Abstract
Genetic rearrangements of the anaplastic lymphoma kinase (ALK) kinase occur in 3% to 13% of non–small cell lung cancer patients and rarely coexist with KRAS or EGFR mutations. To evaluate potential treatment strategies for lung cancers driven by an activated EML4-ALK chimeric oncogene, we generated a genetically engineered mouse model that phenocopies the human disease where this rearranged gene arises. In this model, the ALK kinase inhibitor TAE684 produced greater tumor regression and improved overall survival compared with carboplatin and paclitaxel, representing clinical standard of care. 18F-FDG-PET-CT scans revealed almost complete inhibition of tumor metabolic activity within 24 hours of TAE684 exposure. In contrast, combined inhibition of the PI3K/AKT and MEK/ERK1/2 pathways did not result in significant tumor regression. We identified EML4-ALK in complex with multiple cellular chaperones including HSP90. In support of a functional reliance, treatment with geldanamycin-based HSP90 inhibitors resulted in rapid degradation of EML4-ALK in vitro and substantial, albeit transient, tumor regression in vivo. Taken together, our findings define a murine model that offers a reliable platform for the preclinical comparison of combinatorial treatment approaches for lung cancer characterized by ALK rearrangement.
Introduction
Translocations involving the anaplastic lymphoma kinase (ALK) and nucleophosmin (NPM) were first identified in anaplastic large cell lymphomas (ALCLs) (1). Subsequently, ALK translocations involving novel partners have been identified in other cancers, including lung cancers (2), where the oncogenic event is most commonly due to a small inversion on chromosome 2p that leads to the fusion of ALK, including the entire kinase domain, with Echinoderm Microtubule–associated protein-like 4 (EML4), resulting in constitutive ALK kinase activity (3–6). EML4-ALK translocations are more frequent in adenocarcinomas and in never smokers (7–9). There are several EML4-ALK isoforms, all of which contain virtually identical portions of ALK, and possess potent in vitro transforming activity (3). The most common isoform is variant 1 (V1), fusing exon 13 of EML4 with exon 20 of ALK (3). This fusion oncogene has been detected both in primary lung cancers and in the H3122 cell line (3).
ALK inhibitors, including NVP-TAE684, are effective against the EML4-ALK H3122 cell line both in vitro and in xenografts (3, 10). In H3122 cells, TAE684-mediated ALK inhibition results in downregulation of PI3K/AKT and MEK/ERK1/2 signaling, and apoptosis. The ALK inhibitor crizotinib (PF-02341066), currently in clinical development for ALK-rearranged lung cancer, has demonstrated tumor regressions in approximately 60% of ALK-rearranged lung cancers in an early phase clinical trial (11, 12). These findings suggest that EML4-ALK–driven cancers display features of oncogene dependence or addiction and that ALK inhibitors may be particularly effective for this lung cancer subset.
Despite the therapeutic success of kinase inhibitors in oncogene-addicted tumors, including EGFR mutant lung cancers, chronic myeloid leukemia (CML) and gastrointestinal stromal tumor (GIST), acquired drug resistance develops universally (13–16). Therapeutic strategies to combat drug resistant cancers include the use of second-generation kinase inhibitors, and inhibitors of critical downstream signaling proteins activated by the mutant kinases. Another approach involves disruption of HSP90 function, because many mutant oncoproteins require HSP90 for maturation and conformational stability, and are degraded on HSP90 inhibition (17–19).
To evaluate further therapeutic strategies in ALK-rearranged lung cancer, we have generated a murine lung cancer model driven by inducible expression of the EML4-ALK fusion oncoprotein. Using this model, and the H3122 cell line, we have assessed the efficacy of kinase inhibition, standard chemotherapy, and HSP90 inhibition. These preclinical models provide comprehensive platforms to compare and prioritize potential treatments to evaluate in clinical trials for this lung cancer subpopulation.
Materials and Methods
Mouse drug treatment studies
The generation of genetically engineered mice harboring a doxycycline-inducible EML4-ALK fusion gene was similar to other mouse models we have described (20) and is presented in detail in the Supplementary Methods. These mice were imaged using magnetic resonance imaging (MRI) to document tumor burden after more than 3 weeks of doxycycline exposure (21). Chemotherapy included carboplatin (50 mg/kg in PBS) and paclitaxel (10 mg/kg in PBS) and was delivered by intraperitoneal injection twice weekly. TAE684 (25 mg/kg by oral gavage), 17-DMAG (LC laboratories; 20 mg/kg by daily intraperitoneal injection), AZD6244 (AZD; 25 mg/kg), BEZ235 (BEZ; 45 mg/kg), and WZ4002 (25 mg/kg by oral gavage) were administered as previously described (10, 20, 22). MRI scanning was performed at indicated time points to evaluate treatment effects. Mice were sacrificed after the last imaging time point to harvest tumors, and subjected to pathologic studies (21). Mice used in long-term treatment with different therapies are listed in Supplementary Table S1. In long-term experiments, 17-DMAG was administered 5 days per week, and TAE684 was administered every other day. For pharmacodynamic studies, 2 doses of drugs were administered within 24 hours, with the first dose on day 1 and the second dose on day 2, 3 hours prior to sacrifice and tumor harvest. Xenograft studies using nude mice were performed as previously described (3). For short-term pharmacodynamic studies, mice were given the same dose of 17-DMAG and sacrificed at days 0, 1, 2, 3, 5. Harvested tumors were snap frozen or formalin fixed for further study. Positron emission tomography–computed tomography (PET-CT) and subsequent treatment response measurements were performed as previously described (22). All mice were housed in a pathogen-free animal facility at the Harvard School of Public Health, and all animal experiments were approved by the Institutional Animal Care and Use Committee of Harvard University. Littermates were used as controls in all experiments.
Patient samples
Tumors were snap frozen in liquid nitrogen at the time of surgery. Frozen tumors were embedded in optimal cutting temperature media and sectioned. A hematoxylin and eosin–stained slide was reviewed and only samples without evidence of necrosis and with 60% or more tumor content were chosen for subsequent analyses. Thin sections were used for RNA preparation using Trizol (Invitrogen). EGFR (CTGGGGATCGGCCTCTTC and CCGTAGCTCCAGACAT-CACTCTG) and EML4-ALK (23) genotyping was performed using RT-PCR. All PCR-positive specimens were verified by sequencing. Details of the patient specimens are listed in Supplementary Table S2.
