Abstract
Pilus assembly in gram-positive bacteria occurs by a two-step mechanism, whereby pilins are polymerized and then covalently anchored to the cell wall. In Corynebacterium diphtheriae, the pilin-specific sortase SrtA catalyzes polymerization of the SpaA-type pilus, consisting of the shaft pilin SpaA, tip pilin SpaC and minor pilin SpaB. Cell wall anchoring of the SpaA polymers is triggered when SrtA incorporates SpaB into the pilus base via lysine-mediated transpeptidation; anchoring to the cell wall peptidoglycan is subsequently catalyzed by the housekeeping sortase SrtF. Here we show that SpaB and SpaC formed a heterodimer independent from SpaA polymerization. SrtA was absolutely required for the formation of the SpaBC heterodimer, while SrtF facilitated the optimal cell wall anchoring of this heterodimer. Alanine substitution of the SpaB lysine residue K139 or truncation of the SpaB cell wall sorting signal (CWSS) abolished assembly of the SpaBC heterodimer, hence underscoring SpaB function in transpeptidation and cell wall linkage. Importantly, sortase specificity for the cell wall anchoring step was found to be dependent on the LAFTG motif within the SpaB CWSS. Thus, C. diphtheriae employs a common sortase-catalyzed mechanism involving lysine-mediated transpeptidation to generate both adhesive pilus and simple heterodimeric structures on the bacterial the cell wall.
Introduction
In gram-positive bacteria, a variety of cell wall anchored proteins including the proteinaceous appendages known as pili or fimbriae serve as key virulence determinants that are involved in many facets of bacterial pathogenesis (Mandlik et al., 2008b, Proft & Baker, 2009). A fundamental property of gram-positive bacteria, which distinguishes them from gram-negative bacteria, is their ability to covalently crosslink proteins to the cell wall peptidoglycan following translocation of the protein precursors across the single membrane that envelops the cytoplasm. The gram-positive microbes are able to join proteins to the cell wall through the action of a membrane-bound transpeptidase enzyme named sortase that was first identified in Staphylococcus aureus and shown to be essential for anchoring protein A to the cell wall (Mazmanian et al., 1999). The cell wall anchored proteins are directly recognized by sortase via a cell wall sorting signal (CWSS) present at their carboxyl terminus that is comprised of an LPXTG motif, followed by a short hydrophobic domain and a positively charged tail (Navarre & Schneewind, 1999, Marraffini et al., 2006). The basic mechanism of sortase catalyzed surface protein anchoring is as follows: Once a surface protein precursor is translocated across the cytoplasmic membrane, it is captured by sortase, which forms an acyl-enzyme intermediate by cleaving the LPXTG motif between threonine and glycine and joining the catalytic cysteine residue to the threonine residue of the surface protein. Subsequently, a nucleophilic attack by an amino-group of the stem peptide within the cell wall lipid II precursor resolves this high energy bond intermediate (Perry et al., 2002, Ruzin et al., 2002). As a result, the surface protein is covalently linked to a growing cell wall (Ton-That et al., 2004a). Homologs of S. aureus sortase are ubiquitous in gram-positive microbes and often genetically localized in clusters with genes encoding some of the surface proteins that harbor the CWSS (Ton-That & Schneewind, 2004, Telford et al., 2006, Scott & Zahner, 2006). Depending on their predicted structural features and functions, sortase homologs are divided into several classes, referred to as class A, B, C, or D (Dramsi et al., 2005, Comfort & Clubb, 2004). Forming the largest class are sortases of class C (subgroup 3) or pilin-specific sortases that are involved in crosslinking of monomeric pilins with the LPXTG motif to heteropolymeric pilus structures as first described in Corynebacterium diphtheriae (Ton-That & Schneewind, 2003, Mandlik et al., 2007). Three distinct types of pili, i.e. SpaA-, SpaD and SpaH-type (Gaspar & Ton-That, 2006, Swierczynski & Ton-That, 2006, Ton-That & Schneewind, 2003), are assembled on the surface of C. diphtheriae, which has served as one of the major model organisms for studies of the basic mechanisms of gram-positive pilus assembly (Mandlik et al., 2008b).
Our current view of the mechanism of pilus assembly is largely derived from extensive studies of the SpaA-type pilus of C. diphtheriae, the prototype pilus that specifically mediates the adherence of this organism to pharyngeal epithelial cells (Mandlik et al., 2007). C. diphtheriae harbors five pilin-specific sortases (srtA-E) clustered in three pilus loci, while the housekeeping sortase srtF, which encodes a non-polymerization sortase, resides at a separate location in the chromosome (Ton-That & Schneewind, 2003). The SpaA-type pilus, encoded by the spaA-srtA-spaB-spaC locus, is comprised of SpaA that forms the pilus shaft, SpaC located at the tip, and SpaB incorporated into the pilus structure. In addition to the LPXTG motif present in the CWSS of all pilus precursors, SpaA contains a pilin motif WxxxVxVYPK with a conserved lysine that participates in lysine-mediated transpeptidation (mentioned above), which links individual pilins to form the pilus polymer (Ton-That et al., 2004b). Pilus assembly begins with pilus polymerization catalyzed by the pilin-specific sortase SrtA, whereby SrtA cleaves precursor proteins at sorting signals between threonine and glycine of the LPXTG motif of each pilin, forming acyl-enzyme intermediates with the substrates. Crosslinking of pilins occurs when the lysine residue in the pilin motif of SpaA attacks the thioester of an adjacent acyl-enzyme intermediate (Ton-That & Schneewind, 2003, Mandlik et al., 2008b). Because SpaC does not have a pilin motif, it is thought that the first transpeptidation reaction occurs between SpaC and SpaA via the SpaA pilin motif and the SpaC LPXTG motif (Mandlik et al., 2008b). Mass spectrometry analysis of Bacillus cereus pili supports this mode of crosslinking between the tip and the shaft pilins (Budzik et al., 2009). The cyclic addition of SpaA pilin subunits to the resulting intermediates ensures the extension of pilus polymers, whose length is determined in part by the availability of the shaft pilin (Swierczynski & Ton-That, 2006, Ton-That & Schneewind, 2003).
Pilus extension is terminated when a SpaB molecule enters the pilus base via a similar lysine-mediated transpeptidation reaction using a lysine residue of SpaB (Mandlik et al., 2008a). The nucleophilic attack by the SpaB lysine residue K139 allows the transfer of the pilus polymer from the SrtA polymerization center to the housekeeping sortase SrtF, which proceeds to complete the assembly process by anchoring the polymer to the cell wall peptidoglycan. This biphasic mode of pilus assembly whereby pilus polymerization by a pilin-specific sortase precedes the cell wall anchoring of polymers by the non-polymerization sortase seems to be operative in several genera (Budzik et al., 2007, Nobbs et al., 2008). Consistent with the notion that a base pilin, such as SpaB in C. diphtheriae, functions as the molecular switch for termination of pilus polymerization, structural studies of pili expressed by Streptococcus pyogenes revealed the isopeptide bond formed between the pilin shaft Spy0128 and the basal pilin FctB via Thr311 of Spy0128 and Lys110 of FctB (Linke et al.). Notably, in C. diphtheriae, SpaB is also incorporated into the pilus shaft using the same lysine residue that can trigger the termination reaction (Mandlik et al., 2008a). While SrtA catalyzes the interspersed linkage of SpaB into the SpaA polymer, it is SrtF that joins the SpaB-terminated pilus shaft to the cell wall (see (Mandlik et al., 2008a)).
