Abstract
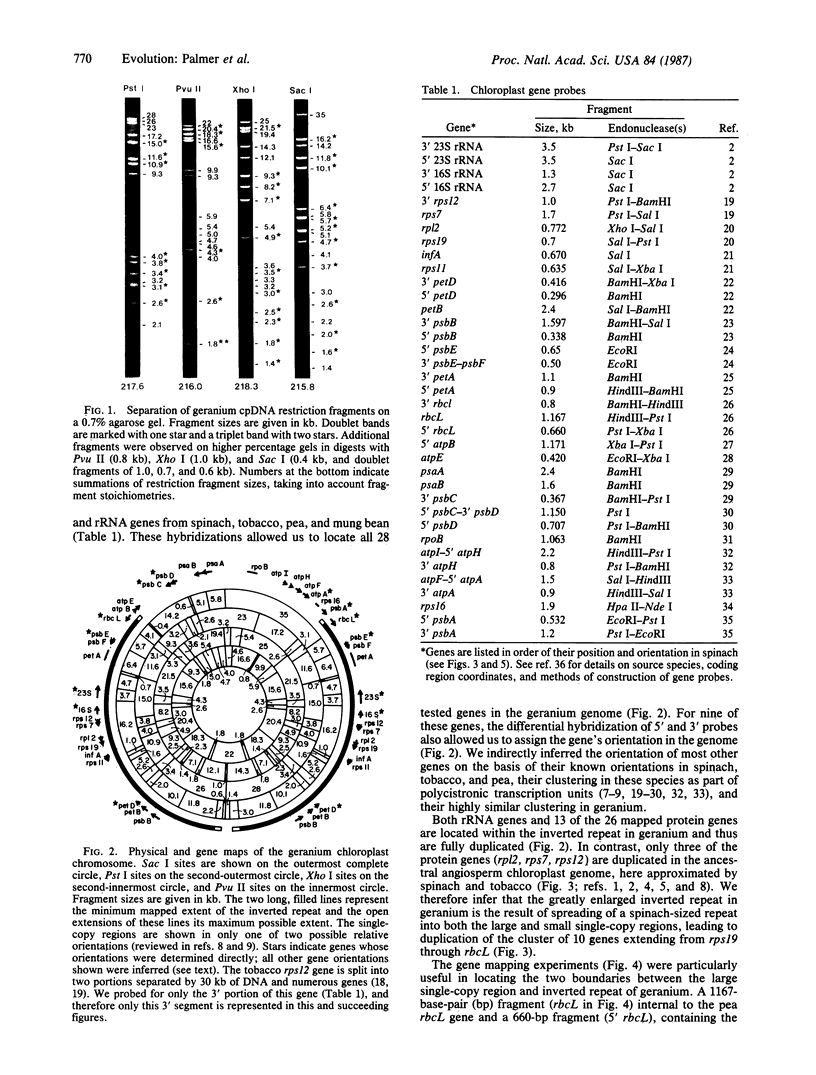
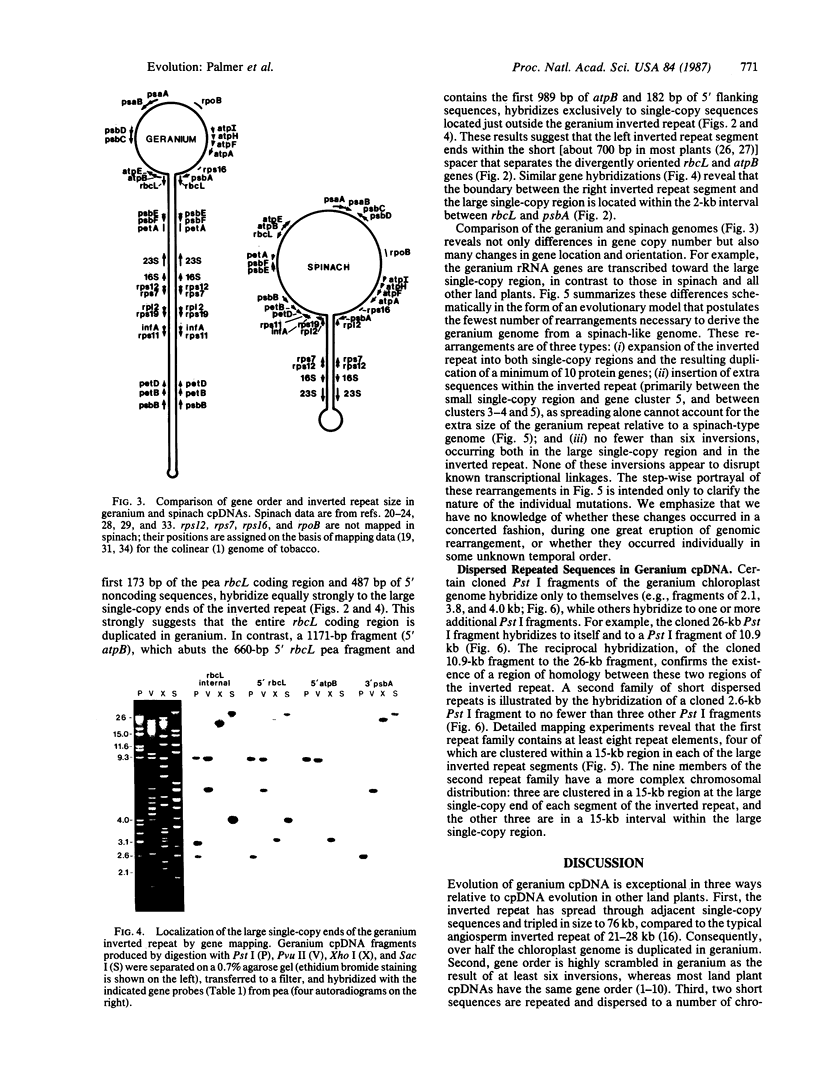
Physical and gene mapping studies reveal that chloroplast DNA from geranium (Pelargonium hortorum) has sustained a number of extensive duplications and inversions, resulting in a genome arrangement radically unlike that of other plants. At 217 kilobases in size, the circular chromosome is about 50% larger than the typical land plant chloroplast genome and is by far the largest described to date, to our knowledge. Most of this extra size can be accounted for by a 76-kilobase inverted duplication, three times larger than the normal chloroplast DNA inverted repeat. This tripling has occurred primarily by spreading of the inverted repeat into regions that are single copy in all other chloroplast genomes. Consequently, 10 protein genes that are present only once in all other land plants are duplicated in geranium. At least six inversions, occurring in both the inverted repeat and large single-copy region, must be postulated to account for all of the gene order differences that distinguish the geranium genome from other chloroplast genomes. We report the existence in geranium of two families of short dispersed repeats and hypothesize that recombination between repeats may be the major cause of inversions in geranium chloroplast DNA.
Keywords: genome evolution, rearrangement, Pelargonium hortorum
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennett M. D., Smith J. B. Nuclear dna amounts in angiosperms. Philos Trans R Soc Lond B Biol Sci. 1976 May 27;274(933):227–274. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- Cozens A. L., Walker J. E., Phillips A. L., Huttly A. K., Gray J. C. A sixth subunit of ATP synthase, an F(0) component, is encoded in the pea chloroplast genome. EMBO J. 1986 Feb;5(2):217–222. doi: 10.1002/j.1460-2075.1986.tb04201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluhr R., Edelman M. Conservation of sequence arrangement among higher plant chloroplast DNAs: molecular cross hybridization among the Solanaceae and between Nicotiana and Spinacia. Nucleic Acids Res. 1981 Dec 21;9(24):6841–6853. doi: 10.1093/nar/9.24.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm H., Edelman M., Koller B., Goloubinoff P., Galun E. The enigma of the gene coding for ribosomal protein S12 in the chloroplasts of Nicotiana. Nucleic Acids Res. 1986 Jan 24;14(2):883–898. doi: 10.1093/nar/14.2.