Gene expression profiling and cross-species Gene Set Enrichment Analysis
Microarray gene expression analysis was performed as described previously (24, 25). Probe level intensity data files in the CEL format were preprocessed using Robust Multi-chip Average program (http://rmaexpress.bmbolstad.com). Gene-expression data were filtered using low stringency, predefined criteria: probe set intensity was 32 in all samples and dynamic variation was more than 2-fold over the entire sample set. After filtering, probes representing the same genes were collapsed into a single value, and standardized by taking the median value for each gene across the sample set. Unsupervised hierarchical clustering was performed using Genepattern suite (http://broad.mit.edu/genepattern). A 2-sided t test was used to determine significant differences in gene expression between mouse tumors harboring EML4-ALK translocation and EGFR mutation. False positives associated with multiple hypothesis testing were calculated with the False Discovery Rate (FDR) method. Genes up- or downregulated by EML4-ALK with fold change greater than 2 and FDR P < 0.05 were considered components of up or downregulated signatures, respectively. Gene Set Enrichment Analysis (GSEA; http://broad.mit.edu/gsea) (26) was used to compare mouse-generated signatures with a rank-ordered gene list obtained from the human dataset. We used the signal-to-noise ratio of EML4-ALK–expressing compared with mutant EGFR-expressing tumors to produce this rank-ordered gene list, and permutation testing and FDR to calculate the significance of enrichment scores.
EML4-ALK variant 1 constructs and retroviral infection
EML4-ALK was cloned into pDNR-Dual (BD Biosciences) as previously described (3). The retroviral vector, JP1536HA was created by inserting a FLAG-tag and an HA-tag before the loxP site of JP1520 vector to allow tagging at the N terminus of the shuttled construct. The EML4-ALK V1 was shuttled into JP1536HA, using the BD Creator System (BD Biosciences). The empty retroviral construct JP1536HA was used as control. H3122 cells were infected with retrovirus according to standard protocols as described previously (13).
Tandem affinity purification, silver staining, and LC-MS/MS
Lysates from H3122-EML4-ALK-JP1536HA or H3122-JP1536HA–expressing cells were prepared in FLAG IP buffer [50 mmol/L Tris-HCl (pH 7.5), 150 mmol/L NaCl, 0.5% NP-40, 1 mmol/L EDTA, 10% glycerol, supplemented with EDTA-free Roche protease inhibitor cocktail]. Lysate (2 mg) was incubated with 20 μL of anti-FLAG agarose. FLAG-tagged protein was purified by affinity purification and eluted with FLAG peptide (Sigma). The eluate was incubated with 20 μL of anti-HA agarose with HA buffer [50 mmol/L Tris-HCl (pH 7.5), 150 mmol/L NaCl, 0.05% NP-40, 1 mmol/L EDTA, 10% glycerol, supplemented with EDTA-free Roche protease inhibitor cocktail]. HA-tagged proteins were eluted with HA peptide (Covance) from HA-agarose. 10% of the final eluate was analyzed by silver staining (SilverQuest Silver Staining Kit; Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions. The remaining 90% of the purified protein complexes were analyzed by a gel-free LC-MS/MS method as described previously (27) with minor modifications (see Supplementary Methods for detailed description).
Cell culture and cell proliferation assays
H3122 cells were maintained as previously described (3) and standard 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assays were used to assess cell proliferation in response to various compounds. (see Supplementary Methods for details of cell culture and compounds used).
Western blotting and immunoprecipitation
Cell lysates were subjected to Western blotting and immunoprecipitation using previously described protocols (19, 28). Antibodies used and experimental procedures are detailed in the Supplementary Methods.
Results
Expression of the EML4-ALK V1 fusion protein in mice leads to lung adenocarcinoma that is similar to the human disease
We generated doxycycline-inducible bitransgenic mice harboring the EML4-ALK (V1) allele (Supplementary Fig. S1A) in combination with the lung epithelial cell–specific reverse transactivator allele. In the absence of doxycycline induction, these mice are healthy with normal lung histology. Doxycycline-dependent lung-specific induction of EML4-ALK expression led to lung tumorigenesis with a latency of less than 10 days (Supplementary Fig. S1B). Lung tumor–bearing mice rapidly lost weight in the first 4 weeks (Supplementary Fig. S1C), and had a median survival of 7 to 8 weeks, confirming that EML4-ALK is a potent oncogene (Supplementary Fig. S1D). Withdrawal of doxycycline led to complete tumor regression within 2 weeks, as revealed by both MRI and histology, indicating that tumor initiation and maintenance are entirely dependent on EML4-ALK expression (Supplementary Fig. S1B). Detailed histologic analysis of the lungs demonstrated these were adenocarcinomas with predominantly bronchioloalveolar carcinoma features and with occasional pleural space and airway invasion by an acinar component (Supplementary Fig. S1B). ALK-rearranged lung cancers in humans are also predominately observed in adenocarcinomas (8). Although signet ring cell features have been observed in human ALK-rearranged lung cancers, we did not observe signet ring cells in the murine cancers (8, 29).
To determine molecular similarities between human and mouse EML4-ALK lung cancer, we performed gene expression studies. In both mice and humans, tumors harboring EML4-ALK and EGFR mutation demonstrated distinctive expression profiles, and tumors driven by the same oncogenic alteration all clustered within the same category, consistent with their genotypic background (Supplementary Fig. S2A and B). We then derived an EML4-ALK–specific expression signature by comparing EML4-ALK–driven tumor samples with EGFR-driven tumor samples in mice. Genes up or downregulated by EML4-ALK with fold change greater than 2 and FDR P < 0.05 were considered components of up- or downregulated signatures, respectively. Subsequent GSEA of these EML4-ALK gene sets (25) indicated significant correlation (P < 0.05) between mouse and human tumor samples (Supplementary Fig. S2C). These findings suggest that the EML4-ALK mouse lung cancers are similar to human lung cancer with the same genotype.
