Abstract
Cardiac troponin – a heterotrimeric protein complex that regulates heart contraction – represents an attractive target for the development of drugs to treat heart disease. Cardiovascular diseases are one of the chief causes of morbidity and mortality worldwide. In France, however, the death rate from heart disease is remarkably low relative to fat consumption. This so called “French paradox” has been attributed to the high consumption of wine in France; and the antioxidant trans-resveratrol is thought to be the primary basis for wine’s cardioprotective nature. It has been demonstrated that trans-resveratrol increases the myofilament Ca2+-sensitivity of guinea-pig myocytes (Liew, R., Stagg, M.A., MacLeod, K.T., and Collins, P., (2005) Eur. J. Pharmacol. 519, 1–8.), however, the specific mode of its action is unknown. In this study, the structure of trans-resveratrol free and bound to the calcium-binding protein, troponin C, was determined by NMR spectroscopy. The results indicate that trans-resveratrol undergoes a minor conformational change upon binding to the hydrophobic pocket of the C-domain of troponin C. The location occupied by trans-resveratrol coincides with the binding site of troponin I – troponin C’s natural binding partner. This has been seen for other troponin C-targeting inotropes and implicates the modulation of the troponin C-troponin I interaction as a possible mechanism of action for trans-resveratrol.
Keywords: Trans-resveratrol, troponin C, NMR spectroscopy, cardiac muscle, contraction, Ca2+-sensitizer
The physiological function of the heart is to pump blood throughout the body in order to fulfill the oxygen and nutrient demands of the organism. The thin filament in heart muscle is made up of actin, tropomyosin, and troponin. Troponin is a heterotrimeric protein complex formed by the Ca2+-binding subunit, troponin C (TnC); the inhibitory subunit, troponin I (TnI); and the tropomyosin-binding subunit, troponin T (TnT). Cardiac TnC (cTnC) has four EF-hand metal binding sites (I–IV) – two in each of its terminal domains. The C-terminal (cCTnC) and N-terminal (cNTnC) domains are connected by a flexible linker, as shown by the NMR (1) and X-ray structures (2). In cNTnC, site I is defunct and site II is a low-affinity Ca2+-binding site; on the other hand, both site III and site IV in cCTnC are functional and can bind either Mg2+ or Ca2+. During the contraction-relaxation cycle, cytosolic Ca2+ concentration dramatically oscillates: at high Ca2+ levels, cNTnC becomes Ca2+-saturated, which primes cNTnC for binding with the “switch” region of cTnI (cTnI147–163) (3, 4); at low Ca2+ concentration, Ca2+ dissociates from cNTnC leading to the release of cTnI147–163. Alternatively, cCTnC remains saturated with either Mg2+ or Ca2+ throughout the contraction cycle and is associated with the “anchoring” region of cTnI (cTnI34–71); an interaction which may play both a structural and regulatory role (5). Association of cTnI147–163 with cNTnC drags the “inhibitory” region of cTnI (cTnI128–147) off of actin, tropomyosin changes its orientation on the thin-filament, the myosin binding site is exposed on actin, and myosin binding to actin leads to contraction (for reviews see (6, 7)).
The strength of heart muscle contraction is regulated by the amount of Ca2+ released from the sarcoplasmic reticulum into the cytosol and by the response of the myofilaments to Ca2+. Many popular cardiotonic agents (such as digitalis or dobutamine) improve contraction in the failing heart by elevating intracellular Ca2+ concentration; however, intracellular Ca2+ modulation carries risks associated with Ca2+ overload such as cardiac arrhythmias, cell injury, or cell death. These limitations have shifted interest to a novel class of cardiotonic drugs: Ca2+-sensitizers. Ca2+-sensitizers induce a positive inotropic effect by modulating the myofilament’s response to cytosolic Ca2+, and consequently may circumvent the risks associated with altering Ca2+ homeostasis (8). The essential role cTnC plays in regulation of contraction makes it a logical target for the development of Ca2+-sensitizers. Several ligands that have been found to have a Ca2+-sensitizing ability through direct interaction with cTnC include trifluoperazine (TFP) (9), bepridil (9, 10), levosimendan (11–13), and EMD 57033 (14–16). While the majority of these compounds target cNTnC to elicit their Ca2+-sensitizing effects, EMD 57033 functions by targeting cCTnC (15). In addition, the natural tea polyphenol, epigallocatechin gallate (EGCg), modulates heart muscle contractility through an interaction with cCTnC (17–19). These results suggest that both domains of cTnC represent targets for the development of Ca2+-sensitizers to treat heart failure.
Cardiovascular diseases are the main cause of death worldwide. Interestingly, the mortality rate from heart disease is significantly lower in France than in other countries with comparable diets rich in fat and other risk factors. It has been suggested that this so called “French paradox” may be attributable to high wine consumption in France (20). Trans-resveratrol (3,4’,5-trihydroxystilbene) is produced in grapevines after fungal infection and exposure to ultraviolet light (21), and Siemann and Creasy proposed that it might be the biologically active ingredient of red wine (22). Trans-resveratrol (resveratrol) has a variety of reported physiological effects including: antiplatelet aggregation, anti-inflammatory, and antioxidant activity linked to longevity (23, 24); protective effects in skin photosensitivity (25); neurodegenerative diseases (26); cancer chemoprevention (27, 28); and cardioprotection (29). Amongst its cardioprotective effects, it has been shown that resveratrol improves recovery of ventricular function including developed pressure in the face of ischemia reperfusion injury (30). It was demonstrated that resveratrol directly affects the contractile function of guinea-pig myocytes, and it increased cell shortening in half the cells tested and decreased shortening in the other half. In the cells where it induced contraction, its relation with the Ca2+ transients was quantitatively determined indicating an increase in myofilament Ca2+-sensitivity (31). These findings indicate a direct relation between resveratrol and the Ca2+ regulated elements in myocytes; however, structural details of this interaction remain unclear.
The present study investigates the interaction between cTnC and resveratrol using the structural technique, NMR spectroscopy. There have been a number of research groups that describe the applications of relatively sparse NMR data for the determination of protein-ligand complexes (32–35). Recently, Hoffman and Sykes described a procedure to determine the structure of W7 bound with cNTnC using a previously determined structure of cNTnC as a template (36). A similar protocol was followed here, and it was discovered that resveratrol binds to cCTnC in a similar manner as EGCg (19) and EMD 57033 (14). Several key hydrophobic interactions between cCTnC and resveratrol stabilize the binary structure. It also appears that resveratrol undergoes only a slight conformational change upon binding cCTnC. The solution structure provides clues into the cardioprotective mechanism of resveratrol, and molecular details of the interface between resveratrol and cCTnC may aid in the design of novel cCTnC-targeting drugs.