As major adhesins of the SpaA pili for corynebacterial binding to pharyngeal cells (Mandlik et al., 2007), SpaB and SpaC pilins are not only incorporated into pilus structures, but also individually anchored to the bacterial cell wall as monomers via their LPXTG motif. Previous work has determined that the housekeeping sortase SrtF catalyzes this pilus-independent cell wall anchoring of these and other minor pilins (Swaminathan et al., 2007). Here, we report the hitherto unsuspected surface display of these adhesive pilins as a covalently crosslinked heterodimer. We show that the pilin-specific sortase SrtA catalyzes the dimerization of SpaB and SpaC and that the housekeeping sortase SrtF catalyzes the efficient anchoring of the SpaBC heterodimer to the cell wall. Remarkably, the specificity for the two sortases is determined by the LPXTG motif of SpaB. The assembly of a bitopic adhesin by joining two disparate pilin adhesins together provides a novel strategy for corynebacteria to craft a lasting intimate zone of adhesion with its target host cell that may facilitate strong host cell signaling, localized toxin delivery and colonization of the pharyngeal tissue. Conceivably, the long-range and intimate interactions of corynebacteria with the host mediated by the pilus and the SpaBC heterodimer, respectively, may also provide a mechanism for gram-positive bacterial motility that is equivalent to the motility of gram-negative bacteria caused by pilus extension and retraction leading to bacterial invasion (Mattick, 2002, Merz et al., 2000).
Results
Cell wall anchoring of heterodimeric minor pilins SpaB and SpaC
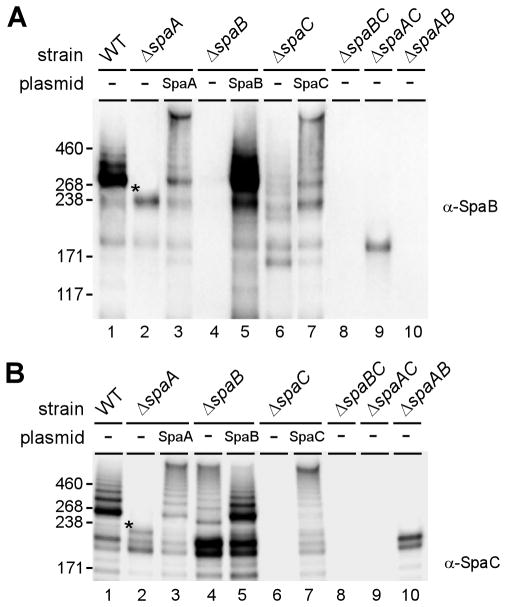
As mentioned above, our previous study showed that SpaB and SpaC are incorporated as individual proteins in the corynebacterial cell wall in addition to their assembly as minor pilins of the SpaA-pilus. The same study also detected two novel high molecular mass (HMM) species of SpaB (around 200 kDa, as compared to the SpaB precursor mass of 22 kDa) in a strain that was devoid of spaA and contained SrtA as the single sortase (Mandlik et al., 2007). To begin to decipher the molecular nature of these novel SpaB complexes, we first analyzed the status of SpaB and SpaC on the cell wall isolated from a series of strains by boiling the cell wall samples in SDS, separating proteins by SDS-PAGE and then blotting with antibodies against the Spa proteins (i.e. α-SpaB and α-SpaC). In a strain only lacking spaA, two predominant species of HMM SpaB were found, one migrating around 220 kDa (asterisk) and the other above 171 kDa, estimated 190 kDa (Fig. 1A, lane ΔspaA). This confirmed our previous observation and indicated that the formation of these HMM complexes is not strain-specific. These two complexes were also detected in the wild type strain but not in a strain lacking spaB (Fig. 1A, lanes WT & ΔspaB). A SpaB-complementing plasmid restored the expression of these specific bands (Fig. 1A, lane 5). Interestingly, when spaC was deleted, the 220 kDa SpaB band disappeared, whereas the 190 kDa SpaB band was still produced (Fig. 1A, lane ΔspaC). The 220 kDa SpaB band was restored by a SpaC-complementing plasmid (Fig. 1A, lane 7), suggesting that the 220 kDa SpaB band may contain SpaC. This inference is consistent with the combined mass of a heterodimer of SpaB (precursor mass of 22 kDa) and SpaC (precursor mass of 202 kDa) and the fact that the 220 kDa SpaB species could not be observed in strains lacking both spaB and spaC, spaA and spaC, or spaA and spaB (Fig. 1A, last three lanes). Indeed, the western blot of these same cell wall samples with α-SpaC antibody confirmed that the 220 kDa band contains SpaC, and that the formation of this band is dependent on the expression of both SpaB and SpaC (see Fig. 1B; lanes 2, 4, 6, 8–10). Attempts to determine the nature of the 190 kDa SpaB-reactive species have so far been unsuccessful.
Fig. 1.
Formation of a high molecular mass (HMM) complex of SpaB and SpaC. Cell wall-linked pilins were isolated by mutanolysin treatment from C. diphtheriae NCTC12139 (WT) and its isogenic derivatives grown on blood agar plates. Protein samples were boiled in SDS and separated on 3–12% Tris-glycine gradient gels and detected by immunoblotting with antibodies against SpaB (α-SpaB) (A) and SpaC (α-SpaC) (B). Asterisk (*) indicates a HMM protein complex of SpaB and SpaC. The positions of molecular mass markers (kDa) are shown.
A SpaB-SpaC complex is recovered by immunoprecipitation and affinity chromatography
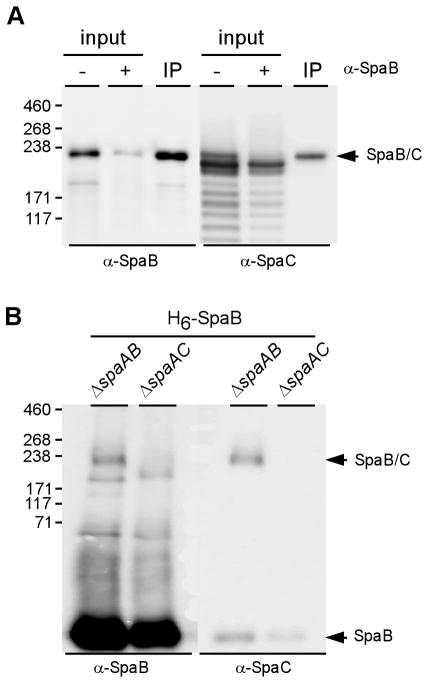
To determine if SpaB and SpaC are indeed crosslinked to form a single molecule, we analyzed secreted and cell wall anchored proteins by immunoprecipitation and affinity chromatography. In the immunoprecipitation experiment, we used culture medium of the strain ΔspaA/ΔsrtF, which is shown below to secrete the SpaBC complex, grown to late log-phase. The supernatant was concentrated (see Experimental procedure) and the concentrated supernatant was incubated with α-SpaB-bound Dynabeads. Equivalent aliquots of the concentrated supernatant (Fig. 2A, input), spent supernatant after immunoprecipitation (Fig. 2A, lane +), and eluted material from the beads (Fig. 2A, lane IP) were analyzed by SDS-PAGE and western blotting with α-SpaB and α-SpaC antibodies (see Experimental procedure). The data in Fig. 2A demonstrate that SpaC is co-immunoprecipitated with SpaB and that the antibody-selected complex does not dissociate upon boiling with SDS.
