-SUnSET and IV-IHC-SUnSET versions of our technique yield quantitatively similar results when they were used to measure relative changes in the rate of protein synthesis in vivo.
In vivo overexpression of constitutively active PKB induces an increase in skeletal muscle protein synthesis
To determine whether our IV-IHC-SUnSET technique could detect an increase in protein synthesis at the single fiber/cell level, we transiently transfected whole TA muscles in vivo with plasmid DNA encoding CA-PKB and the contralateral muscle with DNA encoding GFP as a control for the effects of electroporation. With our approach, we were able to directly compare the rates of protein synthesis in transfected fibers and nontransfected fibers within the same muscle. As shown in Fig. 5A–C, GFP-transfected fibers had a slightly higher rate of protein synthesis (5%) than nontransfected fibers within the same muscle. On the other hand, CA-PKB-transfected fibers had a substantially higher rate of protein synthesis (155%) than nontransfected fibers (Fig. 5D–F). These results confirm, for the first time, that in vivo overexpression of CA-PKB induces a robust increase in skeletal muscle protein synthesis and also demonstrate that our IV-IHC-SUnSET technique can be used to determine relative rates of protein synthesis at the single fiber/cell level.
Figure 5.
In vivo overexpression of CA-PKB induces an increase in protein synthesis as determined with IV-IHC-SUnSET. TA muscles were transfected with plasmid DNA encoding the GFP (A–C) or HA-tagged CA-PKB (D–F). At 3 d after transfection, muscles were subjected to IV-IHC-SUnSET by staining for puromycin and GFP or the HA tag, respectively. A) Representative image of the signal for GFP positive fibers. B) Grayscale image of the signal for puromycin in the same section shown in A. D) Representative image of the signal for CA-PKB positive fibers. E) Grayscale image of the signal for puromycin in the same section shown in D. C, F) Puromycin staining intensity in both transfected (positive) and nontransfected (negative) fibers, expressed relative to the mean value obtained in nontransfected (negative) fibers from the same section; values are plotted on histograms for GFP-transfected muscles (C) and CA-PKB-transfected muscles (F). Values next to group labels in legend are means ± se; n = 180 transfected and nontransfected fibers/group from 3 independent muscles. *P < 0.05 vs. control.
PKB-independent activation of mTOR is sufficient to induce an increase in skeletal muscle protein synthesis and hypertrophy
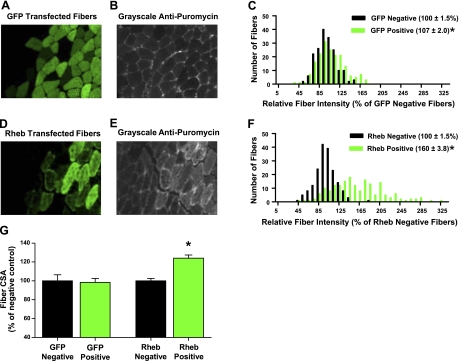
It has been proposed that PKB induces protein synthesis via the activation of signaling by mTOR. However, PKB can also induce signaling through mTOR-independent regulators of protein synthesis such as glycogen synthase kinase 3β (GSK3β). Thus, it is not clear whether PKB-independent activation of mTOR is sufficient to induce an increase in skeletal muscle protein synthesis. To address this issue, we transfected muscles with DNA encoding Rheb, which has consistently been shown to be one of the most proximal activators of mTOR signaling (15). In addition, work from our laboratory has recently demonstrated that Rheb activates mTOR signaling through a PKB-independent mechanism in skeletal muscle (8). As shown in Fig. 6A–F, we have extended this observation by demonstrating that the overexpression of Rheb is also sufficient to induce a 60% increase in skeletal muscle protein synthesis. Furthermore, we simultaneously determined that overexpression of Rheb was sufficient to induce a 24% increase in fiber size within 4 d of transfection (Fig. 6G). Combined, these results indicate that a PKB-independent activation of mTOR is sufficient to induce an increase in protein synthesis and hypertrophy.
Figure 6.