Experimental Procedures
Sample preparation
Labeled 15N- and 15N,13C-cCTnC and 15N-cTnC were obtained from E. coli strains containing the expression vector as previously described (37, 38). Trans-resveratrol was purchased from Sigma-Aldrich Inc (99% purity as determined by gas chromatography). All NMR samples were 500 µL in volume and consisted of 100 mM KCl; 10 mM imidazole, or 8 mM imidazole-d4 and 2 mM imidazole; and 0.5 mM DSS-d6 as an NMR reference standard. Protein concentrations were ~0.2 mM for full length cTnC and ~0.5 mM for cCTnC with 20 mM or 10 mM CaCl2 respectively. Sample pH was maintained at ~6.9 for all NMR experiments. Although resveratrol naturally exists in cis and trans isomeric forms, trans-resveratrol was used in this study due to its higher concentrations in red wine (0–15 µg/ml) in contrast with cis-resveratrol (0–5 µg/ml) (24). Stock solutions of 20 mM trans-resveratrol (resveratrol) in a 100 mM tris(2-carboxyethyl)phosphine (TCEP) - DMSO-d6 solution were prepared. TCEP was used to allay oxidation of resveratrol as done for EGCg and Ascorbic Acid (19, 39). The concentration ratio of resveratrol:cCTnC was ~4:1 for NMR experiments of the complex. All the stock solutions were prepared fresh prior to each experiment and were wrapped with aluminum foil to prevent photo-degradation.
NMR Spectroscopy and data processing
The NMR data used for this study was collected on Varian Inova 500-MHz and Unity 600-MHz spectrometers at 30 °C, or a Varian Inova 800-MHz spectrometer at 25 °C. All spectrometers have triple resonance probes with Z-pulsed field gradients. One-dimensional 1H; and two-dimensional 1H,1H-NOESY and 1H,1H-ROESY experiments were acquired of resveratrol in D2O with mixing times of 100 ms for the NOESY and 50, 150, 200, and 300 ms mixing times for the ROESY experiments. Two-dimensional 1H,13C-HSQC and 1H,15N-HSQC experiments were acquired to monitor the titration of resveratrol into 13C,15N- or 15N-labeled cCTnC. Intramolecular NOEs of resveratrol when in complex with cCTnC were measured with the two-dimensional 13C,15N-filtered NOESY experiment with a mixing time of 100 ms (40, 41); intermolecular distance restraints between resveratrol and cCTnC were derived from two-dimensional 13C-edited/filtered NOESY-HSQC (mixing times: 150, 200, 250 ms) (42) and three-dimensional 13C-edited/filtered HMQC-NOESY (mixing times: 200, 250 ms) (43, 44) experiments. In order to check whether the conformation of cCTnC was perturbed by the presence of resveratrol, a two-dimensional 1H,1H-NOESY was acquired of cCTnC•resveratrol in D2O with a mixing time of 150 ms. VNMRJ (Varian Inc.) was used for the analysis of one-dimensional NMR spectra, all two-dimensional and three-dimensional NMR data were processed with NMRPipe (45) and analyzed with NMRView (46). Chemical shifts of cCTnC were assigned using those deposited for cTnC (1) and the methionine methyls were assigned from assignments previously determined by mutagenesis (47). The interaction between resveratrol and cCTnC is in fast exchange, so the resonances from cCTnC could be followed throughout the titration of resveratrol and the assignments translated to the chemical shifts of the cCTnC•resveratrol complex.
Resveratrol assignment and assessment of stability
The proton chemical shifts of resveratrol were assigned in D2O by the use of one-dimensional 1H, two-dimensional 1H,1H-NOESY, and two-dimensional 1H,1H-ROESY NMR experiments. In order to test the stability of resveratrol in aqueous solution, 500 µL samples of ~0.3 mM resveratrol were prepared in D2O. 1H NMR spectra were acquired at one-hour increments to test for oxidative degradation. Another sample of resveratrol was prepared with 10 mM TCEP and reassessed the sample stability. In both samples, the pD was ~7.0.
NMR structure and Ab initio calculations of resveratrol free in solution
The PRODRG web server (48) was used to create initial coordinates of resveratrol and XPLO-2D (49) was used to generate topology and parameter files for resveratrol. The structure of resveratrol was determined by ROE intensities of resveratrol at mixing time of 200 ms. XPLOR-NIH (50) was used to calculate the solution structure of resveratrol. Four NOEs contributed to the structure calculation, which were binned from strong (2.00-1.80 Å), medium (2.80-1.80 Å), and weak (3.60-1.80 Å). The simulated annealing comprised square-well potentials for interproton distances and a patch to keep the aromatic rings and olefin bond planar; the aromatic rings of resveratrol were allowed to freely rotate around the C1’-Cα’ and C1-Cα bonds. 100 structures were calculated, and the lowest 10 in energy were kept. The lowest energy solution structure of free resveratrol was used as an input structure in Gaussian03 (51) to calculate the total density and electrostatic potential (ESP). The energy calculation was performed in aqueous solvent with Becke’s three-parameter Lee, Yang, Parr (B3LYP) hybrid functional with a split-valence basis set and polarization d- and p-orbitals added (B3LYP/6-311++G(d, p)). Final contour surfaces were represented using GaussView 3.0.
Resveratrol titrations into cTnC and cCTnC
The 20 mM resveratrol stock solution was titrated into an NMR tube containing full length 15N-labeled cTnC, 15N-, and 15N,13C-labeled cCTnC. Resveratrol additions were made to final concentrations of 62, 185, 367,665, 956, 1517 and 2050 µM for the titration with cTnC and to final concentrations of 43, 171, 379, 581, 975, 1346 and 1701 µM for the titration with cCTnC. One-dimensional 1H and two-dimensional 1H,15N-HSQC spectra were acquired at each titration point. Additionally, 1H,13C-HSQC spectra were acquired throughout the titration of resveratrol into 13C,15N-labeled cCTnC. At each titration point, the HSQC spectra were assigned, and residue specific chemical shift perturbations (CSPs) that were well resolved and underwent a large change were kept for the dissociation constant calculation. Chemical shift changes (Δδ) were calculated using the following equation:
The dissociation constants (KD) for resveratrol were calculated by using a global fitting approach with the program xcrvfit (www.bionmr.ualberta.ca/bds/software/xcrvfit), as described previously (52). Briefly, the set of kept titration curves were fit using a fixed KD range and a floating final shift value (since residues were perturbed to a varying extent). The sum of squared error (SSE) was minimized by optimizing the KD, and thus a global KD was calculated that best fit all the CSPs. The binding of resveratrol to cTnC and cCTnC was fit with a 1:1 stoichiometry:
Concentrations of cTnC and cCTnC were calculated by integrating the one-dimensional-slice of the 1H,15N-HSQC spectrum, and comparing the spectral intensity with that of a sample with a protein concentration determined by amino acid analysis. The concentrations of the resveratrol stock solutions were determined by comparing the proton spectral intensity of resveratrol peaks with the DSS proton intensity. Concentrations of cTnC, cCTnC, and resveratrol were then corrected for dilution that occurred during the titrations. Since the addition of resveratrol slowly decreased the pH of the sample, the pH was adjusted to ~6.9 with 1M NaOH when necessary.