Fig. 2.
Covalent crosslinking of SpaB and SpaC. (A) Co-immunoprecipitation of SpaB- SpaC. The culture medium fraction of a strain lacking spaA and srtF was incubated with α-SpaB-bound Dynabeads (2:1 ratio). Protein samples were boiled in SDS and separated on 3–12% Tris-glycine gradient gels and α-SpaB-bound pilins were analyzed by immunoblotting with α-SpaB and α-SpaC. (B) Co-purification of SpaC by His-tagged SpaB. His-tagged SpaB from cell wall lysates of various strains expressing His-tagged SpaB from a multi-copy plasmid was purified by affinity chromatography. Purified protein samples were analyzed by immunoblotting with α-SpaB and α-SpaC.
We also isolated His-tagged SpaB expressed from a multi-copy plasmid by affinity chromatography of the cell wall fractions obtained from various corynebacterial strains all lacking spaA. The expected HMM SpaB-SpaC complex was again detected in the SpaB-complemented ΔspaAB strain but not in the SpaB-complemented ΔspaAC strain (Fig. 2B). Altogether, these results demonstrate the existence of a SpaB-SpaC heterodimer.
Surface assembly of the SpaBC heterodimer requires both the pilus-specific sortase SrtA and the housekeeping sortase SrtF
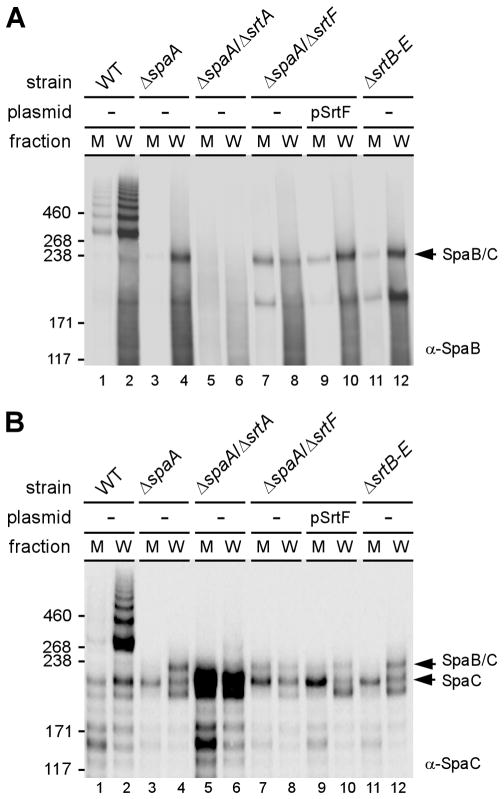
As mentioned above, pilins are crosslinked by the pilin-specific sortase SrtA and the anchoring of pilus polymers to the cell wall is predominantly catalyzed by the housekeeping sortase SrtF (Mandlik et al., 2008b). To examine how the SpaB-SpaC heterodimer is formed, and how this novel structure is linked on the bacterial surface, we analyzed culture medium (M) and cell wall (W) fractions of a series of mutant strains grown to mid-log phase. Equivalent protein samples concentrated by TCA precipitation were boiled in SDS and analyzed by western blotting with α-SpaB and α-SpaC (Fig. 3). With the wild type and ΔspaA strains, the SpaB-SpaC heterodimer reactive to both antibodies was mostly found in the cell wall fraction (Fig. 3, lanes 1–4). When srtA was deleted, the SpaB-SpaC heterodimer was completely abrogated (Fig. 3, lanes 5 and 6). By contrast, the SpaB-SpaC heterodimer still formed in the strain devoid of SrtF, but the complex was mostly secreted in the culture medium (compare lanes 7 and 8), unlike that observed with the wild type and the srtF+ ΔspaA parental strain (lanes 1 and 3). A SrtF-complementing plasmid restored cell wall anchoring of the SpaB-SpaC heterodimer in the ΔspaA/ΔsrtF mutant strain (lanes 9 and 10). Finally, when only SrtA and SrtF sortases were present (ie. in the ΔsrtB-E strain), the heterodimer was found predominantly in the cell wall fraction (lanes 11 and 12). Together, these results demonstrate that SrtA is absolutely required for the formation of the SpaB-SpaC heterodimer and that SrtF plays a predominant role in anchoring this heterodimeric adhesin to the cell wall.
Fig. 3.
Requirement of pilin-specific sortase SrtA and the housekeeping sortase SrtF for the formation of the SpaB-SpaC complex. Equivalent cultures of corynebacteria were fractionated into culture medium (M) and cell wall (W) fractions. Protein samples were boiled in SDS and separated on 3–12% Tris-glycine gradient gels and detected by immunoblotting with α-SpaB (A) and α-SpaC (B). The position of a HMM protein complex of SpaB and SpaC is shown with an arrow. Molecular mass markers (kDa) are indicated.
The SpaB-SpaC heterodimer is formed by a transpeptidation reaction mediated by lysine 139 of SpaB
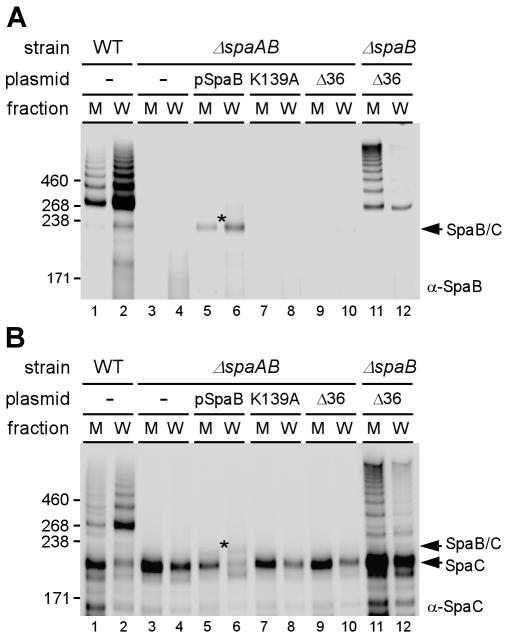
In a previous study, the incorporation of SpaB at the pilus base was shown to act as the molecular switch that terminates pilus polymerization by SrtA and initiates the cell wall anchoring of the pilus by SrtF (Mandlik et al., 2008a). Importantly, the transpeptidation reaction that incorporates SpaB into the pilus base was shown to require the lysine residue 139 of SpaB, which is postulated to join to the threonine residue of the SpaA LPXTG motif. In turn, the SpaB LPXTG motif joins the pilus to the cell wall. To determine whether the assembly of the SpaB-SpaC heterodimer also involves these two elements of SpaB, we next analyzed how mutations of these elements affect SpaB-SpaC heterodimerization and cell wall assembly (Fig. 4).