IV-IHC-SUnSET reveals that PKB-independent activation of mTOR is sufficient to induce an increase in skeletal muscle protein synthesis and hypertrophy. TA muscles were transfected with plasmid DNA encoding the GFP (A–C) or HA-tagged Rheb (D–F). At 4 d after transfection, muscles were subjected to IV-IHC-SUnSET by staining for puromycin and GFP or the HA tag, respectively. A) Representative image of the signal for GFP-positive fibers. B) Grayscale image of the signal for puromycin in the same section shown in A. D) Representative image of signal for Rheb-positive fibers. E) Grayscale image of the signal for puromycin in the same section shown in D. C, F) Puromycin staining intensity in both transfected (positive) and nontransfected (negative) fibers, expressed relative to the mean value obtained in nontransfected (negative) fibers from the same section; values are plotted on histograms for GFP-transfected muscles (C) and Rheb-transfected muscles (F). G) Cross-sectional area (CSA) of the transfected fibers was measured and expressed relative to the CSA of nontransfected fibers in the same section. All values are means ± se; n = 180 transfected and nontransfected fibers/group from 3 independent muscles. *P < 0.05 vs. nontransfected fibers..
Skeletal muscle fiber type-dependent differences in protein synthesis
To further highlight the conceptually important types of data that can be obtained with our IV-IHC-SUnSET technique, we investigated the uniformity of protein synthesis rates among individual muscle fibers within whole skeletal muscles. We were motivated to perform these experiments because it has been hypothesized, but never clearly demonstrated, that different muscle fiber types have different rates of protein synthesis (16). This issue has not been successfully addressed because radioactive-based techniques lack the sensitivity needed to perform measurements on single fibers/cells. Our IV-IHC-SUnSET technique enabled us to overcome this limitation, and, consequently, we determined for the first time that type 2B and type 2X fibers have significantly lower rates of protein synthesis than type 2A fibers in two anatomically and functionally different mouse skeletal muscles (Fig. 7A–F and Supplemental Fig. S3A–F, respectively). In addition, we determined that only very minor differences in protein synthesis rates exist between type 2A and type 1 fibers in the mouse PLT and SOL muscles (Fig. 7G–I and Supplemental Fig. S3G–I). Furthermore, it has recently been proposed, although never demonstrated, that there is a negative correlation between muscle fiber size and the rate of protein synthesis (16). To investigate this issue, we performed a correlation analysis on fiber size and protein synthesis rates and found only a weak negative correlation between these variables (r=0.32; Supplemental Fig. S4A). Moreover, we performed the same analysis within each PLT fiber type group, in which there is ≥2-fold variation in both fiber size and protein synthesis rate, and surprisingly found either positive correlations (type 1 fibers, r=0.52; type 2A, r=0.2; Supplemental Fig. S4B, C) or no correlation at all (type 2X, r=0.14; type 2B, r=0.1; Supplemental Fig. S4D, E). Thus, the initial negative correlation between size and rate that was observed when all fiber types were analyzed as a single group was not found when the same analyses were performed within each specific fiber type. This finding suggested that the small negative correlation between fiber size and protein synthesis rate might simply be a result of fiber type-dependent differences in size (type 1≈2A<2X<2B). In support of this conclusion, we found that when we plotted the mean protein synthesis rate and mean fiber size for each of the 4 fiber types, a very strong negative correlation existed (r=0.99, P<0.007; Supplemental Fig. S4F). Combined, these observations indicate that there are differences in the rate of protein synthesis among the different fiber types found in mouse skeletal muscle, and these differences are more related to fiber type rather than to fiber size.
Figure 7.
IV-IHC-SUnSET reveals skeletal muscle fiber type-dependent differences in protein synthesis. A, D, G) PLT muscles were subjected to IV-IHC-SUnSET by triple staining for puromycin (Puro, red), type 2A fibers (green), and either type 2B (A), type 2X (D), or type 1 fibers (blue; G). B, E, H) Grayscale image of the puromycin signal in the same section as shown in A, D, and G, respectively. C, F, I) Puromycin staining intensity in type 2A fibers and either type 2B, type 2X, or type 1 fibers, expressed relative to the mean value obtained in type 2A fibers from a given section; values are plotted on histograms for type 2B (C), type 2X (F), and type 1 fibers (I). Values are means ± se; n = 87–120 fibers/group from 4 independent muscles. *P < 0.05 vs. type 2A fibers.