J-surface mapping
The program Jsurf was used to localize the binding site of resveratrol on cCTnC. Jsurf approximates the origin of the CSP as a single point-dipole in the center of an aromatic ring from a ligand (53). With the coordinates of cTnC as input, Jsurf depicts the coordinates of a ligand ring as a dot based on the magnitude and sign of the CSPs. The region that shows the highest dot density is referred to as the j-surface, and is where the aromatic constituents of a ligand are most likely to reside. The chemical shifts in the 1H,13C-HSQC spectrum of the final point in the titration of cCTnC (CSPL) with resveratrol were subtracted from the initial chemical shifts of cCTnC (CSP).
Only peaks that were well resolved and underwent a concentration-dependant CSP of ≥ 0.012 ppm in the proton dimension were used in the analysis; results were displayed in PyMOL.
Structure calculation of cCTnC•resveratrol
The structure of cCTnC•resveratrol was calculated using restraints for cCTnC from the complex of cCTnC•EGCg because the CSPs induced by both ligands are quite similar. A similar data-driven protocol has been used for the cNTnC•W7 complex (36). Distance restraints for cCTnC were calibrated with CYANA (54) using an upper limit of 6 Å. Dihedral angle restraints from TALOS (55) were used as well as 12 distance restraints from X-ray crystallographic data of chelating oxygen atoms to the two Ca2+ ions. CYANA was used to calculate 100 structures of cCTnC, of which the 30 conformers with the lowest target function were used to further refine the structure with XPLOR-NIH. The restraints were converted from CYANA format into XPLOR-NIH format. The simulated annealing protocol of XPLOR-NIH, with 10 000 high temperature steps and 6000 cooling steps, was used in the structure calculation. The NOEs were averaged using the R-6 with a soft well potential. Spin-diffusion was a concern because the mixing times of 200 ms and 250 ms for the measurement of intermolecular NOEs were high. Therefore, only NOEs that had an NOE intensity≥μ-1/2σ were kept for the structure calculation, and all were calibrated with the same distance. A total of 23 intermolecular NOEs (6.0-1.8 Å) and four intramolecular NOEs (2.6-1.8 Å) were used in the structure calculation. 100 structures were calculated, from which the lowest 50 in energy were kept for refinement in explicit solvent with a water box edge length of 18.8 Å. It has been shown that refinement in explicit solvent including electrostatic potentials can improve the quality of structures (56). Atomic charges calculated by Gaussian03 for resveratrol (see above) were included at this point in the structure calculation. The final ensemble is represented by the 20 lowest energy structures after water refinement and was validated by Procheck (57) available with the online Protein Structure Validation Software (PSVS) suite (http://psvs-1_4-dev.nesg.org/); and the structural statistics are available in supplementary table 1.
Results
Stability of Resveratrol
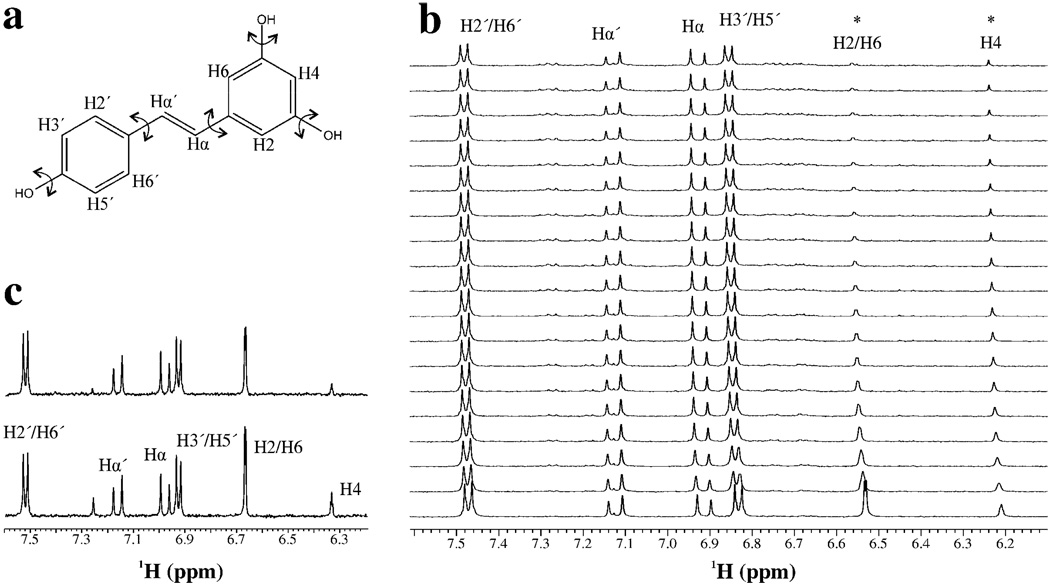
NMR chemical shift assignments of trans-resveratrol (resveratrol), or trans-resveratrol derivatives have been previously reported in acetone (58), chloroform (59), DMSO-d6 (60), and in a DMSO-d6/D2O mixture (61). Resveratrol (figure 1) in D2O was assigned by one-dimensional 1H, two-dimensional 1H,1H-NOESY, and two-dimensional 1H,1H-ROESY NMR experiments. The aromatic protons were assigned using three and four bond couplings. H2’/H6’ were distinguished from H3’/H5’ by the acquisition of a two-dimensional NOESY and ROESY experiments; only the H2’/H6’ made ROE contacts with the ethylene protons (Hα and Hα’). The unambiguous assignments of Hα and Hα’ were facilitated with previously published assignments for resveratrol (58–61). The ethylene protons had a vicinal coupling constant of 16.4 Hz; consistent with a trans-ethylene bond (62).
Fig. 1. Stability of resveratrol.
a. Chemical structure of resveratrol. Torsion angles that were allowed to rotate during structure calculations are indicated. b. Degradation of resveratrol as a function of time. One-dimensional 1H-NMR spectra acquired every 1 hour (bottom, first spectrum; top, last spectrum). c. The spectrum of resveratrol in the presence of 10 mM TCEP. Bottom spectrum is at time=0 and the top spectrum is of resveratrol after 24 hours. The unlabeled peak downfield from Hα’ is from 1H-imidazole which exchanged with D2O over time.
The stability of resveratrol has been addressed previously (63). The authors found that under acidic conditions, resveratrol was stable for at least 42 hours; while at pH 10, resveratrol had a half-life of 1.6 Hrs. Since three-dimensional NMR experiments require a stable sample for 2–3 days, and the samples are at neutral pH; the necessity of a reducing agent was investigated. The degradation of resveratrol was measured by acquiring one-dimensional 1H-NMR spectra at one hour time points. Significant degradation of resveratrol was observed, primarily in the di-meta-hydroxyl (di-m-OH) ring (figure 1b), and a half-life of 5–6 hours was estimated by monitoring the chemical shift change of the H2/H6 proton pair (supplementary figure 1). The sample also changed color from clear to brown over the course of the 24 hours. Next, 10 mM of TCEP was added to a sample of resveratrol and no significant degradation occurred after one day (figure 1c). There was some decrease in peak intensities; however, there was no change in chemical shift or sample color; even after 72 hours.