ALK kinase inhibitor is a more effective therapy than chemotherapy in EML4-ALK murine lung adenocarcinoma
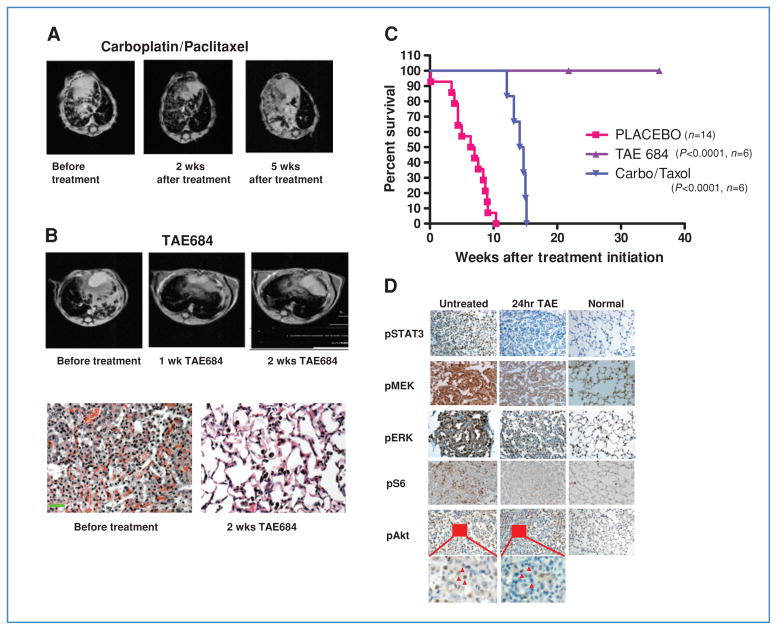
The current standard of care for advanced lung cancer is cytotoxic chemotherapy. However, for subsets of lung cancer, defined by an activated kinase oncogenic driver, kinase inhibitors may be more effective, as recently demonstrated for gefitinib in EGFR mutant disease (30). We therefore investigated whether a similar therapeutic paradigm would apply to EML4-ALK lung cancer in our preclinical model. We compared the efficacy of TAE684 (n = 9) to carboplatin/paclitaxel (n = 3) in mice with MRI-confirmed tumors following doxycycline induction. Carboplatin/paclitaxel treatment resulted in only a modest reduction in tumor volume (17%–27%) by 2 weeks as measured by MRI (Fig. 1A). Continuous treatment did not result in further tumor regression. Instead, resistance rapidly developed, and the tumors progressed and exceeded the original tumor burden by 5 weeks of treatment (Fig. 1A). In contrast, all of the TAE684-treated mice achieved complete regression within 2 weeks. Histologic analysis showed grossly normal lung structure without evidence of tumor cells (Fig. 1B). Additionally, the clinical condition of tumor-bearing TAE684-treated mice improved rapidly (Supplementary Fig. S1C), and they remained healthy without notable side effects. 18F-Fluorodeoxyglucose (18F-FDG), uptake in lung tumors by PET-CT scan was substantially diminished after only 2 doses of TAE684 within 24 hours, consistent with potent reduction of tumor metabolic activity, whereas no metabolic response was seen following treatment using an EGFR kinase inhibitor (Supplementary Fig. S3). In some of the mice, TAE684 treatment was then continued over an extended period of time (~75 weeks). To date, drug-resistant tumors have not developed. Withdrawal of TAE684 caused rapid tumor relapse, whereas reapplication of TAE684 reinduced complete regression. In the context of this model, TAE684 afforded superior survival (P < 0.0001; log rank test) compared with carboplatin/paclitaxel (Fig. 1).
Figure 1.
ALK-targeted therapy shows significant advantage over carboplatin/paclitaxel against murine EML-ALK lung cancer. A, representative MRI scans from a mouse harboring EML4-ALK–driven lung adenocarcinoma taken before treatment, and after 2 and 5 weeks of carboplatin/paclitaxel. B, MRI imaging and pathologic analysis demonstrate complete tumor regression following TAE684 for 2 weeks. Scale bar, 100 μm. C, survival curves following continuous treatment with vehicle, TAE684, or carboplatin/paclitaxel. D, tumor-bearing mice untreated or treated with TAE684 for 24 hours. Tumors were harvested and analyzed by immunohistochemistry for the indicated phosphoproteins, demonstrating significant downregulation of ALK signaling pathways. Small areas (insets) of pAkt staining are enlarged to compare signals before and after TAE684 treatment. Red arrows, tumor cells. Staining and histology of lungs from nontumor bearing mice are shown for comparison.
We next evaluated the effects of TAE684 treatment on downstream signaling proteins. Mice were treated with either vehicle or TAE684, sacrificed 2 hours following treatment and tumors examined by immunohistochemistry. In the TAE684-treated mice, there was significant downregulation of p-AKT, p-ERK1/2, p-S6, and p-STAT3, all of which have been previously identified in signaling pathways engaged by NPM-ALK (31–34) (Fig. 1D).
Inhibition of PI3K and MEK, but not STAT3, suppresses growth of an EML4-ALK-expressing lung cancer cell line and modestly inhibits tumor progression in vivo
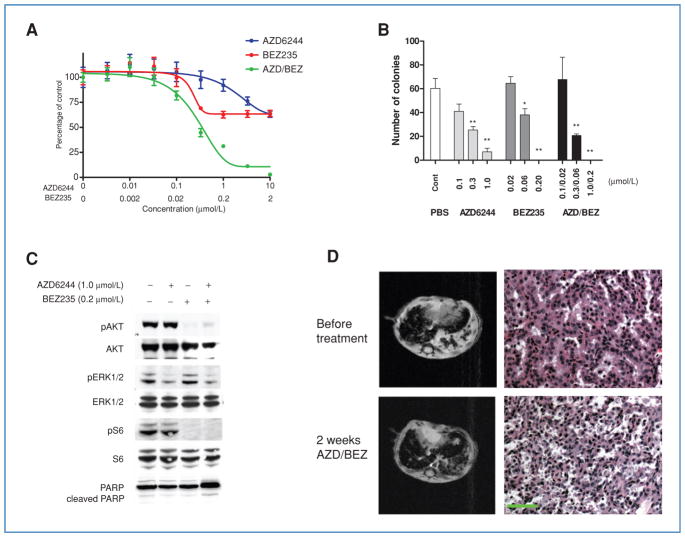
Simultaneous inhibition of the PI3K/Akt/mTOR and MEK/ERK1/2 pathways has been successful in preclinical models of KRAS and EGFR mutant non–small cell lung carcinoma (NSCLC) (21, 32), prompting us to evaluate a similar strategy in EML4-ALK–driven murine lung cancer and in the H3122 cells. Additionally, previous studies in ALCL harboring NPM-ALK rearrangement demonstrated the importance of STAT3 activation (32). In these cells, STAT3 is mainly activated by JAK3, a client of NPM-ALK (35). Because the expression of JAK3 is largely limited to hematopoietic tissues (36, 37), whether STAT3 activation plays a critical role in EML4-ALK lung tumor cells is unknown. The STAT3 inhibitor, S3i-201 (38) was not effective in H3122 cells. In contrast, the MEK inhibitor AZD and the PI3K/mTOR inhibitor NVP-BEZ suppressed H3122 proliferation either as single agents or in combination (Fig. 2A and B). The treated cells demonstrated downregulation of phospho-AKT and phospho-ERK 1/2 (Fig. 2C). mTOR activity was also sharply reduced with BEZ (Fig. 2C). The concentration levels of AZD and BEZ are comparable with the effective concentrations used previously in EGFR mutant NSCLC cell lines (39). However, in the murine model of EML4-ALK lung cancer, combined inhibition of MEK and PI3K/mTOR signaling, using previously established treatment conditions and dosing schedules (22), resulted in only modest reduction in tumor burden after 2 weeks of treatment. Among 4 treated mice, we detected tumor regression of approximately 20%, comparable with the efficacy achieved by chemotherapy, but far less effective than TAE684 (Fig. 2D). We were not able to perform experiments utilizing the triple combination of AZD/BEZ and S3i-201 due to unacceptable systemic toxicity. We also employed a JAK3 inhibitor, CP-690550, at established doses (40, 41) in combination with AZD/BEZ, but did not observe enhanced tumor regressions.