Fig. 4.
Requirement of the SpaB lysine residue 139 and cell wall sorting signal for the formation of SpaB-SpaC dimers. Cell fractionation of various strains was analyzed as previously described in Fig. 3. Plasmids expressing wild type SpaB (pSpaB), SpaB with mutation at K139 (K139A) and SpaB without the CWSS (Δ36) are shown.
As described above, both medium and cell wall fractions of a series of strains were analyzed by western blot with α-SpaB and α-SpaC. A control experiment confirmed that a complementing plasmid that expressed wild type SpaB restored the production of the SpaB-SpaC heterodimer in the Δ spaAB strain (Fig. 4AB, compare lanes 3–4 with lanes 5–6). More of the SpaB-SpaC heterodimer was recovered in the cell wall fraction compared to the medium. Strikingly, substitution of the SpaB K139 with alanine abolished the formation of the SpaB-SpaC heterodimer (Fig. 4AB- lanes 7–8). This is not due to a gross structural abnormality of the SpaB mutant protein; however, as we show below that this K139A mutant SpaB is still efficiently linked to the cell wall as monomers via its intact LPXTG motif. The SpaB-SpaC heterodimer was hardly detected when the SpaB cell wall sorting signal (CWSS) was deleted (Δ36 spaB mutant, lanes 9–10). Note that some SpaB-SpaC dimers were weakly observed with prolonged exposure during immunoblotting (data not shown). Interestingly, when SpaA was present, the absence of the sorting signal of SpaB did not prevent its incorporation into pilus polymers, but rather prevented the cell wall anchoring step, thereby leading to the secretion of polymers into the culture medium (lanes 11–12) as shown previously (Mandlik et al., 2008a). We conclude that the K139 of SpaB joins it to SpaC and that the CWSS of SpaB joins the SpaBC heterodimer to the cell wall.
The SpaB K139A mutant is defective in crosslinking but not in cell surface assembly
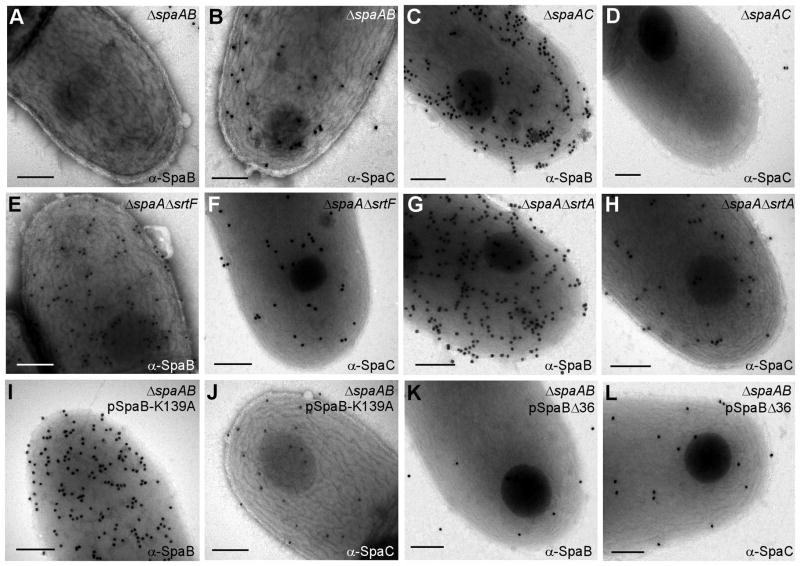
To further establish the specific role of the SpaB K139 residue in the heterodimerization of SpaB-SpaC, we next analyzed selected corynebacterial strains by immune-electron microscopy. Corynebacteria were reacted with a specific antibody against SpaB (α-SpaB) or SpaC (α-SpaC) and then reacted with 18 nm-gold particles conjugated to IgG. The samples were viewed by an electron microscope after staining with uranyl acetate. Consistent with previous observations (Mandlik et al., 2007), SpaB and SpaC signals were abundantly observed in the absence of spaA and independent of each other (Fig. 5A–5D), and SpaB signal was consistently more frequent than that of SpaC on the bacterial surface (compare Fig. 5B with 5C). In agreement with biochemical data presented above (Fig. 3), the absence of SrtF significantly reduced the cell surface SpaB signals (compare Fig. 5E with 5C). In contract, SpaB signal was not affected when SrtA was also deleted (compare Fig. 5G with 5C). Importantly, although the K139A mutation in SpaB abolished the formation of the SpaB-SpaC heterodimer (Fig. 4), it did not affect the cell surface assembly of the mutant SpaB monomers (compare Fig. 5I with 5C and 5G). In contrast, the CWSS was essential for surface assembly of SpaB as truncation of this domain abolished the SpaB reactive signals from the bacterial surface (Fig. 5K). The few gold particles seen in the image are likely due to missorted SpaB molecules that resisted rinsing during sample preparation. Note that SpaC signal was consistently detected when SpaA and SpaB were missing, when SpaB crosslinking was blocked by K139A mutation or when SpaB linkage to the cell wall was abolished by truncating its CWSS (Fig. 5B, 5J and 5L). This SpaC signal was specific as it was not observed in strain that lack SpaC, SpaC mutant that has mutations in the LPXTG motif or in a sortase-less mutant ((Mandlik et al., 2007) and data not shown).
Fig. 5.
Cell surface assembly of SpaB and SpaC analyzed by immune-electron microscopy. Corynebacteria were immobilized on nickel carbon-coated grids, stained with α-SpaB or α-SpaC, followed by goat anti-rabbit IgG conjugated to 18 nm gold particles. Samples were viewed by an electron microscopy after stained with 1% uranyl acetate. A scale bar of 200 nm.