Structure of Resveratrol in D2O
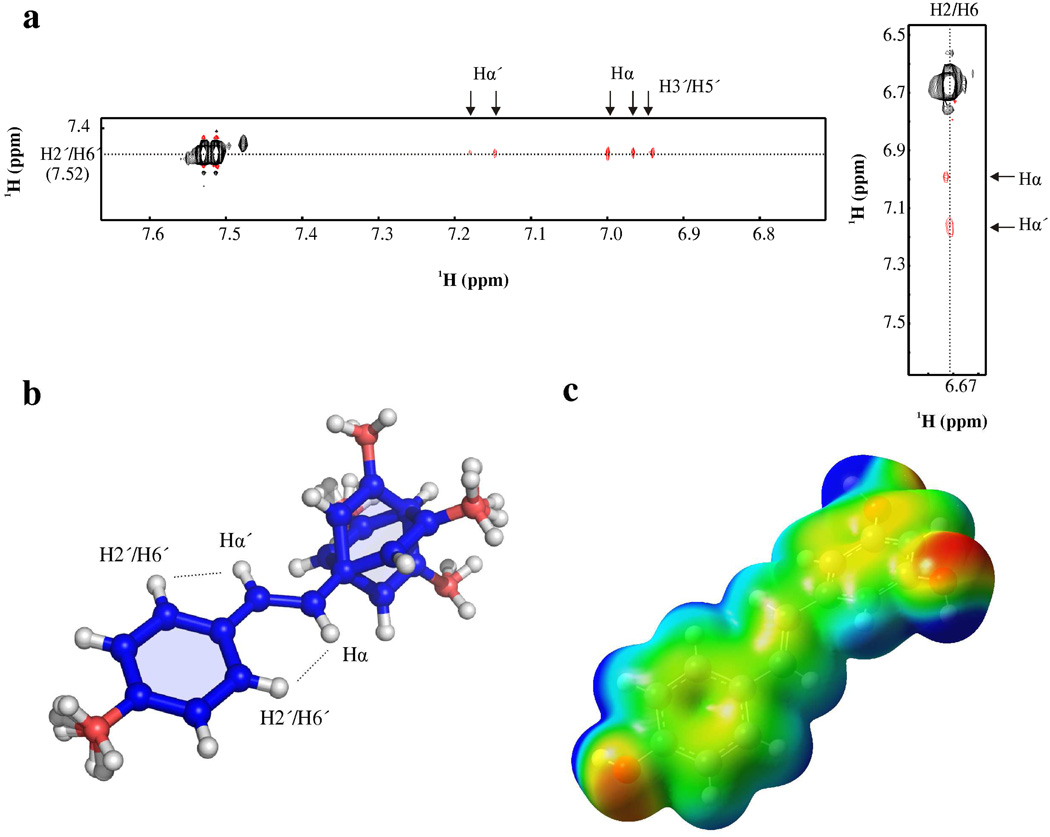
In order to determine the structure of resveratrol in solution, two-dimensional ROESY spectra were acquired with different mixing times (50, 150, 200, and 300 ms); the ROE build-up curve is shown in supplementary figure 2. Two slices from the ROESY spectrum (mixing time = 200 ms) are shown in figure 2a. The H2’/H6’ protons make much stronger ROEs with the ethylene protons than the H2/H6 protons do (table 1). The strong ROE between H2’/H6’ and Hα is only consistent with a coplanar orientation of the para-hydroxyl (p-OH) ring and olefin. On the other hand, weak, approximately uniform ROEs between the ethylene protons and H2/H6 is indicative of roughly equal distance between H2/H6 and Hα and Hα’; consistent with a tilted and/or flexible di-m-OH ring. The ensemble of the 10 lowest energy structures is shown in figure 2b. The torsion angles formed by Cα’-Cα-C1-C6 and Cα-Cα’-C1’-C6’ can be used to describe the orientations of the two phenolic rings with respect to the olefin. In table 2 the torsion angles for a number of structures of resveratrol are indicated. The p-OH ring is coplanar (0.5 ± 0.3°) with the olefin, whereas the di-m-OH ring is tilted by 43.9 ± 0.4°. The structure of free resveratrol has also been solved by NMR spectroscopy in DMSO (60) and by X-ray crystallography (64); both structures revealed an overall planar structure. The X-ray structure of a trimethoxy-derivative of resveratrol was less planar; particularly in respect to the orientation of the di-m-O-CH3 ring (65). In the crystal structure of resveratrol, extensive hydrogen bonds contribute to the planarity of resveratrol, while the inability of 3,4’,5-trimethoxystilbene to form intermolecular hydrogen bonds, resulted in a slight twist of the rings. It is possible that the presence of TCEP in the sample decreased the amount of intermolecular hydrogen-bonding of resveratrol by perturbing stacking interactions, and may explain the somewhat tilted conformation of the di-m-OH ring. The lowest energy structure in the ensemble of resveratrol was used to calculate its electronic properties with the quantum chemistry program Gaussian03 (51) (figure 2c).
Fig. 2. Structure of resveratrol.
a. Two-dimensional-ROESY spectrum of resveratrol in D2O. b. Ensemble of resveratrol aligned to the olefin atoms: Hα’, Cα’, Hα, and Cα. ROEs measured between H2’/H6’ and Hα and Hα’ are drawn on the structure to illustrate that the intensity of the ROE between H2’/H6’ and Hα requires a planar orientation of phenol ring. c. Gaussian calculation from lowest energy structure of resveratrol. The electrostatic potential of resveratrol was mapped with an isovalue of 0.004 e/Å3 (−7e−2 eV, red; 7e2+ eV, blue).
Table 1. Comparison of Relative ROE or NOE intensities.
| Contact | Free resveratrola | Bound Resveratrol |
|---|---|---|
| H2’/H6’ - Hα’ | 0.51 | 0.67 |
| H2’/H6’ - Hα | 1.00 | 0.84 |
| H2/H6 – Hα | 0.17 | 0.71 |
| H2/H6 – Hα’ | 0.16 | 1.00 |
At mixing time of 200 ms.
Table 2. Comparison of resveratrol ring orientation with other structures.