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe C. J. The endpoints of an inversion in wheat chloroplast DNA are associated with short repeated sequences containing homology to att-lambda. Curr Genet. 1985;10(2):139–145. doi: 10.1007/BF00636479. [DOI] [PubMed] [Google Scholar]
- Morris J., Herrmann R. G. Nucleotide sequence of the gene for the P680 chlorophyll alpha apoprotein of the photosystem II reaction center from spinach. Nucleic Acids Res. 1984 Mar 26;12(6):2837–2850. doi: 10.1093/nar/12.6.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme M., Tanaka M., Chunwongse J., Shinozaki K., Sugiura M. A tobacco chloroplast DNA sequence possibly coding for a polypeptide similar to E. coli RNA polymerase beta-subunit. FEBS Lett. 1986 May 5;200(1):87–90. doi: 10.1016/0014-5793(86)80516-6. [DOI] [PubMed] [Google Scholar]
- Oishi K. K., Shapiro D. R., Tewari K. K. Sequence organization of a pea chloroplast DNA gene coding for a 34,500-dalton protein. Mol Cell Biol. 1984 Nov;4(11):2556–2563. doi: 10.1128/mcb.4.11.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. D. Comparative organization of chloroplast genomes. Annu Rev Genet. 1985;19:325–354. doi: 10.1146/annurev.ge.19.120185.001545. [DOI] [PubMed] [Google Scholar]
- Palmer J. D., Jorgensen R. A., Thompson W. F. Chloroplast DNA variation and evolution in pisum: patterns of change and phylogenetic analysis. Genetics. 1985 Jan;109(1):195–213. doi: 10.1093/genetics/109.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. D., Thompson W. F. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell. 1982 Jun;29(2):537–550. doi: 10.1016/0092-8674(82)90170-2. [DOI] [PubMed] [Google Scholar]
- Quigley F., Weil J. H. Organization and sequence of five tRNA genes and of an unidentified reading frame in the wheat chloroplast genome: evidence for gene rearrangements during the evolution of chloroplast genomes. Curr Genet. 1985;9(6):495–503. doi: 10.1007/BF00434054. [DOI] [PubMed] [Google Scholar]