The sorting signal of SpaB governs sortase specificity in cell wall anchoring
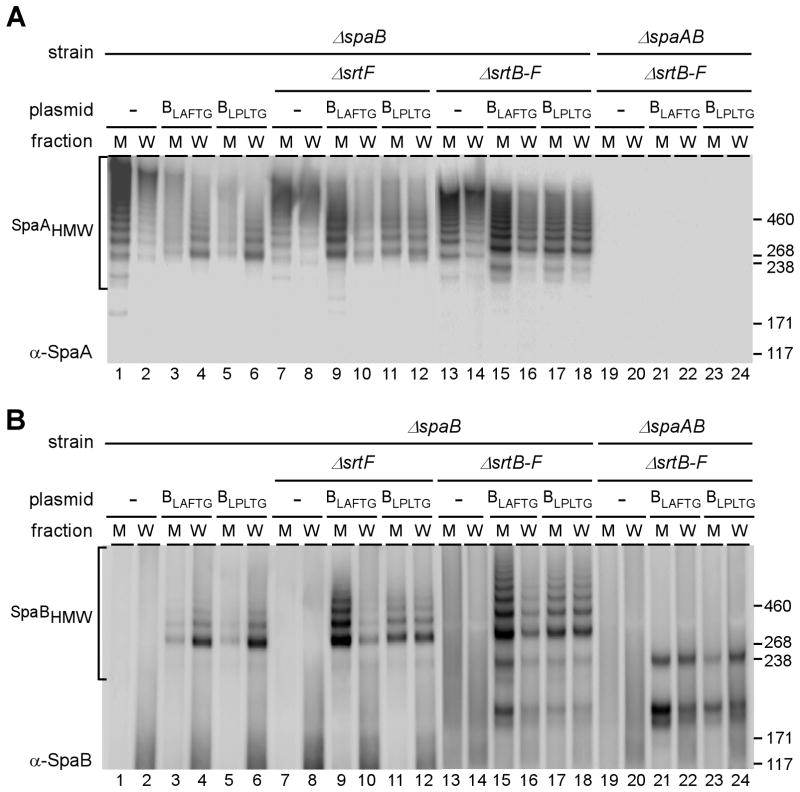
To further examine the function of SpaB CWSS in the cell wall assembly of the heterodimer, we substituted the LAFTG motif within the SpaB CWSS with the SpaA LPLTG motif by site-directed mutagenesis. A plasmid expressing this “hybrid” SpaB protein was introduced in strains that lack spaB and srtF, spaB and srtB-F, or spaB, spaA and srtB-F. Cell surface assembly of the SpaA pili was analyzed as described in Fig. 4 above. Western blots of cell wall and culture medium fractions with SpaA antibody showed that the Δ spaB mutant secretes SpaA polymers into the culture medium (Fig. 6A, lanes 1–2), thus confirming our previous report (Mandlik et al., 2008a). Introduction into this strain the plasmid that expresses wild type SpaB (BLAFTG) resulted in an efficient cell wall anchoring of the SpaA polymers (Fig. 6AB, lanes 3–4). Surprisingly, the substitution of SpaB’s LAFTG motif with the LPLTG motif of SpaA did not cause an alteration in cell wall anchoring efficiency (Fig. 6AB, lanes 5–6). An important question now arises as to which sortase is catalyzing the cell wall anchoring of this mutant form of SpaB.
Fig. 6.
Sortase specificity determined by the LPXTG motif of SpaB. Cell fractionation of various strains was analyzed as described in Fig. 3. Plasmids expressing wild type SpaB (BLAFTG) and SpaB with the LPLTG motif of SpaA (BLPLTG) are shown.
To probe specific sortase function, we next examined cell wall anchoring of SpaA pili in strain lacking both spaB and srtF. In the absence of SrtF, the SpaA polymers formed without SpaB or with wild type SpaB-complementing plasmid were largely secreted into the culture medium (Fig. 6AB- compare lanes 7–10). This supports our previous suggestion that SpaB is the preferred substrate of SrtF (Mandlik et al., 2008a). Remarkably, when SpaB is modified to contain the LPLTG motif of SpaA, the deficiency of cell wall anchoring of SpaAB polymers in the absence of SrtF was vastly compensated (Fig. 6AB, lanes 11–12). That SrtA is the catalyst of this improved cell wall anchoring step was demonstrated by the fact that a similar efficiency of cell wall anchoring of the “hybrid” SpaB was observed in a strain that contains SrtA as the sole sortase (lanes 13–18).
Finally, we probed whether SrtA can also catalyze both crosslinking and cell wall anchoring of the SpaB-SpaC heterodimer with SpaB having the LPLTG motif. As shown in lanes 19–24 of Fig. 6B, in a strain that contains SrtA as the sole sortase and no SpaA, the SpaB-SpaC heterodimer is still formed. Importantly, when SpaB contained the LPLTG motif, more SpaBC heterodimers were cell wall-anchored (lanes 23–24) as compared to that with wild type SpaB (lanes 21–22).
Therefore, similar to the mechanism of pilus assembly, SrtA catalyzes the heterodimerization of SpaB-SpaC in which SpaB participates in the transpeptidation reaction, and SrtF subsequently links SpaB-SpaC to cell wall via the sorting signal of SpaB.
Discussion
The prototype SpaA pilus is constructed with SpaA making up the pilin shaft as the shaft pilin, while SpaC is located at its tip and SpaB at the pilus base (Ton-That & Schneewind, 2003). In previous in vitro work, we have established that it is the SpaA-type pilus that mediates the specific, high affinity interaction of corynebacteria to cultured pharyngeal epithelial cells (Mandlik et al., 2007). Remarkably, it turns out that the two minor pilins SpaB and SpaC, not the shaft pilin SpaA, are the critical adhesins for this key host interaction, which underlies pharyngitis caused by C. diphtheriae. Furthermore, we showed that the SpaB and SpaC adhesins are individually anchored to the cell surface in addition to their display as components of the pilus fibers (Mandlik et al., 2007), which led us to propose that the monomeric adhesins on the bacterial surface may permit an intimate association of the bacteria with the human host cells with implications for toxin delivery, host cell signaling and colonization (Mandlik et al., 2008a). The studies reported here have uncovered yet another novel feature in the cell surface display of these adhesive pilins: SpaB and SpaC are crosslinked in the form of a SpaB-SpaC heterodimer that is displayed on the corynebacterial cell surface as a bitopic adhesin molecule.
All gram-positive pilins harbor an LPXTG motif as part of their cell wall sorting signal (Mandlik et al., 2008b), which serves two distinct functions. During pilus assembly, the threonine residue in the LPXTG motif of one pilin is joined to another pilin through a threonine-lysine bond, involving a nucleophilic lysine residue of the latter that participates in the transpeptidation reaction catalyzed by a pilus-specific sortase. The other function of the pilin’s LPXTG motif is to permit the joining of the protein to the cell wall, which also involves a transpeptidation reaction mediated by a nucleophilic amine of the lipid-II precursor of the peptidoglycan. What then dictates whether pilus polymerization continues or the polymer is incorporated in the cell wall? Numerous studies have now pinpointed the specialized function of the housekeeping sortase (or a non-polymerizing sortase) in the cell wall anchoring step of pilus assembly, and a biphasic model of pilus assembly has emerged in which pilus polymerization by a pilus-specific sortase is followed by the cell wall anchoring of pilus polymers by the housekeeping sortase (Mandlik et al., 2008a, Swaminathan et al., 2007, Budzik et al., 2007, Nobbs et al., 2008, Smith et al., 2010). Our work has revealed further that SpaB acts as the molecular switch that terminates pilus polymerization and facilitates cell wall linkage (Mandlik et al., 2008a, Swaminathan et al., 2007). Pilus polymerization is terminated when SpaB enters the pilus base via a transpeptidation reaction that utilizes a specific nucleophilic lysine residue (K139) of SpaB. The housekeeping sortase SrtF housing this terminal SpaB subunit then catalyzes the transfer of the polymer-linked SpaB to the cell wall peptidoglycan. The present work revealed that SpaB carries out a separate transpeptidation reaction using its K139 that joins SpaB to SpaC and that this SpaB-terminated heterodimer is then transferred to the cell wall by a mechanism that utilizes SrtF and parallels the last step of pilus assembly.