Ring twist illustrates the orientation of the ring with comparison to the central olefin of resveratrol. The p-OH ring twist is the Cα-Cα’-C1’-C6’ torsion angle; the di-m-OH ring twist is the Cα’-Cα-C1-C6 torsion angle. For the NMR structures, both positive and negative torsions were measured so they are reported in absolute degrees. Multiple values are given when more than one resveratrol is present in the X-ray structures.
| Structure |
p-OH ring twist (°) |
di-m-OH ring twist (°) |
|---|---|---|
| X-ray structure of resveratrol (64) | 8.02 | −2.98 |
| X-ray structure of 3,5,4’-Trimethoxystilbene (65) | 3.53/−9.16 | 21.13/−16.32 |
| NMR structure of resveratrol (D2O) a | 0.5 ± 0.3 | 43.9 ± 0.4 |
| NMR structure of cCTnC a | 18.6 ± 10.6 | 35.2 ± 8.7 |
| X-ray structure of Chalcone Synthase (pdb: 1CGZ)(70) | −9.21 | −32.00 |
| X-ray structure of Transthyretin (pdb: 1DVS)(71) | 0.7/1.06 | 0.55/1.13 |
| X-ray structure of Quinone Reductase 2 (pdb: 1SG0)(73) | 0.23/0.23 | 3.11/2.15 |
| X-ray structure of mutant Chalcone Synthase (pdb: 1U0W)(72) | −0.16/−2.98 | 0.18/−1.49 |
| X-ray structure of Stilbene Synthase (pdb: 1Z1F)(74) | 62.92 | −66.97 |
| X-ray structure of Cytosolic Sulfotransferase (pdb: 3CKL) | 19.84/20.51 | 5.80/19.16 |
| X-ray structure of Leukotriene A4 Hydrolase (pdb: 3FTS)(76) | −27.28 | −17.65 |
| X-ray structure of F1-ATPase (pdb: 1JIZ) (75)b | 27.68/19.90/−35.06/42.25 | −9.25/7.60/45.69/−21.39 |
This study.
The authors could not distinguish between two binding modes, and therefore concluded that resveratrol may bind to F1-ATPase in multiple poses.
Resveratrol binding to cTnC
To characterize resveratrol’s interaction with troponin it was titrated into a sample containing 15N-labeled cTnC. 1H,15N-HSQC NMR spectra were acquired throughout the titration and most of the large chemical shift perturbations (CSPs) were of backbone amides in the C-domain of cTnC (supplementary figure 3). In addition, the residues that typically experience large perturbations upon cTnI or ligand binding to the N-domain (4, 52) such as Gly34, Gly42, Glu66, and Asp73 remained relatively unperturbed. The linear nature of the CSPs is indicative of a 1:1 stoichiometry for resveratrol binding to cTnC. Multiple binding would lead to non-linear CSPs, as was observed for TFP and bepridil binding to cTnC (9). Using the program xcrvfit, the CSPs of the backbone amides of Thr124, Gly125, Ile128, Thr129, Gly140, Gly159, and Glu161 were plotted as a function of resveratrol-to-cTnC concentration; a global dissociation constant (KD) of 243 µM (SSE = 0.02) was determined (supplementary figure 3b).
Resveratrol binding to cCTnC
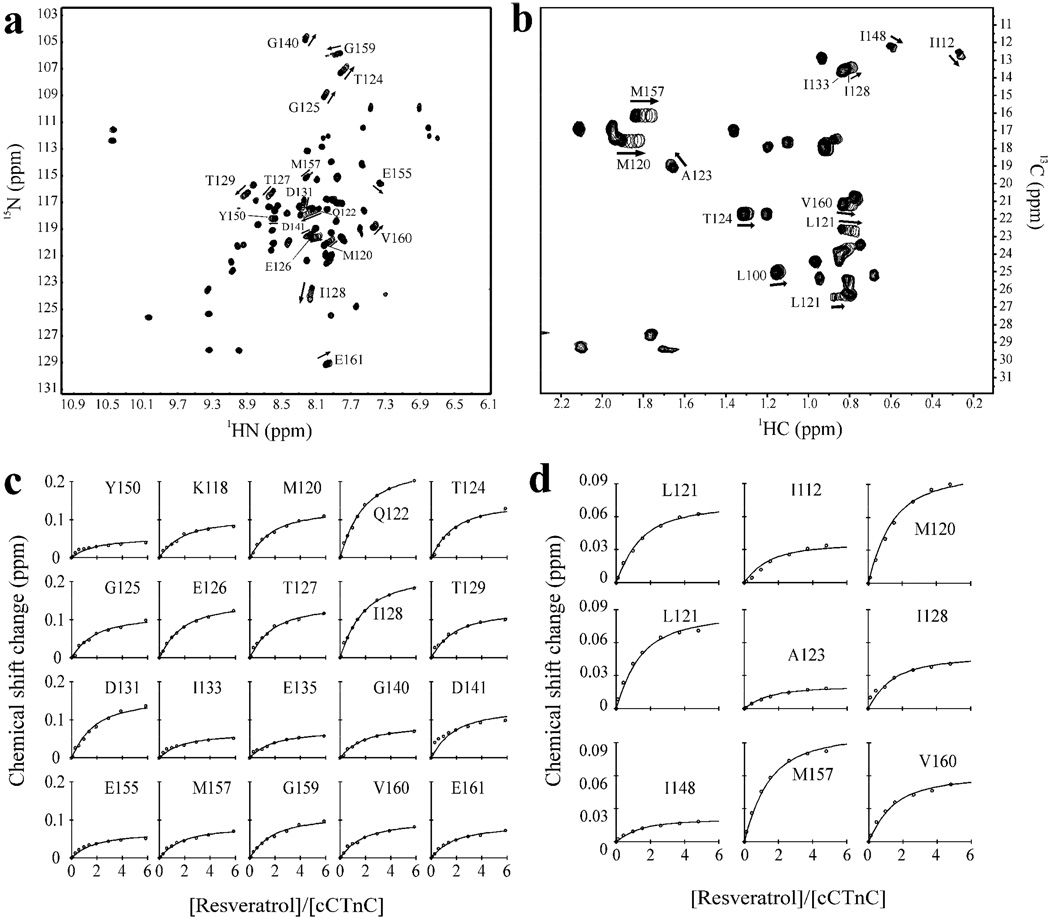
To test whether the interaction between resveratrol and cTnC was the same as in the isolated C-domain, resveratrol was titrated into cCTnC. Two-dimensional 1H,13C and 1H,15N HSQC experiments were used to monitor the titration of resveratrol into a sample containing cCTnC (figure 3a and 3b). The 1H,13C-HSQC (without resveratrol) was assigned using chemical shift assignments deposited for cTnC (1). Since resveratrol interacts with cCTnC in fast exchange, most assigned resonances could be easily followed throughout the titration. Global dissociation constants of 240 µM (SSE = 0.08); from the backbone amides, and 301 µM (SSE = 0.02); from the methyl groups of cCTnC were calculated (figure 3). The residue specific CSPs are almost identical in both pattern and amplitude between the titrations of resveratrol to cCTnC and cTnC (supplementary figure 3c). The comparable dissociation constants and CSP patterns suggest that resveratrol interacts with cCTnC the same as it does with full-length cTnC.
Fig. 3. Binding of resveratrol to cCTnC.
a. 1H,15N-HSQC and b. 1H,13C-HSQC (methyl-region) spectra of cCTnC acquired throughout the titration with resveratrol. The first point in the titration is represented with all 20 contours; whereas, each titration point with resveratrol is represented by a single contour. Direction of CSPs are indicated by arrows for several peaks. c. fitting of 1H,15N-HSQC and d. 1H,13C-HSQC NMR data with xcrvfit to determine the dissociation constant.