Figure 2.
PI3K/mTOR and MEK/ERK1/2 inhibition reduce viability of EML4-ALK-dependent lung cancer cells in vitro but not in vivo. A, H3122 cells were treated with the indicated concentrations of the MEK inhibitor AZD6244, the PI3K-mTOR inhibitor BEZ235, or the combination of both drugs. Cell viability was determined after 72 hours by MTS assay. Data are presented as the percentage of viable cells compared with untreated cells. B, H3122 cells were treated with indicated doses of AZD6244, BEZ235, or both for 14 days, and colonies counted. Bars denote SD. Student’s t tests were performed comparing PBS with AZD6244, BEZ235, or AZD/BEZ. *, P < 0.01, **, P < 0.001. C, H3122 cells were treated with PBS, AZD6244 (1 μmol/L), BEZ235 (0.2 μmol/L), or both for 6 hours. Lysates were subjected to Western blotting with the indicated antibodies, demonstrating expected effects on signal transduction by these compounds. D, EML4-ALK tumor-bearing mice were treated with the combination of AZD6244 and BEZ235. Tumor volumes were documented by MRI imaging. Mice were sacrificed after 2 weeks of treatment for pathologic analysis. Scale bar, 100 μm.
EML4-ALK interacts with HSP family members
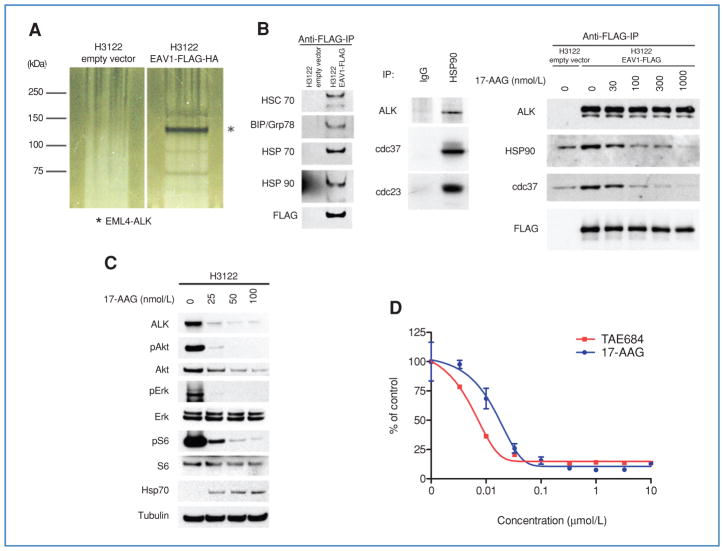
To further study ALK signaling and to identify additional potential therapeutic targets, we examined EML4-ALK–associated proteins using tandem affinity purification coupled with mass spectrometry. We constructed an EML4-ALK V1 expression vector tagged with FLAG and HA, introduced it into H3122 cells followed by 2 rounds of immunoprecipitation-based purification (Fig. 3A). We identified several interacting proteins (Fig. 3A) and analyzed these by MS (Supplementary Table S3A). As anticipated, we detected EML4 (both full length and EML4-ALK) and ALK (from EML4-ALK only) as among the most abundant proteins (detected by 13 and 8 unique peptides, respectively; Supplementary Fig. S4A and B). Additionally, we also detected heat shock protein (HSP) family members (HSPA5 (BIP/grp78) and HSPA8 (HSC70)) as abundant interacting proteins (both detected by 5 unique peptides; Supplementary Fig. S4C and D). Neither protein was detected in the control affinity purification.
Figure 3.
Association of EML4-ALK proteins with the HSP complex. A, silver staining of Tandem Affinity-purified proteins associated with EML4-ALK V1. *, EML4-ALK; other bands represent novel proteins. B, EML4-ALK is a sensitive HSP90 client. Left, validation of EML4-ALK–associated proteins. Lysates from H3122 cells expressing FLAG-HA–tagged EML4-ALK V1 or empty vector were subjected to immunoprecipitation with anti-FLAG antibodies followed by Western blotting with antibodies against the indicated proteins, demonstrating the association of multiple HSP family members with ectopic EML4-ALK. Middle, association between endogenous EML4-ALK and HSP90 in H3122 cells. Lysates were subjected to immunoprecipitation with IgG or an anti-HSP90 antibody followed by Western blotting for ALK, cdc37, and p23. Right, the association of EML4-ALK with HSP90 and cdc37 is disrupted by 17-AAG treatment. Cells were treated with the indicated concentrations of 17-AAG for 1 hour. Lysates were subjected to immunoprecipitation with an anti-FLAG antibody followed by Western blotting for the indicated proteins. C, H3122 cells were treated with the indicated concentrations of 17-AAG for 24 hours and lysates analyzed by Western blotting for the indicated proteins, demonstrating depletion of ALK and downstream signaling proteins at a 17-AAG concentration as low as 25 nmol/L. D, H3122 cells were treated with TAE684 or 17-AAG for 72 hours, and analyzed using the MTS assay.
To validate the physical association of the HSP protein complex and EML4-ALK, we performed coimmunoprecipitation experiments using the FLAG/HA-tagged EML4-ALK expression construct. Both HSPA5 and HSPA8, which were identified by MS, coprecipitated with EML4-ALK (Fig. 3B). Moreover, additional HSP family members, including HSPA1A (HSP70) and HSP90, were also detected in association with EML4-ALK (Fig. 3B). We further confirmed the endogenous association of HSP90 in the H3122 cells with ALK by immunoprecipitation with an HSP90 antibody. ALK and 2 other known HSP90 partners, cdc37 and p23, were detected in complex with HSP90 (Fig. 3B). The association of EML4-ALK and HSP90 was disrupted by 17-AAG–mediated HSP90 inhibition (Fig. 3B). These findings suggest that HSP family members may play a critical role in protein folding and structural stability of EML4-ALK.