The SpaB-SpaC heterodimer uncovered in this study revealed itself as a novel band produced in a SpaA-deletion strain that corresponded to the combined mass of SpaB and SpaC, and also crossreacted with both anti-SpaB and anti-SpaC antibodies (see Fig. 1). We established that this SpaB-SpaC heterodimer is also expressed in the wild type corynebacteria but it appeared to be more abundant and most clearly visible when the shaft pilin SpaA was absent (Fig. 1). The nature and existence of the SpaB-SpaC heterodimer was then established by independent immunoprecipitation and His-SpaB affinity purification experiments (Fig. 2). We then established that the formation of the SpaB-SpaC heterodimer is absolutely dependent on SrtA, the pilin-specific sortase (Fig. 3), as well as the K139 residue of SpaB that normally participates in the transpeptidation reaction catalyzed by SrtA during pilus assembly (Fig. 4). Finally, the cell wall anchoring of the SpaB-SpaC heterodimer was shown to require the LAFTG motif of SpaB, and the catalysis of this reaction was found to be dependent upon SrtF, the housekeeping sortase, as might be predicted from its documented role in the final step of pilus assembly (Fig. 5 & 6).
A conundrum that arose from these studies is how SpaB acts as the pilin for executing the polymerization-termination switch in conjunction with SrtF, and conversely, how SpaB favors SrtF in lieu of SrtA for catalysis of the cell wall anchoring step. One obvious solution to this conundrum comes from our observation that SpaB has the LAFTG motif that is slightly different from the LPLTG motif of SpaA and SpaC, and thus may serve as a specificity element by differential recognition between SrtF and SrtA (Mandlik et al., 2008a, Swaminathan et al., 2007). We envisioned that the SpaB LAFTG motif might have evolved as a preferred substrate for SrtF so as to enhance the cell wall anchoring step. This model has a clear prediction: switching the LAFTG motif of SpaB to the LPLTG motif of SpaA might make the hybrid SpaB a preferred substrate for SrtA, and hence release the SrtF dependency of both the pilus anchoring step and the cell wall assembly of the SpaB-SpaC heterodimer. The prediction turned out to be true. First, the LPLTG-containing SpaB is incorporated in pilus polymers as well as the SpaB-SpaC heterodimer. Second, the cell wall incorporation of the polymers and the SpaB-SpaC heterodimer is less dependent on SrtF and is catalyzed by SrtA preferentially (Fig. 6). It is intriguing that a SpaB mutant lacking its CWSS (i.e. Δ36) was able to incorporated into the pilus polymers and heterodimers (Fig. 4 and (Mandlik et al., 2008a)). This raises another conundrum that how the engineered SpaB pilin without its CWSS would be embedded into the membrane before sortase recognition and catalysis. We posited that pilus assembly must be coupled to the general secretion machinery for efficiency, coordination, and regulation of pilus biogenesis (Mandlik et al., 2008a). Consistent with this conjecture, we and others have shown that the Sec machinery is closely associated with sortase and pilin substrates (Guttilla et al., 2009, Kline et al., 2009). Furthermore, a pilin-specific sortase mutant with its truncated transmembrane domain was shown to be able to catalyze some forms of pilus polymers (Guttilla et al., 2009).
Clearly, C. diphtheriae has evolved a simple but elegant mechanism to display on its surface three distinct forms of adhesive structures built from pilins (Fig. 7). On the one hand, long pilus fibers are built to contain two distinct adhesins (SpaC and SpaB) at the tip of these fibers, within fibers and at the fiber-cell wall interface (Ton-That & Schneewind, 2003). Such geometry is useful in facilitating a long range interaction of corynebacteria and its host, at least upon their initial encounter, and later bringing bacteria closer to the host cell surface by a zippering mechanism. On the other hand, the pilin adhesins are also displayed as individual entities that are closely apposed to the bacterial envelop and thus may provide a broad surface for a close contact with the host cell. As we speculated before, the zone of adherence formed between these elements may strengthen signaling of some host cell receptors and allow a localized delivery of the exotoxin to the host cell. In this regard, we find the third type of structure, the heterodimeric pilin uncovered in this study most exciting. The bitopic nature of this heterodimeric pilin adhesin provides a mechanism for intimate host-bacterial interaction that may last longer than may be possible otherwise with monomeric pilin-receptor interactions. We are currently designing experiments to test this model in vitro and its relevance to colonization and pathogenicity in vivo using an animal model. The presence of these three types of adhesive structures on the bacterial surface also raises an intriguing possibility for pilus mediated bacterial motility. Differential expression of these structures during bacterial infection may mimic the mechanism of motility of gram-negative bacteria mediated by pilus extension and retraction as an invasive strategy (Mattick, 2002, Merz et al., 2000, Naumann et al., 1999). It will be important to investigate whether the relative amounts of these adhesive structures change during bacterial growth and infection.
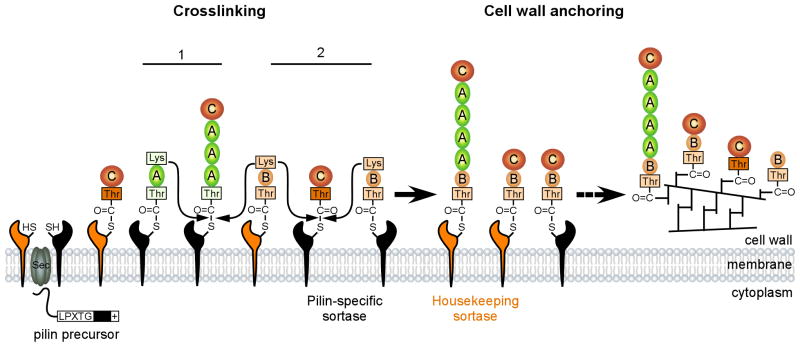
Fig. 7.
Lysine-mediated transpeptidation leading to assembly of SpaABC pili and monomeric and heterodimeric SpaB/SpaC. SpaB pilin precursors are synthesized in the cytoplasm and translocated across the membrane by the Sec machinery. The pilins are captured and formed acyl-enzyme intermediates by pilin-specific sortase SrtA (black) and the housekeeping sortase SrtF (orange). Pilus polymerization (1) begins with a nucleophilic attack by the lysine residue within the SpaA pilin motif to the thioester of an adjacent acyl-SrtA enzyme intermediate. SrtA also catalyzes dimerization (2) of SpaB and SpaC via a nucleophilic attack by the SpaB lysine residue (curved arrows) to the thioester bond of an acyl- enzyme intermediate formed between the threonine residue of the SpaC LPXTG motif and the cysteine residue of SrtA. While cell wall anchoring of SpaA polymers is efficiently catalyzed by SrtF, cell wall anchoring of monomeric and heterodimeric SpaB and SpaC is catalyzed by both sortases. Dash arrow indicates multiple steps. Pilins, sortases, and the threonine residues are color-coded. Pilus polymerization and dimerization are indicated in numbers.