- Sijben-Müller G., Hallick R. B., Alt J., Westhoff P., Herrmann R. G. Spinach plastid genes coding for initiation factor IF-1, ribosomal protein S11 and RNA polymerase alpha-subunit. Nucleic Acids Res. 1986 Jan 24;14(2):1029–1044. doi: 10.1093/nar/14.2.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmann A., Stutz E. Nucleotide sequence of soybean chloroplast DNA regions which contain the psb A and trn H genes and cover the ends of the large single copy region and one end of the inverted repeats. Nucleic Acids Res. 1983 Oct 25;11(20):7157–7167. doi: 10.1093/nar/11.20.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torazawa K., Hayashida N., Obokata J., Shinozaki K., Sugiura M. The 5' part of the gene for ribosomal protein S12 is located 30 kbp downstream from its 3' part in tobacco chloroplast genome. Nucleic Acids Res. 1986 Apr 11;14(7):3143–3143. doi: 10.1093/nar/14.7.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey D. L., Auffret A. D., Gray J. C. Structure and topology of cytochrome f in pea chloroplast membranes. Cell. 1984 Feb;36(2):555–562. doi: 10.1016/0092-8674(84)90248-4. [DOI] [PubMed] [Google Scholar]
- Zurawski G., Bottomley W., Whitfeld P. R. Junctions of the large single copy region and the inverted repeats in Spinacia oleracea and Nicotiana debneyi chloroplast DNA: sequence of the genes for tRNAHis and the ribosomal proteins S19 and L2. Nucleic Acids Res. 1984 Aug 24;12(16):6547–6558. doi: 10.1093/nar/12.16.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G., Bottomley W., Whitfeld P. R. Sequence of the genes for the beta and epsilon subunits of ATP synthase from pea chloroplasts. Nucleic Acids Res. 1986 May 12;14(9):3974–3974. doi: 10.1093/nar/14.9.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G., Bottomley W., Whitfeld P. R. Structures of the genes for the beta and epsilon subunits of spinach chloroplast ATPase indicate a dicistronic mRNA and an overlapping translation stop/start signal. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6260–6264. doi: 10.1073/pnas.79.20.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G., Whitfeld P. R., Bottomley W. Sequence of the gene for the large subunit of ribulose 1,5-bisphosphate carboxylase from pea chloroplasts. Nucleic Acids Res. 1986 May 12;14(9):3975–3975. doi: 10.1093/nar/14.9.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ee J. H., Vos Y. J., Planta R. J. Physical map of chloroplast DNA of Spirodela oligorrhiza; analysis by the restriction endonucleases PstI, xhoI, SacI. Gene. 1980 Dec;12(3-4):191–200. doi: 10.1016/0378-1119(80)90101-8. [DOI] [PubMed] [Google Scholar]