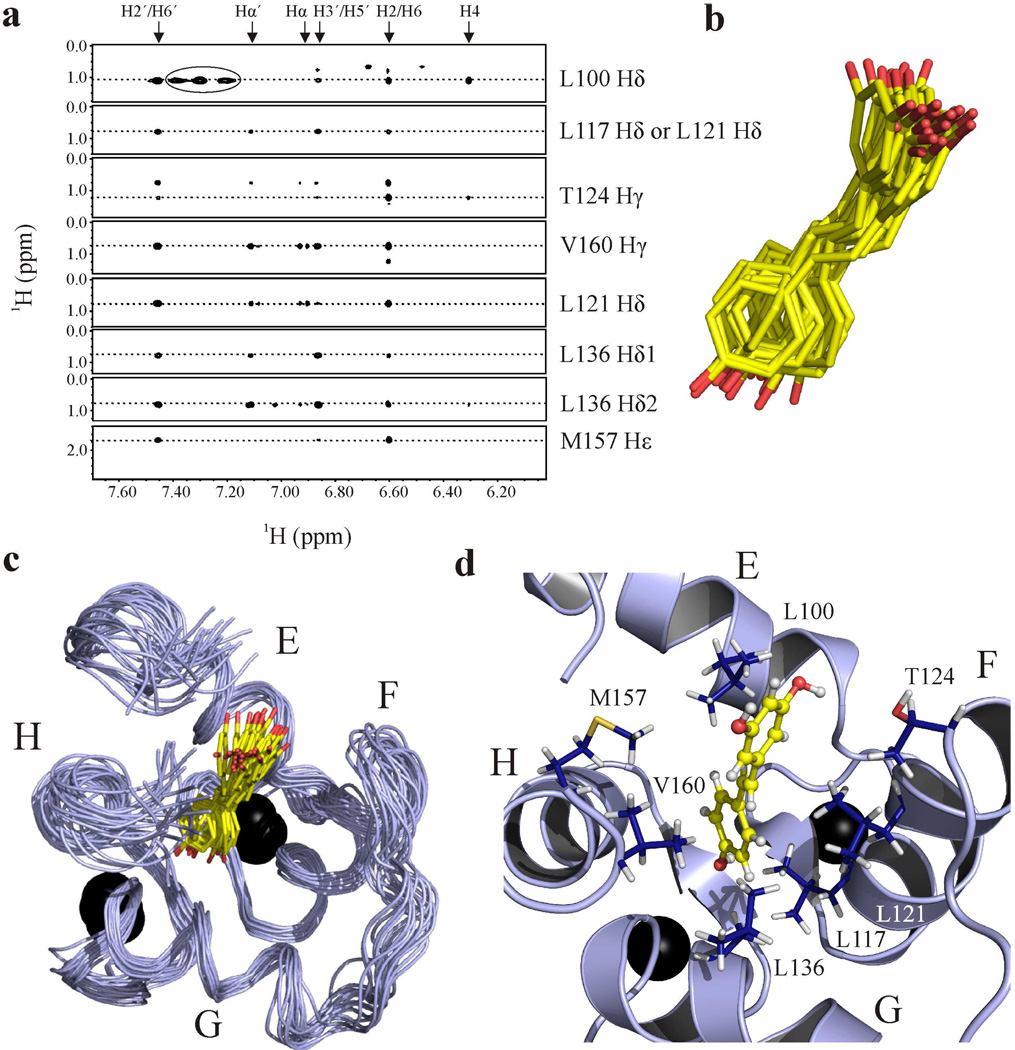
Following determination of the affinity and stoichiometry of resveratrol binding to cCTnC, a j-surface mapping was executed to predict resveratrol’s binding location on cCTnC. J-surface mapping works to predict a binding site by assuming that the ring current from an aromatic constituent of a ligand is the primary source of ligand-induced CSPs (53). Typically, amide protons are used in the calculation; however, it is well-established that cTnC undergoes a large conformational change upon ligand binding. Thus, most of the backbone CSPs are indicative of a global change in its structure rather than direct contact with a given ligand (66). In order to circumvent this difficulty, the CSPs of methyl protons were used in the j-surface calculation – as changes in these chemical shifts are more likely indicative of direct interactions with resveratrol. Methyl resonances used in the calculation were those that underwent ≥ |0.012| ppm in the proton dimension. These included: Ile112, Met120, Leu121, Ala123, Ile128, Ile148, Met157, and Val160. The structure of cTnC (pdb: 1aj4) (1) was used in the j-surface calculation, and the results localize the binding of resveratrol to the cleft formed by the four helices of cCTnC (figure 4). There appears to be two j-surfaces: one near the surface of cCTnC and the other deeper in the core. Because the j-surface calculation is unable to identify the specific pose resveratrol adopts when bound to cCTnC, NMR spectroscopy was used to identify intermolecular contacts between resveratrol and cCTnC.
Fig. 4. J-surface representation.
Results are mapped on the structure of cTnC (pdb: 1aj4). The backbone atoms are depicted in cartoon representation (light blue), Ca2+ ions are represented by black spheres, and the j-surface dot density is depicted using yellow spheres. Residues used in the j-surface calculation were: Leu121, Val160, Ala123, Ile148, Ile128, Met157, Met120, and Ile112.
Structure of cCTnC•resveratrol complex
The structure of cCTnC free or in complex with a number of binding partners has been solved by both X-ray crystallography and NMR spectroscopy (1, 2, 14, 19, 67–69). Instead of determining the structure of cCTnC again, a data-driven docking approach was pursued. The 1H,15N-HSQC CSPs induced by resveratrol were compared with those induced by ligands that interact with cCTnC: cTnI34–71, EMD 57033, and EGCg (supplementary figure 4). The CSPs are most similar between resveratrol and EGCg (in both magnitude and pattern); hence the intramolecular NOEs and dihedrals restraints of cCTnC used in the structure calculation were taken from the cCTnC•EGCg complex (19). The only new restraints used in the structure calculation were those that defined the structure and pose of resveratrol in complex with cCTnC.