Experimental Procedure
Bacterial strains, plasmids and media
Corynebacterium diphtheriae strain NCTC13129 (WT), obtained from the American Type Culture Collection, was used to generate deletion mutant strains according to a previously published protocol (Ton-That & Schneewind, 2003) (Table 1). Corynebacteria were grown in heart infusion broth (HIB) or on trypticase soy agar supplemented with 5% sheep blood (TSA II). Kanamycin was used at a final concentration of 25 μg/ml if needed. Reagents were purchased from Fisher Scientific or Sigma-Aldrich unless otherwise indicated.
Table 1.
Corynebacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and description | Reference |
|---|---|---|
| Strain | ||
| NCTC13129 | Wild type | (Ton-That & Schneewind, 2003) |
| HT10 | ΔspaB | (Ton-That et al., 2004b) |
| HT11 | ΔspaA | (Ton-That & Schneewind, 2003) |
| HT13 | ΔspaC | (Ton-That & Schneewind, 2003) |
| HT27 | ΔspaBC | (Ton-That et al., 2004b) |
| HT52 | ΔspaA, ΔsrtA | (Guttilla et al., 2009) |
| HT62 | ΔspaA, ΔsrtF | (Swaminathan et al., 2007) |
| HT67 | ΔspaA, srtBCDE | This study |
| AM5 | ΔspaAB | This study |
| AM6 | ΔspaAC | This study |
| JC3 | ΔspaB, ΔsrtF | This study |
| JC4 | ΔspaB, ΔsrtA | This study |
| Plasmid | ||
| pCGL0243 | E. coli/Corynebacterium shuttle vector | (Ankri et al., 1996) |
| pSpaA | pCGL0243 containing wild-type SpaA from C. diphtheriae | (Ton-That & Schneewind, 2003) |
| pSpaB | pCGL0243 containing wild-type SpaB from C. diphtheriae | (Mandlik et al., 2007) |
| pSpaC | pCGL0243 containing wild-type SpaC from C. diphtheriae | (Mandlik et al., 2007) |
| pSpaB-K139A | pCGL0243 containing SpaB with mutation in the lysine 139. | (Mandlik et al., 2008a) |
| PSpaBΔ36 | pCGL0243 containing SpaB with deletion of 36 aa residues from C-terminus. | (Mandlik et al., 2008a) |
| pSrtF | pCGL0243 containing wild-type SrtF from C. diphtheriae | (Swaminathan et al., 2007) |
| pH6-SpaB | pCGL0243 containing SpaB with MH6 inserted upstream of the SpaB LAFTG motif | This study |
| pSpaBLPLTG | Same as pSpaB except that the LAFTG motif was mutated to LPLTG | This study |
Plasmid Construction
pH6-SpaB
Primers P1 (5′-aaaagatcttgagttcgattggctttttttc-3′ ) a n d P 2 (5′-aaagaattcccgtgcagtcatacaatccc-3′) were used to amplify the spaA promoter sequence from C. diphtheriae NCTC13129 chromosomal DNA, while appending BglII and EcoRI sites (underlined). Primers B1 (5′-aaagaattcggaagaggacaaccaccca-3′) and B2 (5′-aaaggtaccctttttcggagccggagaa-3′) were used to amplify the 5′ spaB sequence, coding the first 178 amino acids, with EcoRI and KpnI sites appended, whereas primers B3 (5′-aaaggtaccatgcatcaccatcaccatcacacaccaccgcgactggc-3′ ) a n d B 4 ( 5′-aaaagatcttaggttactcacgtcttatcc-3′) were for the 3′ spaB sequence coding the last 39 amino acids, while inserting a sequence for MHHHHHH and KpnI and BglII sites. The generated fragments were digested with appropriate restriction enzymes and ligated into the cleaved BglII sites of the E. coli/Corynebacterium shuttle vector pCGL0243 (Ankri et al., 1996). The recombinant plasmid was confirmed by DNA sequencing and transformed into corynebacteria by electroporation (Ton-That & Schneewind, 2003).
pSpaBLPLTG
To generate pSpaBLPLTG or BLPLTG primers B5 (5′-aagacaccaccgcgactgcccttgaccggcgcaagcatcctc-3′) and B6 (5′-gaggatgcttgcgccggtcaagggcagtcgcggtggtgtc-3′) were used for PCR-based site-directed mutagenesis as described previously (Mandlik et al., 2007), with pSpaB (Mandlik et al., 2008a) as a template.
Western blot analysis
Corynebacterial cultures of various strains grown in HIB to OD600 ~ 0.7 were normalized based on their optical density. Cell fractionation of all aliquots with the same number of cells and subsequent treatment with mutanolysin were followed by a published protocol (Guttilla et al., 2009) with a slight modification. Briefly, the supernatants of the harvested cell aliquots were subjected to 7.5% TCA precipitation as medium fractions (M). The cell pellets were washed once with SMM buffer (0.5M sucrose, 10 mM MgCl2, and 10 mM maleate, pH 6.8) before digested with mutanolysin (300 U/ml) in SMM buffer in the presence of 1 mM PMSF at 37°C for 5 hr. After digestion, cell wall fractions (W) were obtained by centrifugation and TCA precipitation. For corynebacteria grown on blood agar plates, cells were scraped, washed and resuspended into SMM before digestion with mutanolysin for cell wall fractions. All samples were boiled in sodium dodecyl sulfate (SDS) containing sample buffer, separated by 3–12% Tris-Glycine gradient gels, and blotted with specific antisera.
Immunoprecipitation
Supernatants prepared from late-log growth phase of strain HT62 lacking spaA and srtF were concentrated 100-fold by Amicon Ultracel 100 kDa concentrators (Millipore). To 100 μl of the concentrated supernatants, 50 μl of α-SpaB-bound Dynabeads (Invitrogen), prepared according to the manufacturer’s protocol, was added. After incubation at room temperature with shaking, the beads were separated from the supernatants and α-SpaB-bound proteins were eluted from the beads by elution buffer (supplied by the manufacturer). Equivalent samples before and after adding α-SpaB-bound Dynabeads as well as eluted samples were analyzed by western blot using α-SpaB and α-SpaC.