To determine a functional role for HSP family members in maintaining stability of EML4-ALK, we treated H3122 cells with 17-AAG. EML4-ALK was efficiently depleted following 17-AAG treatment, with concomitant extinguishing of downstream signaling, evident by reduced p-AKT, p-ERK1/2, and p-S6 (Fig. 3C). HSP70 expression increased following 17-AAG treatment, a pharmacodynamic marker of effective HSP90 inhibition (42). Furthermore, 17-AAG inhibited H3122 proliferation with an IC50 of 20 nmol/L (Fig. 3D). Taken together, our findings indicate that EML4-ALK is a sensitive HSP90 client.
HSP90 inhibition causes regression of EML4-ALK–driven H3122 xenografts and murine lung adenocarcinomas
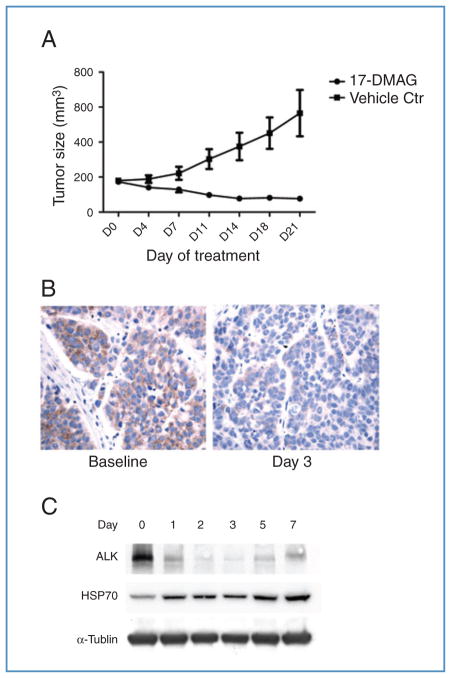
To confirm a potential therapeutic effect of HSP90 inhibition on H3122 cells in vivo, we established xenografts and treated the mice with either vehicle or the water-soluble geldanamycin 17-DMAG. As demonstrated in Fig. 4A, 17-DMAG caused tumor regression in this model. Additionally, short-term treatment with 2 doses of 17-DMAG within 24 hours confirmed marked reduction in total ALK expression, as demonstrated by immunohistochemical staining (Fig. 4B) and Western blotting (Fig. 4C) of harvested xenografts. We further observed HSP70 induction in the xenografts, consistent with the pharmacodynamic effects of 17-DMAG treatment (Fig. 4C).
Figure 4.
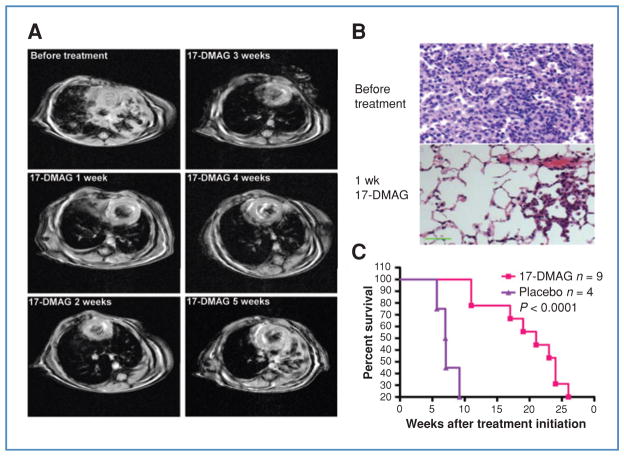
17-DMAG–mediated HSP90 inhibition is effective against EML4-ALK–driven lung cancer in H3122 xenograft. A, mice bearing H3122 xenografts were treated with vehicle or 17-DMAG, and tumor volumes were plotted over time. Bars denote SE. B, immunohistochemical analysis for ALK. Left, H3122 xenografts express EML4-ALK in the cytoplasm. Right, following 3 days of treatment with 17-DMAG, the expression of ALK was markedly reduced. C, Western blots of tumor lysates from mice treated with 17-DMAG at the indicated time points, demonstrating reduced ALK expression and induction of HSP70. Lane 0, untreated.
We next treated tumor-bearing EML4-ALK transgenic mice with 17-DMAG. Similar to the results with H3122 xenografts, we observed an average of 84% tumor regression (n = 5, Supplementary Table S4) within 1 week of treatment (Fig. 5A, mouse 292). Histologic analysis showed remnant cancer cells and dramatic restoration of normal lung structure (Fig. 5B). We continued to treat these mice for an extended period of time, and documented tumor volume each week by MRI (Fig. 5A; Supplementary Table S4). Our results showed that tumor response was not durable, and varied significantly among mice during treatment (Supplementary Table S4).
Figure 5.
17-DMAG is effective against EML4-ALK–driven lung cancer in genetically engineered mouse model. A, representative MRI images from mouse 292, harboring EML4-ALK lung adenocarcinoma, treated for 5 consecutive weeks with 17-DMAG, demonstrating an initial response that was not durable (see Supplementary Table S4 for quantification of tumor volumes). B, hematoxylin and eosin (H&E) staining of tumor sections from mice left untreated or treated with 17-DMAG for 1 week, demonstrating only small residual tumor and restoration of normal lung architecture. Scale bar, 100 μm. C, Kaplan–Meier survival analysis of mice with EML4-ALK–driven lung cancer treated with 17-DMAG or placebo. Median survival of placebo control is 7.05 weeks whereas that for the 17-DMAG treated group is 21 weeks after treatment initiation.
To determine whether 17-DMAG impacted survival, we compared treatment with 17-DMAG to placebo. Median survival increased from 7 weeks in the placebo group to 21 weeks in the 17-DMAG–treated group, (P < 0.0001) (Fig. 5C). This improvement in overall survival was noted even though the durability of response did not match that achieved with TAE684 (Fig. 6A).
Figure 6.