DISCUSSION
This study describes the first nonradioactive technique for measuring changes in protein synthesis, ex vivo and in vivo, in whole tissues via WB and at the single-cell level via IHC. This novel methodology is important because it allows for the visualization and quantification of protein synthesis and eliminates the need for generating radioactive tissues/animals.
To demonstrate that our technique was capable of detecting increases in protein synthesis, we used the SA model of muscle hypertrophy. The results show that the EV-SUnSET technique was indistinguishable from a standard radioactive-based technique for detecting the SA-induced increase in protein synthesis. The ex vivo SA-induced increase in protein synthesis observed in this study is also very similar to that previously reported in the SOL muscle ex vivo, using radioactive labeling (13). Notably, when applied in vivo, our IV-SUnSET technique also detected the SA-induced increase in protein synthesis, and the magnitude of this effect was not significantly different from that detected ex vivo. Our SA experiments also showed that our IV-SUnSET technique can detect increases in protein synthesis that occur concomitant with increases in rRNA. Furthermore, using insulin stimulation of C2C12 myoblasts, we provided evidence that the basic principles of SUnSET can also be used to detect acute increases in protein synthesis that occur in the absence of changes in rRNA. Combined, these results indicate that our technique can accurately detect an increase in skeletal muscle protein synthesis under both ex vivo and in vivo conditions.
To establish that our IV-SUnSET technique could also be used to detect decreases in protein synthesis, we subjected mice to 48 h of food deprivation. The results demonstrated that food deprivation significantly reduced protein synthesis in the heart, kidney, and several different skeletal muscles. These results are important because they demonstrate that our IV-SUnSET technique can detect not only increases but also decreases in protein synthesis in vivo. Furthermore, the results demonstrate that our technique can successfully be used in a variety of different tissues.
Because of limitations in traditional radioactive techniques, investigators have not been able to examine in vivo protein synthesis rates at the single fiber/cell level. The IV-IHC-SUnSET version of our technique overcomes this limitation and allowed us to measure the effect of 48 h of food deprivation on protein synthesis in muscle cross sections. Notably, the results with our IV-IHC-SUnSET technique showed excellent quantitative agreement compared with results obtained in whole-tissue homogenates with IV-SUnSET (53 vs. 65%, P=0.33). Thus, these results demonstrate that, under conditions affecting the whole tissue, protein synthesis measurements made on individual cells with IV-IHC-SUnSET accurately reflect the changes that occur at the whole-tissue level when measured with IV-SUnSET.
To determine whether our IV-IHC-SUnSET technique could detect changes that occur specifically within a subset of cells of a given tissue, we performed in vivo transfection of skeletal muscle with CA-PKB. With this approach, we were able to compare protein synthesis rates between transfected fibers and nontransfected fibers within the same skeletal muscle section. We chose CA-PKB for this analysis because it has been well documented that transfection with CA-PKB induces a large hypertrophic response at the single fiber level (17). However, it has never been demonstrated that this response is due to an increase in the rate of protein synthesis. Because in vivo transfection efficiencies are often relatively low, measurements of the effect of CA-PKB at the whole-tissue level could severely underestimate the effect present specifically within the transfected fibers. Using our IV-IHC-SUnSET technique, we demonstrated that protein synthesis rates are in fact dramatically elevated (155% increase) in muscles fibers transfected with CA-PKB compared with nontransfected fibers. Furthermore, we transfected fibers with GFP as an alternative control condition and show that the effects of CA-PKB are not simply due to the effects of electroporation/transfection. Thus, these results demonstrate that our IV-IHC-SUnSET technique can be combined with other in vivo molecular perturbations to determine the effects of various signaling molecules on protein synthesis at the single fiber/cell level.