Purification of His-tagged SpaB
Corynebacteria from 100 ml of late-log phase cell cultures of strain AM5 or AM6 harboring pH6-SpaB from large quantities (100 ml) were harvested by centrifugation and washed in SMM buffer before mutanolysin treatment for 5 hs. After digestion, cell wall fractions (supernatants) were obtained by centrifugation and dialyzed overnight at 4°C against EQ buffer (150mM NaCl, 50 mMTris.HCl, pH 7.5). Cell wall linked His-tagged SpaB was purified by affinity chromatography using nickel-NTA sepharose (Qiagen). Unbound proteins were washed extensively with 10 columns of wash buffer (150mM NaCl, 50 mMTris.HCl, pH 7.5, 10% glycerol), followed by 1 column of wash buffer containing 10 mM imidazole and 30 mM imidazole, successively. Eluted proteins were TCA-precipitated for western blotting analysis using α-SpaB and α-SpaC.
Immunoelectron microscopy
Corynebacteria grown on blood agar plates were scraped, washed in phosphate buffered saline (PBS) and resuspended in 100 mM NaCl before immunolabeling according to a previous protocol (Swierczynski & Ton-That, 2006). Briefly, a drop of bacterial suspension was spotted onto a carbon-coated nickel grid, which was washed three times by PBS containing 2% BSA and blocked for 1 h in PBS containing 0.1% gelatin. Grids were incubated with primary antibodies diluted 1:100 in PBS with 2% BSA for 1 hour, followed by washing and blocking. Samples were then treated with 18-nm gold-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) diluted at 1:20 in PBS with 2% BSA for 1 h. The grids were washed five times with water before staining with 1% uranyl acetate. Samples were viewed in a JEOL JEM1400 electron microscope.
Acknowledgments
We thank members of our laboratory for critical review of the manuscript and discussion. This work was supported by the U.S. Public Health Service grant AI061381 from the National Institute of Allergy and InfectiousDiseases to H.T.-T.
References
- Ankri S, Reyes O, Leblon G. Electrotransformation of highly DNA-restrictive corynebacteria with synthetic DNA. Plasmid. 1996;35:62–66. doi: 10.1006/plas.1996.0007. [DOI] [PubMed] [Google Scholar]
- Budzik JM, Marraffini LA, Schneewind O. Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol Microbiol. 2007;66:495–510. doi: 10.1111/j.1365-2958.2007.05939.x. [DOI] [PubMed] [Google Scholar]
- Budzik JM, Oh SY, Schneewind O. Sortase D Forms the Covalent Bond That Links BcpB to the Tip of Bacillus cereus Pili. J Biol Chem. 2009;284:12989–12997. doi: 10.1074/jbc.M900927200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comfort D, Clubb RT. A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect Immun. 2004;72:2710–2722. doi: 10.1128/IAI.72.5.2710-2722.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dramsi S, Trieu-Cuot P, Bierne H. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res Microbiol. 2005;156:289–297. doi: 10.1016/j.resmic.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Gaspar AH, Ton-That H. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J Bacteriol. 2006;188:1526–1533. doi: 10.1128/JB.188.4.1526-1533.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttilla IK, Gaspar AH, Swierczynski A, Swaminathan A, Dwivedi P, Das A, Ton-That H. Acyl enzyme intermediates in sortase-catalyzed pilus morphogenesis in gram-positive bacteria. J Bacteriol. 2009;191:5603–5612. doi: 10.1128/JB.00627-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline KA, Kau AL, Chen SL, Lim A, Pinkner JS, Rosch J, Nallapareddy SR, Murray BE, Henriques-Normark B, Beatty W, Caparon MG, Hultgren SJ. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J Bacteriol. 2009;191:3237–3247. doi: 10.1128/JB.01837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke C, Young PG, Kang HJ, Bunker RD, Middleditch MJ, Caradoc-Davies TT, Proft T, Baker EN. Crystal structure of the minor pilin FctB reveals determinants of Group A Streptococcal pilus anchoring. J Biol Chem. 2010 doi: 10.1074/jbc.M109.089680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandlik A, Das A, Ton-That H. The molecular switch that activates the cell wall anchoring step of pilus assembly in gram-positive bacteria. Proc Natl Acad Sci U S A. 2008a;105:14147–14152. doi: 10.1073/pnas.0806350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandlik A, Swierczynski A, Das A, Ton-That H. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol Microbiol. 2007;64:111–124. doi: 10.1111/j.1365-2958.2007.05630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandlik A, Swierczynski A, Das A, Ton-That H. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008b;16:33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- Merz AJ, So M, Sheetz MP. Pilus retraction powers bacterial twitching motility. Nature. 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- Naumann M, Rudel T, Meyer TF. Host cell interactions and signalling with Neisseria gonorrhoeae. Curr Opin Microbiol. 1999;2:62–70. doi: 10.1016/s1369-5274(99)80011-3. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Rosini R, Rinaudo CD, Maione D, Grandi G, Telford JL. Sortase A utilizes an ancillary protein anchor for efficient cell wall anchoring of pili in Streptococcus agalactiae. Infect Immun. 2008;76:3550–3560. doi: 10.1128/IAI.01613-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry AM, Ton-That H, Mazmanian SK, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J Biol Chem. 2002;277:16241–16248. doi: 10.1074/jbc.M109194200. [DOI] [PubMed] [Google Scholar]
- Proft T, Baker EN. Pili in Gram-negative and Gram-positive bacteria - structure, assembly and their role in disease. Cell Mol Life Sci. 2009;66:613–635. doi: 10.1007/s00018-008-8477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin A, Severin A, Ritacco F, Tabei K, Singh G, Bradford PA, Siegel MM, Projan SJ, Shlaes DM. Further evidence that a cell wall precursor [C(55)-MurNAc-(peptide)-GlcNAc] serves as an acceptor in a sorting reaction. J Bacteriol. 2002;184:2141–2147. doi: 10.1128/JB.184.8.2141-2147.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JR, Zahner D. Pili with strong attachments: Gram-positive bacteria do it differently. Mol Microbiol. 2006;62:320–330. doi: 10.1111/j.1365-2958.2006.05279.x. [DOI] [PubMed] [Google Scholar]
- Smith WD, Pointon JA, Abbot E, Kang HJ, Baker EN, Hirst BH, Wilson JA, Banfield MJ, Kehoe MA. Roles of minor pilin subunits Spy0125 and Spy0130 in the serotype M1 Streptococcus pyogenes strain SF370. J Bacteriol. 2010;192:4651–4659. doi: 10.1128/JB.00071-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan A, Mandlik A, Swierczynski A, Gaspar A, Das A, Ton-That H. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol Microbiol. 2007;66:961–974. doi: 10.1111/j.1365-2958.2007.05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swierczynski A, Ton-That H. Type III pilus of corynebacteria: Pilus length is determined by the level of its major pilin subunit. J Bacteriol. 2006;188:6318–6325. doi: 10.1128/JB.00606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in Gram-positive pathogens. Nat Rev Microbiol. 2006;4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Marraffini LA, Schneewind O. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim Biophys Acta. 2004a;1694:269–278. doi: 10.1016/j.bbamcr.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Marraffini LA, Schneewind O. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol Microbiol. 2004b;53:251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Schneewind O. Assembly of pili in Gram-positive bacteria. Trends Microbiol. 2004;12:228–234. doi: 10.1016/j.tim.2004.03.004. [DOI] [PubMed] [Google Scholar]