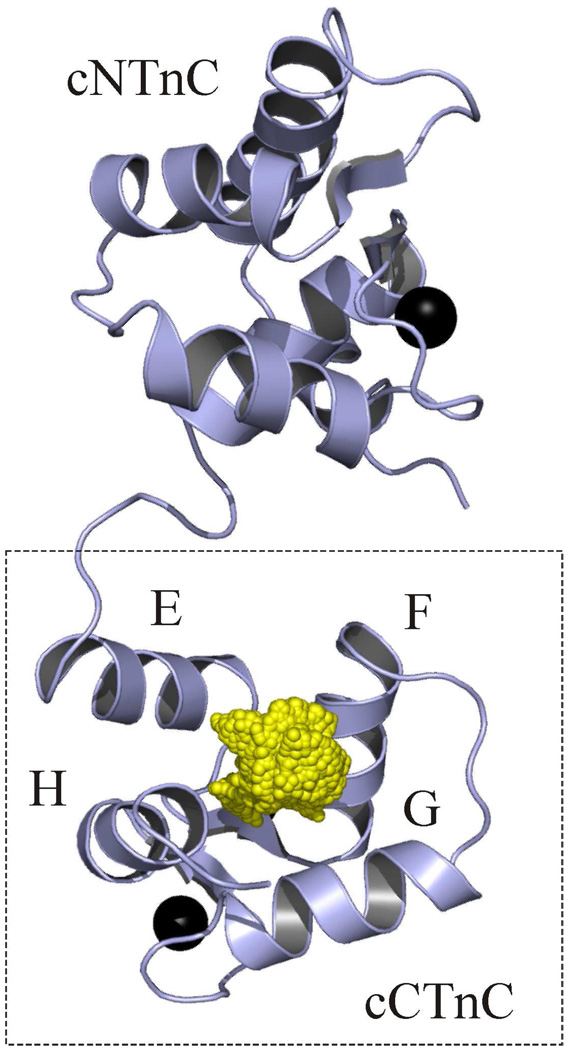
Intermolecular NOEs were measured between cCTnC and resveratrol with 13C,15Nedited/filtered NOESY experiments. These experiments measure solely NOEs between a 13C/15N-labeled molecule and an unlabeled molecule. Two-dimensional 13C,15N- filtered/edited NOESYHSQC (42) experiments were acquired with 150, 200, and 250 ms mixing times to establish the optimal mixing times to acquire the three-dimensional experiments. Since most of the intermolecular contacts observed were between methyl groups from cCTnC and the aromatic ring protons of resveratrol, the three-dimensional 13C-edited/filtered HMQCNOESY experiment (43, 44) was acquired. This NMR experiment is optimized for the identification of intermolecular NOEs involving 13C-methyls (44). Two three-dimensional 13C-edited/filtered HMQCNOESY experiments were acquired: one with a mixing time of 250 ms and one with a mixing time of 200 ms. Intermolecular NOEs that had intensities ≥ 0.04 were included in the structure calculation. This is because of the long experimental mixing times of the intermolecular NOESY experiments and the tightly coupled nature of resveratrol increased the possibility of spin diffusion. When all NOEs were included in the structure run, many violations occurred, presumably because of spin diffusion. The intermolecular NOEs from a mixing time of 200 ms, used in the structure calculation are shown in figure 5a. Initially, a structure calculation followed as was done for the EGCg calculation. The quality of the structure was subsequently improved by running a structure refinement in water; at this stage electrostatic potentials and atomic charges were included (56). The 20 lowest energy structures of the complex are shown in figures 5b and 5c. The structure indicates that, congruent with the prediction by j-surface mapping, resveratrol is localized to the hydrophobic pocket of cCTnC. The resveratrol-cCTnC interaction site is populated primarily by non-polar contacts. Resveratrol contacts the side chains of Leu100, Leu117, Leu121, Thr124, Leu136, Met157, and Val160 (figure 5d). Weak NOEs between resveratrol and Ile148 and Ile112 (both β-sheet residues) – not used in the structure run because they did not meet the intensity cutoff criteria – were also consistent with the final structure. In addition to the methyl-resveratrol contacts, the structure indicates that arene-arene contacts made between resveratrol and Phe104, Phe153, and Phe156 contribute to the binding energy of resveratrol.
Fig. 5. Structure of the binary complex of cCTnC•resveratrol.
a. Strip plots showing the NOEs between resveratrol and cCTnC. The resveratrol assignments are at the top of the spectrum; cCTnC assignments are indicated on the right. Peaks that are circled represent intramolecular cCTnC NOEs that were not adequately filtered. b. Ensemble of resveratrol from the binary complex. The carbon atoms are shown in yellow and the oxygen atoms in red. c. 20 lowest energy structures of the cCTnC•resveratrol complex with the backbone trace of cCTnC in light blue; helices E to H are labeled. d. Resveratrol’s binding pocket on the lowest energy structure of cCTnC. Residues for which NOEs were measured are labeled and depicted in stick representation with carbon atoms colored in dark blue; resveratrol is shown in ball-and-stick representation (hydrogen atoms in white). Calcium ions are shown as black spheres.
In addition to measuring intermolecular NOEs between resveratrol and cCTnC to determine their relative positions, the conformation of bound resveratrol was also determined. The intramolecular NOEs of resveratrol were measured with the 13C,15N-filtered NOESY experiment (40, 41), which works by removing signals from a 13C,15N-labeled molecule, keeping only signals from an unlabeled molecule. All structure-defining intramolecular NOEs were close to the same intensity, with the strongest being the H2/H6-Hα’ contact (see table 1 and supplementary figure 5). This is in contrast with free resveratrol, where the strongest ROE was between H2’/H6’ and Hα; and suggests that resveratrol undergoes a slight conformational change upon binding cCTnC. In order to limit biasing the structure of resveratrol during the calculation, one distance (2.6-1.8 Å) for the four intramolecular contacts was used; and the final structure was checked against the raw NOE data. Indeed, the results are consistent with the relative NOE intensities: the closest proton pair was the H2/H6-Hα’ proton pair (2.24 ± 0.14 Å). The other distances are: H2/H6-Hα; 2.57 ± 0.05 Å, H2’/H6’-Hα’; 2.28 ± 0.06 Å, and H2’/H6’-Hα; 2.32 ± 0.07 Å. The torsion angles were measured for resveratrol when in complex with cCTnC and were compared to those determined for free resveratrol. The p-OH ring is more twisted than what was observed for free resveratrol (from 0.5 ± 0.3° to 18.6 ± 10.6°), whereas the torsion angle of the di-m-OH ring is not significantly different (from 43.9 ± 0.4° to 35.2 ± 8.7°).
There have been a number of crystal structures of resveratrol in complex with proteins solved, including: alfalfa chalcone synthase (CHS) (70), the fibril-forming transthyretin (TTR) (71), a variant of alfalfa CHS (72), quinone reductase 2 (QR2) (73), peanut stilbene synthase (STS) (74), bovine F1-ATPase (75), Leukotriene A4 hydrolase (76), and human cytosolic sulfotransferase (not published; but deposited in the PDB with the ID: 3ckl). In most of these structures resveratrol is planar or slightly distorted from a planar conformation with torsion angles no greater than 45°; the only exception is in the peanut STS-resveratrol complex, where both rings of resveratrol are twisted > 60° (74) (see table 2). These results, in conjunction with the structures free resveratrol and resveratrol derivatives, point to a relatively rigid resveratrol framework. In accordance with the X-ray and NMR structures, Caruso et al. calculated single-point energy versus the torsion angle of the p-OH ring, and found the lowest energy structure is a planar conformation (64).