It has been proposed that PKB induces protein synthesis by promoting mTOR signaling to downstream substrates that regulate cap-dependent translation (e.g., ribosomal S6 kinase and eukaryotic initiation factor 4E binding protein; ref. 18). However, PKB can also induce signaling through mTOR-independent regulators of protein synthesis such as GSK3β, which could, in turn, affect other potential regulators of translation such as the eukaryotic initiation factor 2B (19) and β-catenin (20). Thus, to date, it has not been established whether a PKB-independent activation of mTOR is sufficient to induce a global increase in protein synthesis. By combining our IV-IHC-SUnSET technique with transient in vivo transfection, we determined that overexpression of Rheb, one of the most proximal and PKB-independent activators of mTOR (8, 15), induces a 60% increase in protein synthesis and a 24% increase in muscle fiber size within 4 d. Based on these results, it can now be concluded that a PKB-independent activation of mTOR is indeed sufficient to induce an increase in protein synthesis, as well as hypertrophy in vivo. This observation is particularly significant because some physiologically relevant types of muscle growth-promoting stimuli, such as mechanical loading, are known to induce mTOR signaling via a PKB-independent mechanism (9, 10).
It has been known for some time that protein synthesis rates differ between various skeletal muscles (21). However, whole skeletal muscles are heterogeneous tissues made up of muscle fiber types composed of different contractile protein isoforms (e.g., MHC isoforms), with varying proportions of membranous systems (sarcoplasmic reticulum and t-tubular system), and enzymes and substrates associated with energy metabolism (e.g., mitochondria and glycogen; ref. 22). Thus, whole-muscle studies have not been able to answer whether differences in protein synthesis exist between individual muscle fiber types within the same whole muscle. In a recent review, it has been hypothesized that smaller highly oxidative fiber types (e.g., types 1 and 2A) have greater rates of protein synthesis than larger glycolytic fiber types (e.g., types 2X and 2B; ref. 16). Using our IV-IHC-SUnSET technique on mouse PLT muscle sections, which contain all four limb muscle fiber types, we have shown for the first time that type 2B and 2X fast-twitch fibers do indeed have lower rates of protein synthesis than type 2A fibers within the same muscle (∼34 and ∼22%, respectively). Interestingly, however, there was no difference between the type 1 and type 2A fibers. Although our findings could be influenced by fiber type differences in perfusion (23) and thus differential kinetics of puromycin uptake, they are supported by the data of Habets et al. (24), who showed that rat muscle fiber 28S rRNA content, an indicator of translational capacity, decreased in the following order of fiber type: 2A > 2X > 2B. Thus, our results indicate that muscle fiber types differ, not only with respect to contractile properties and energy metabolism but also in their rates of protein synthesis.
It has also recently been proposed that muscle fiber protein synthesis rates are negatively related to muscle fiber size (16). When we examined the relationship between fiber size and protein synthesis rates across all fibers in the PLT muscle, we found that fiber size explained only a small proportion of the variance in protein synthesis rate. Furthermore, when we split these fibers into their fiber type groups and performed the same correlation analysis on each fiber type, we actually found positive correlations or none at all. These data suggested that, when all fibers are analyzed as a single group, the small negative correlation between fiber size and protein synthesis rate might simply be a result of fiber type-dependent differences in size (type 1≈2A<2X<2B). In support of this hypothesis, we found a strong negative correlation (r=0.99) when the mean data for fiber size and protein synthesis rate were plotted according to fiber type (Supplemental Fig. S4F). Taken together, these results suggest that muscle fiber protein synthesis rates are related more to fiber type per se than to fiber size. This relationship might be explained by use-dependent differences in protein turnover due to differences in the frequency of recruitment during normal activity (25). For example, motor units containing type 1 and 2A fibers are, in general, activated more frequently than motor units composed of 2X and 2B fibers, and this result could lead to the higher protein synthesis rates that occur in response to higher use-dependent protein degradation rates.