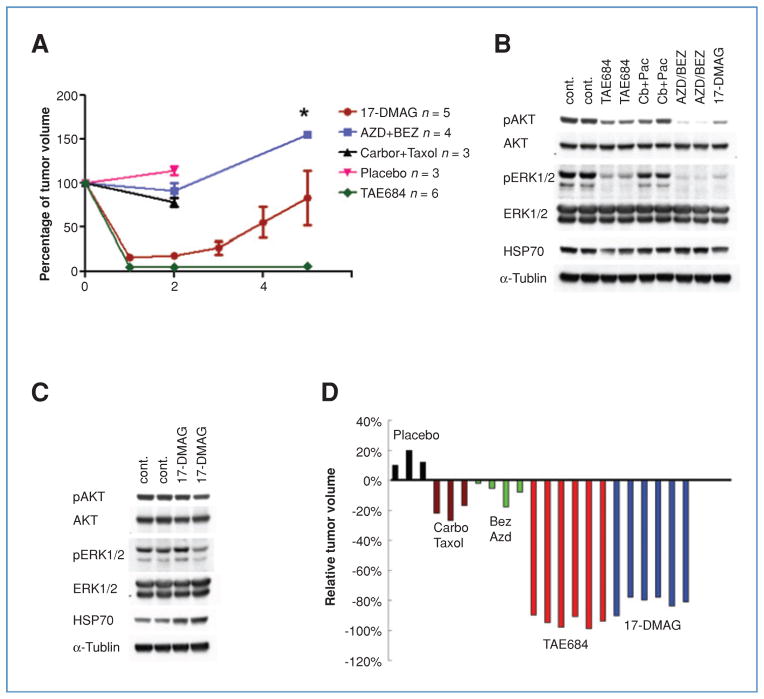
Comparison of different therapeutic strategies in the murine EML4-ALK lung cancer model. A, dynamics of tumor volume change with different treatment strategies. Time points for tumor volume sampling are indicated by colored dots. Bars denote SE. *, n = 2, no SE calculated. B, comparison of ALK downstream signaling in mice with EML4-ALK–driven lung cancer treated for 1 day with the indicated drug(s). Tumor lysates were analyzed by Western blot with the indicated antibodies. C, ALK downstream signaling following 40 days of treatment with vehicle or 17-DMAG. Tumor lysates were analyzed by Western blot with the indicated antibodies. D, tumor volume changes by MRI (Image J software) 2 weeks after the indicated treatments. Each color represents a different treatment approach; each bar represents tumor volume change after treatment in 1 mouse.
We also performed pharmacodynamic studies using tumors from the 17-DMAG–treated animals. After short-term treatment (24 hours), 17-DMAG treatment results in reduced expression of p-AKT and p-ERK1/2, similar to tumors from mice treated with TAE684 and AZD/BEZ (Fig. 6B). However, in recurrent tumors harvested after long-term treatment (40 days), signaling was restored, as demonstrated by p-AKT and p-ERK 1/2 levels similar to vehicle-treated mice (compare Fig. 6B and Fig. 6C). Nonetheless, HSP70 induction was noted in recurrent tumors, consistent with continued inhibition of HSP90 during the treatment course (Fig. 6C).
Discussion
ALK-rearranged lung cancers are a subset of cancers that are clinically sensitive to ALK inhibitors (11, 12). The ALK inhibitor crizotinib (PF-02341066) is currently undergoing clinical development in a randomized phase III trial and is being compared with standard chemotherapy. However, much remains to be understood about EML4-ALK biology, and the identification of alternative strategies to treat these cancers remains a clinical priority, because acquired resistance to targeted ALK inhibition is likely to emerge.
A recently published study described a mouse lung cancer model initiated by constitutively over-expressed EML4-ALK driven by lung-specific surfactant C promoter. This transgenic model also showed responses to an ALK-specific inhibitor (43). However, the short life span of these mice after birth, due to early expression of EML4-ALK in the late stage of embryonic development, potentially limits its use in performing comparative studies of different treatment strategies. We thus developed a new EML4-ALK mouse lung cancer model that phenocopies the molecular features of human ALK-rearranged lung cancer, and allows us to compare and prioritize therapeutic approaches.
Using this model, we demonstrate that inhibition of ALK activity, using TAE684, is more effective than conventional chemotherapy (Fig. 1C). The degree of tumor regression is analogous to that of EGFR kinase inhibitors used to treat mutant EGFR-driven murine lung cancers (44). However, in contrast to EGFR mutant lung cancer, the combination of PI3K and MEK inhibitors, although effective in vitro, was not effective in our EML4-ALK mouse model (39). These discrepancies attest to the importance of preclinical in vivo disease modeling in evaluating potential efficacy of individual treatment approaches. Our pharmacodynamic results indicated that both pAkt and pERK1/2 are efficiently suppressed by BEZ and AZD, suggesting that other potential EML4-ALK effector(s) may act to promote tumor survival in vivo, and could serve as important therapeutic target(s). It is possible that the strong expression of the EML4-ALK fusion protein in our model system may also require higher drug concentrations or more potent compounds for complete pathway inhibition. Further work will be required to address this issue and determine whether combined PI3K/MEK inhibition is a worthwhile strategy in EML4-ALK–driven lung cancer.
To identify other potential therapeutic targets, we demonstrate the association of EML4-ALK with several intracellular chaperones, including HSP90 (Fig. 3B). Previous studies suggested that NPM-ALK is also a client of HSP90 and HSP70 (45–47). We further demonstrated that geldanamycin compounds caused dissociation of HSP90 from EML4-ALK, and were effective in vitro, and in a xenograft model and in our murine adenocarcinoma model in vivo. In fact, 17-DMAG ranked second of the 4 treatments evaluated in the EML4-ALK–driven murine lung adenocarcinomas, and was more effective than chemotherapy and combined PI3K/mTOR/MEK inhibition (Fig. 6A and D).
Despite impressive initial responses to 17-DMAG responses were not durable. This result is similar to those observed with geldanamycins used to treat murine adenocarcinomas harboring EGFR mutation (19, 48). The mechanism by which resistance develops is presently not defined. However, we detected upregulation of HSP70 in mice that have developed resistance to 17-DMAG (Fig. 6C) suggesting continued HSP90 inhibition. Possible mechanisms of resistance to 17-DMAG could involve alterations in ALK, changes in expression pattern of intracellular chaperones or emergence of an oncogenic driver not dependent on HSP90 for conformational stability. Nonetheless, HSP90 inhibition tripled the survival of treated mice, indicating the importance of the initial tumor response. Of note, the geldnamycin IPI-504 has demonstrated preliminary activity in NSCLC in a phase III trial; 2 of 5 patients who achieved partial response had tumors harboring EML4-ALK translocations (49). These clinical findings further highlight the similarities of our mouse model to human EML4-ALK NSCLC. Further assessment of HSP90 inhibition, both with geldanamycin and new, potent nongeldanamycin HSP90 inhibitor compounds, is warranted (28, 50) and may represent an alternative approach to targeted ALK inhibition.
In summary, we have developed a model of EML4-ALK NSCLC that is similar both in molecular features (gene expression profile) and treatment response (ALK TKI and HSP90 inhibitor) to human EML4-ALK NSCLC. This preclinical model will be a useful tool for evaluating future therapies in this subset of NSCLC.
Supplementary Material
Acknowledgments
We thank Dr. Jing Chen (Emory University) for providing NPM-ALK plasmid and Dr. Hans-Juergen Heidebrecht (University of Kiel, Germany) for providing EML4 plasmid.
Grant Support
This study is supported by grants from the National Institutes of HealthR01 CA136851 (P.A. Jänne and N.S. Gray), R01 CA090687 (G.I. Shapiro), R01 AG2400401 (K-K. Wong), R01 CA122794 (K-K. Wong), P50 CA090578 [Dana-Farber/Harvard Cancer Center (DF/HCC) Specialized Program of Research Excellence (SPORE) in Lung Cancer (G.I. Shapiro, P.A. Jänne, and K-K. Wong)], the Kahn Family Charitable Foundation Fund for Lung Cancer Research (K-K. Wong and P.A. Jänne), and the Hazel and Samuel Bellin research fund (P.A. Jänne).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science. 1994;263:1281–4. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 2.Palmer RH, Vernersson E, Grabbe C, Hallberg B. Anaplastic lymphoma kinase: signalling in development and disease. Biochem J. 2009;420:345–61. doi: 10.1042/BJ20090387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, Holmes AJ, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–83. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perner S, Wagner PL, Demichelis F, Mehra R, Lafargue CJ, Moss BJ, et al. EML4-ALK fusion lung cancer: a rare acquired event. Neoplasia. 2008;10:298–302. doi: 10.1593/neo.07878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shinmura K, Kageyama S, Tao H, Bunai T, Suzuki M, Kamo T, et al. EML4-ALK fusion transcripts, but no NPM-, TPM3-, CLTC-, ATIC-, or TFG-ALK fusion transcripts, in non-small cell lung carcinomas. Lung Cancer. 2008;61:163–9. doi: 10.1016/j.lungcan.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi T, Sonobe M, Kobayashi M, Yoshizawa A, Menju T, Nakayama E, et al. Clinicopathologic features of non-small-cell lung cancer with EML4-ALK fusion gene. Ann Surg Oncol. 2010;17:889–97. doi: 10.1245/s10434-009-0808-7. [DOI] [PubMed] [Google Scholar]
- 8.Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–53. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong DW, Leung EL, So KK, Tam IY, Sihoe AD, Cheng LC, et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009;115:1723–33. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]
- 10.Galkin AV, Melnick JS, Kim S, Hood TL, Li N, Li L, et al. Identification of NVP-TAE684, a potent, selective, and efficacious inhibitor of NPM-ALK. Proc Natl Acad Sci USA. 2007;104:270–5. doi: 10.1073/pnas.0609412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bang Y, Kwak EL, Shaw AT, Camidge DR, Iafrate AJ, Maki RG, et al. Clinical activity of the oral ALK inhibitor PF-02341066 in ALK-positive patients with non-small cell lung cancer (NSCLC) [abstract] J Clin Oncol. 2010;28:3. [Google Scholar]
- 12.Kwak EL, Camidge DR, Clark J, Shapiro GI, Maki RG, Ratain MJ, et al. Clinical activity observed in a phase I dose escalation trial of an oral c-met and ALK inhibitor, PF-02341066 [abstract] J Clin Oncol. 2009;27(Suppl):3509. [Google Scholar]
- 13.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 14.Gajiwala KS, Wu JC, Christensen J, Deshmukh GD, Diehl W, DiNitto JP, et al. KIT kinase mutants show unique mechanisms of drug resistance to imatinib and sunitinib in gastrointestinal stromal tumor patients. Proc Natl Acad Sci USA. 2009;106:1542–7. doi: 10.1073/pnas.0812413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–80. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 17.Basso AD, Solit DB, Chiosis G, Giri B, Tsichlis P, Rosen N. Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function. J Biol Chem. 2002;277:39858–66. doi: 10.1074/jbc.M206322200. [DOI] [PubMed] [Google Scholar]
- 18.Grbovic OM, Basso AD, Sawai A, Ye Q, Friedlander P, Solit D, et al. V600E B-Raf requires the Hsp90 chaperone for stability and is degraded in response to Hsp90 inhibitors. Proc Natl Acad Sci USA. 2006;103:57–62. doi: 10.1073/pnas.0609973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimamura T, Lowell AM, Engelman JA, Shapiro GI. Epidermal growth factor receptors harboring kinase domain mutations associate with the heat shock protein 90 chaperone and are destabilized following exposure to geldanamycins. Cancer Res. 2005;65:6401–8. doi: 10.1158/0008-5472.CAN-05-0933. [DOI] [PubMed] [Google Scholar]
- 20.Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–4. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li D, Ji H, Zaghlul S, McNamara K, Liang MC, Shimamura T, et al. Therapeutic anti-EGFR antibody 806 generates responses in murine de novo EGFR mutant-dependent lung carcinomas. J Clin Invest. 2007;117:346–52. doi: 10.1172/JCI30446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–6. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeuchi K, Choi YL, Soda M, Inamura K, Togashi Y, Hatano S, et al. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res. 2008;14:6618–24. doi: 10.1158/1078-0432.CCR-08-1018. [DOI] [PubMed] [Google Scholar]
- 24.Carretero J, Shimamura T, Rikova K, Jackson AL, Wilkerson MD, Borgman CL, et al. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer Cell. 2010;17:547–59. doi: 10.1016/j.ccr.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, et al. An oncogenic KRAS2 expressionsignature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ficarro SB, Adelmant G, Tomar MN, Zhang Y, Cheng VJ, Marto JA. Magnetic bead processor for rapid evaluation and optimization of parameters for phosphopeptide enrichment. Anal Chem. 2009;81:4566–75. doi: 10.1021/ac9004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimamura T, Borgman CL, Chen L, Li D, Foley KP, Sang J, et al. The novel Hsp90 inhibitor STA-9090 has potent anticancer activity in in vitro and in vivo models of lung cancer [abstract] Proc Am Assoc Cancer res. 2010:4679. [Google Scholar]