As demonstrated in this study, the use of SUnSET offers several methodological advantages over the use of traditional radioactive-based techniques. Furthermore, the SUnSET methodology avoids several assumptions that are typically made when radioactive measurements of protein synthesis are performed. For example, with traditional radioactive measurements, the charged tRNA pool represents the precursor pool. However, measurement of tRNA specific activity is technically difficult, and consequently the specific activity of the tRNA pool is usually assumed to be equal to the specific activity measured for the amino acid pool in the blood or incubation medium. This assumption is important because the specific activity of the charged tRNA pool can be influenced by several factors, including the concentrations of both labeled and endogenous amino acid pools, which, in turn, are influenced by the rate at which the amino acid is synthesized, metabolized, and degraded by the tissue of interest. Puromycin, however, is not a naturally occurring molecule in eukaryotes, and thus the problem of the specific activity of the precursor pool being affected by endogenous puromycin levels is not relevant. Second, because puromycin is a tRNA analog, unlike amino acids, the intracellular pool of puromycin is the equivalent of the activated tRNA pool. Nevertheless, despite many of the advantages of the SUnSET methodology, it is still limited by the fact that it cannot measure absolute or fractional rates of protein synthesis. Although this is a disadvantage, it does eliminate the need to assume that the precursor pool (free puromycin) reaches equilibrium in a near instantaneous amount of time. Instead, the SUnSET methodology, like traditional radioactive measurements of protein synthesis, relies on the primary assumption that the time for the precursor pool to reach equilibrium is not altered by the experimental paradigm. Because both techniques rely on this assumption, it can be argued that the SUnSET technique is an excellent nonradioactive alternative for measuring relative rates of protein synthesis.
In summary, this study describes an innovative nonradioactive technique that uses standard WB and IHC technologies to visualize and quantify relative ex vivo and in vivo rates of protein synthesis. We have demonstrated that this technique can be used in a variety of different tissues, and we have used it to obtain novel conceptual information that has not been possible to obtain with traditional radioactive-based methods. Thus, it is anticipated that this technique will allow for substantial advancement in research with the aim of understanding the mechanisms that regulate protein synthesis in numerous tissue and cell types.
Supplementary Material
Acknowledgments
The type 1, 2a, and 2b MHC monoclonal antibodies developed by S. Schiaffino and the type 2x MHC monoclonal antibody developed by C. Lucas were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the Eunice Kennedy Shriver National Institute of Child Health and Human Development and maintained by the University of Iowa Department of Biology (Ames, IA, USA).
This work was supported by U.S. National Institutes of Health grant AR057347 to T.A.H. and Agence Nationale de la Recherche DC-trans and Human Frontier Science Program Organization grant RGP0045/2005 to P.P.
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1. Wolfe R. R., Chinkes D. L. (2005) Isotope Tracers in Metabolic Research, John Wiley & Sons, Hoboken, NJ, USA [Google Scholar]
- 2. Schmidt E. K., Clavarino G., Ceppi M., Pierre P. (2009) SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 [DOI] [PubMed] [Google Scholar]
- 3. Nathans D. (1964) Puromycin inhibition of protein synthesis: incorporation of puromycin into peptide chains. Proc. Natl. Acad. Sci. U. S. A. 51, 585–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nakano K., Hara H. (1979) Measurement of the protein-synthetic activity in vivo of various tissues in rats by using [3H]puromycin. Biochem. J. 184, 663–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sandri M. (2008) Signaling in muscle atrophy and hypertrophy. Physiology (Bethesda) 23, 160–170 [DOI] [PubMed] [Google Scholar]