- 29.Rodig SJ, Mino-Kenudson M, Dacic S, Yeap BY, Shaw A, Barletta JA, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res. 2009;15:5216–23. doi: 10.1158/1078-0432.CCR-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 31.Bai RY, Ouyang T, Miething C, Morris SW, Peschel C, Duyster J. Nucleophosmin-anaplastic lymphoma kinase associated with anaplastic large-cell lymphoma activates the phosphatidylinositol 3-kinase/Akt antiapoptotic signaling pathway. Blood. 2000;96:4319–27. [PubMed] [Google Scholar]
- 32.Chiarle R, Simmons WJ, Cai H, Dhall G, Zamo A, Raz R, et al. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat Med. 2005;11:623–9. doi: 10.1038/nm1249. [DOI] [PubMed] [Google Scholar]
- 33.Marzec M, Kasprzycka M, Liu X, El-Salem M, Halasa K, Raghunath PN, et al. Oncogenic tyrosine kinase NPM/ALK induces activation of the rapamycin-sensitive mTOR signaling pathway. Oncogene. 2007;26:5606–14. doi: 10.1038/sj.onc.1210346. [DOI] [PubMed] [Google Scholar]
- 34.Marzec M, Kasprzycka M, Liu X, Raghunath PN, Wlodarski P, Wasik MA. Oncogenic tyrosine kinase NPM/ALK induces activation of the MEK/ERK signaling pathway independently of c-Raf. Oncogene. 2007;26:813–21. doi: 10.1038/sj.onc.1209843. [DOI] [PubMed] [Google Scholar]
- 35.Amin HM, Medeiros LJ, Ma Y, Feretzaki M, Das P, Leventaki V, et al. Inhibition of JAK3 induces apoptosis and decreases anaplastic lymphoma kinase activity in anaplastic large cell lymphoma. Oncogene. 2003;22:5399–407. doi: 10.1038/sj.onc.1206849. [DOI] [PubMed] [Google Scholar]
- 36.Gurniak CB, Berg LJ. Murine JAK3 is preferentially expressed in hematopoietic tissues and lymphocyte precursor cells. Blood. 1996;87:3151–60. [PubMed] [Google Scholar]
- 37.Thomis DC, Gurniak CB, Tivol E, Sharpe AH, Berg LJ. Defects in B lymphocyte maturation and T lymphocyte activation in mice lacking Jak3. Science. 1995;270:794–7. doi: 10.1126/science.270.5237.794. [DOI] [PubMed] [Google Scholar]
- 38.Siddiquee K, Zhang S, Guida WC, Blaskovich MA, Greedy B, Lawrence HR, et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci USA. 2007;104:7391–6. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faber AC, Li D, Song Y, Liang MC, Yeap BY, Bronson RT, et al. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proc Natl Acad Sci USA. 2009;106:19503–8. doi: 10.1073/pnas.0905056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Changelian PS, Flanagan ME, Ball DJ, Kent CR, Magnuson KS, Martin WH, et al. Prevention of organ allograft rejection by a specific Janus kinase 3 inhibitor. Science. 2003;302:875–8. doi: 10.1126/science.1087061. [DOI] [PubMed] [Google Scholar]
- 41.Kudlacz E, Conklyn M, Andresen C, Whitney-Pickett C, Changelian P. The JAK-3 inhibitor CP-690550 is a potent anti-inflammatory agent in a murine model of pulmonary eosinophilia. Eur J Pharmacol. 2008;582:154–61. doi: 10.1016/j.ejphar.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 42.Sharp S, Workman P. Inhibitors of the HSP90 molecular chaperone: current status. Adv Cancer Res. 2006;95:323–48. doi: 10.1016/S0065-230X(06)95009-X. [DOI] [PubMed] [Google Scholar]
- 43.Solomon B, Varella-Garcia M, Camidge DR. ALK gene rearrangements: a new therapeutic target in a molecularly defined subset of non-small cell lung cancer. J Thorac Oncol. 2009;4:1450–4. doi: 10.1097/JTO.0b013e3181c4dedb. [DOI] [PubMed] [Google Scholar]
- 44.Ji H, Li D, Chen L, Shimamura T, Kobayashi S, McNamara K, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell. 2006;9:485–95. doi: 10.1016/j.ccr.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 45.Bonvini P, Dalla Rosa H, Vignes N, Rosolen A. Ubiquitination and proteasomal degradation of nucleophosmin-anaplastic lymphoma kinase induced by 17-allylamino-demethoxygeldanamycin: role of the co-chaperone carboxyl heat shock protein 70-interacting protein. Cancer Res. 2004;64:3256–64. doi: 10.1158/0008-5472.can-03-3531. [DOI] [PubMed] [Google Scholar]
- 46.Bonvini P, Gastaldi T, Falini B, Rosolen A. Nucleophosmin-anaplastic lymphoma kinase (NPM-ALK), a novel Hsp90-client tyrosine kinase: down-regulation of NPM-ALK expression and tyrosine phosphorylation in ALK(+) CD30(+) lymphoma cells by the Hsp90 antagonist 17-allylamino,17-demethoxygeldanamycin. Cancer Res. 2002;62:1559–66. [PubMed] [Google Scholar]
- 47.Georgakis GV, Li Y, Rassidakis GZ, Medeiros LJ, Younes A. The HSP90 inhibitor 17-AAG synergizes with doxorubicin and U0126 in anaplastic large cell lymphoma irrespective of ALK expression. Exp Hematol. 2006;34:1670–9. doi: 10.1016/j.exphem.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 48.Regales L, Balak MN, Gong Y, Politi K, Sawai A, Le C, et al. Development of new mouse lung tumor models expressing EGFR T790M mutants associated with clinical resistance to kinase inhibitors. PloS One. 2007;2:e810. doi: 10.1371/journal.pone.0000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sequist LV, Natale RB, Senzer NN, Martins R, Lilenbaum R, Gray JE, et al. Association between anaplastic lymphoma kinase rearrangements (rALK) and the clinical activity of IPI-504 (retaspimycin hydrochloride), a novel Hsp90 inhibitor, in patients with non-small cell lung cancer (NSCLC) [Abstract] J Clinl Oncol. 2010;28 (Suppl):7517. [Google Scholar]
- 50.Sessa C, Sharma SK, Britten CD, Vogelzang NJ, Bhalla KN, Mita MM, et al. A phase I dose escalation study of AUY922, a novel HSP90 inhibitor, in patients with advanced solid malignancies [Abstract] J Clin Oncol. 2009;27(Suppl):3532. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.