- 6. Erbay E., Park I. H., Nuzzi P. D., Schoenherr C. J., Chen J. (2003) IGF-II transcription in skeletal myogenesis is controlled by mTOR and nutrients. J. Cell Biol. 163, 931–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Y., Inoki K., Guan K. L. (2004) Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol. Cell. Biol. 24, 7965–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goodman C. A., Miu M. H., Frey J. W., Mabrey D. M., Lincoln H. C., Ge Y., Chen J., Hornberger T. A. (2010) A phosphatidylinositol 3-kinase/protein kinase B-independent activation of mammalian target of rapamycin signaling is sufficient to induce skeletal muscle hypertrophy. Mol. Biol. Cell 21, 3258–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hornberger T. A., Stuppard R., Conley K. E., Fedele M. J., Fiorotto M. L., Chin E. R., Esser K. A. (2004) Mechanical stimuli regulate rapamycin-sensitive signalling by a phosphoinositide 3-kinase-, protein kinase B- and growth factor-independent mechanism. Biochem. J. 380, 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. O'Neil T. K., Duffy L. R., Frey J. W., Hornberger T. A. (2009) The role of phosphoinositide 3-kinase and phosphatidic acid in the regulation of mammalian target of rapamycin following eccentric contractions. J. Physiol. 587, 3691–3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garlick P. J., McNurlan M. A., Preedy V. R. (1980) A rapid and convenient technique for measuring the rate of protein synthesis in tissues by injection of [3H]phenylalanine. Biochem. J. 192, 719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Adams G. R., Caiozzo V. J., Haddad F., Baldwin K. M. (2002) Cellular and molecular responses to increased skeletal muscle loading after irradiation. Am. J. Physiol. Cell Physiol. 283, C1182–C1195 [DOI] [PubMed] [Google Scholar]
- 13. Augert G., Monier S., Le Marchand-Brustel Y. (1986) Effect of exercise on protein turnover in muscles of lean and obese mice. Diabetologia 29, 248–253 [DOI] [PubMed] [Google Scholar]
- 14. Garlick P. J., Millward D. J., James W. P., Waterlow J. C. (1975) The effect of protein deprivation and starvation on the rate of protein synthesis in tissues of the rat. Biochim. Biophys. Acta 414, 71–84 [DOI] [PubMed] [Google Scholar]
- 15. Avruch J., Long X., Lin Y., Ortiz-Vega S., Rapley J., Papageorgiou A., Oshiro N., Kikkawa U. (2009) Activation of mTORC1 in two steps: Rheb-GTP activation of catalytic function and increased binding of substrates to raptor. Biochem. Soc. Trans. 37, 223–226 [DOI] [PubMed] [Google Scholar]
- 16. Van Wessel T., de Haan A., van der Laarse W., Jaspers R. (2010) The muscle fiber type-fiber size paradox: hypertrophy or oxidative metabolism? Eur. J. Appl. Physiol. 110, 665–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bodine S. C., Stitt T. N., Gonzalez M., Kline W. O., Stover G. L., Bauerlein R., Zlotchenko E., Scrimgeour A., Lawrence J. C., Glass D. J., Yancopoulos G. D. (2001) Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 3, 1014–1019 [DOI] [PubMed] [Google Scholar]
- 18. Nader G. A. (2005) Molecular determinants of skeletal muscle mass: getting the “AKT” together. Int. J. Biochem. Cell Biol. 37, 1985–1996 [DOI] [PubMed] [Google Scholar]
- 19. Jefferson L. S., Fabian J. R., Kimball S. R. (1999) Glycogen synthase kinase-3 is the predominant insulin-regulated eukaryotic initiation factor 2B kinase in skeletal muscle. Int. J. Biochem. Cell Biol. 31, 191–200 [DOI] [PubMed] [Google Scholar]
- 20. Schakman O., Kalista S., Bertrand L., Lause P., Verniers J., Ketelslegers J. M., Thissen J. P. (2008) Role of Akt/GSK-3β/β-catenin transduction pathway in the muscle anti-atrophy action of insulin-like growth factor-I in glucocorticoid-treated rats. Endocrinology 149, 3900–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goldberg A. L. (1967) Protein synthesis in tonic and phasic skeletal muscles. Nature 216, 1219–1220 [DOI] [PubMed] [Google Scholar]
- 22. Schiaffino S., Sandri M., Murgia M. (2007) Activity-dependent signaling pathways controlling muscle diversity and plasticity. Physiology (Bethesda) 22, 269–278 [DOI] [PubMed] [Google Scholar]
- 23. McAllister R. M., Amann J. F., Laughlin M. H. (1993) Skeletal muscle fiber types and their vascular support. J. Reconstr. Microsurg. 9, 313–317 [DOI] [PubMed] [Google Scholar]
- 24. Habets P. E., Franco D., Ruijter J. M., Sargeant A. J., Pereira J. A., Moorman A. F. (1999) RNA content differs in slow and fast muscle fibers: implications for interpretation of changes in muscle gene expression. J. Histochem. Cytochem. 47, 995–1004 [DOI] [PubMed] [Google Scholar]
- 25. Armstrong R. B., Laughlin M. H. (1985) Metabolic indicators of fibre recruitment in mammalian muscles during locomotion. J. Exp. Biol. 115, 201–213 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.