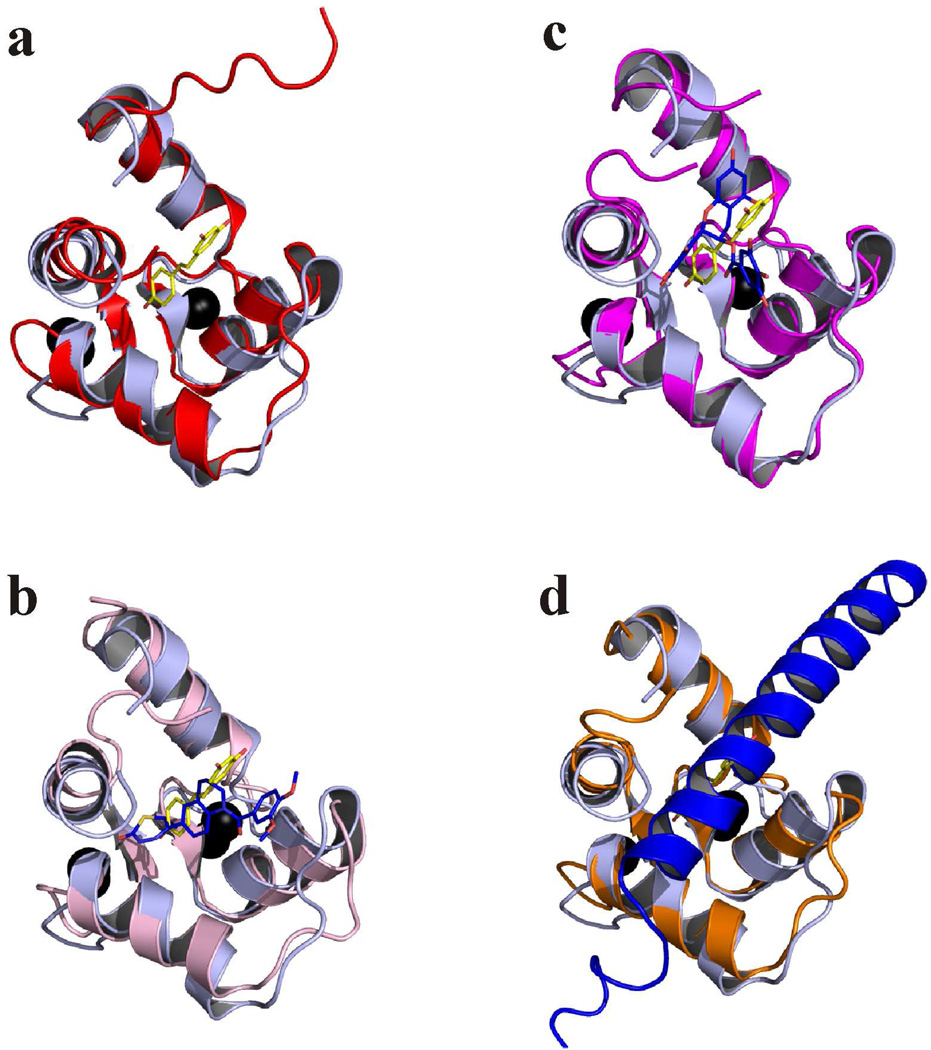
The structure of cCTnC in the cCTnC•resveratrol complex is not much different than that of cCTnC in other complexes. The Cα from residues in secondary structure elements of cCTnC•resveratrol were superimposed with cCTnC (pdb: 3ctn), cCTnC•EMD57033 (pdb: 1ih0), cCTnC•EGCg (pdb: 2kdh), and cCTnC•cTnI34–71 (pdb: 1j2d) and rmsds of: 1.17 Å, 1.50 Å, 1.02 Å, and 1.26 Å were determined, respectively. These values indicate that resveratrol does not significantly perturb the structure of cCTnC. The E-helix of cCTnC•resveratrol is shifted away from the hydrophobic cleft, similar to the cCTnC•EMD5033 complex. The position of the F-helix of cCTnC•resveratrol is in almost an identical position as in the cCTnC•EGCg and cCTnC•cTnI34–71 complexes; however, is shifted further away from the E-helix of cCTnC and not as far away as in the cCTnC•EMD57033 structure.
Discussion
The regulatory role of the Ca2+-dependant interaction between cTnI147–163 and cNTnC is well established, and has been the interaction site primarily concentrated on for the development of Ca2+-sensitizers (77, 78). However, the exclusive development of drugs that target cNTnC has been scrutinized as growing evidence indicates that cCTnC is also involved in contraction regulation. The dilated cardiomyopathy mutation of cCTnC, Gly159Asp, decreases myofilament Ca2+-sensitivity(79) via its modulation of the cTnI34–71-cCTnC interaction(67). The hypertrophic cardiomyopathy mutation, Asp145Glu, increases Ca2+-sensitivity(80), presumably by decreasing Ca2+ and cTnI binding to cCTnC(81). The ablation of the Ca2+-binding ability of the C-domain of cTnC increases the Ca2+-sensitivity of muscle contraction(82). There are two isoforms of TnC in insect flight muscle: the F1 isoform which regulates stretch-activated force, and the F2 isoform which is responsible for Ca2+-activated contraction. Although it is not clear how F1 regulates stretch-activation, the interaction between the C-domain of F1 and TnH (an ortholog of cTnI) may play a role in regulation of stretch-activation(83). Finally, as previously mentioned, the cardiotonic agents EMD 57033 (15, 84) and EGCg (17, 18) target cCTnC to modulate heart muscle contractility.
In this study, resveratrol was found to interact with the C-domain of cTnC, and the structure was determined by NMR spectroscopy. The p-OH group of resveratrol lies in the hydrophobic core of cCTnC, whereas the di-m-OH ring points towards the exterior of the protein. The stabilizing contacts between resveratrol and cCTnC are predominantly hydrophobic. In the other resveratrol-protein complexes, the binding site of resveratrol is also dominated by hydrophobic interactions; however, unlike the cCTnC•resveratrol structure, hydrogen bonds between the hydroxyls of resveratrol and the amino acids that line the binding pockets also contribute to the binding energy (70–76). The design of resveratrol with the p-OH converted to a hydrophobic constituent may therefore increase its affinity for cCTnC; for example, converting the hydroxyl to a fluorine atom would increase the lipophilicity of resveratrol without dramatically decreasing its size (85, 86). On the other hand, removing the p-OH would undoubtedly reduce the antioxidant ability of resveratrol, especially given that the p-OH has been implicated as being the principal hydroxyl responsible for resveratrol’s antioxidant nature (64, 87–89).
The comparison of the cCTnC•resveratrol structure with the structures of cCTnC•EMD57033 and cCTnC•EGCg yields insights into several key functional groups. In the structure, the p-OH aromatic ring of resveratrol is positioned in a similar manner as the thiadiazinone ring of EMD 57033 – with the para hydroxyl pointed towards the cleft formed by helices G and H. The di-m-OH ring of resveratrol faces away from the hydrophobic cavity of cCTnC – much like the benzendiol of EGCg – which leaves its hydroxyl moieties free to hydrogen bond with the surrounding aqueous milieu. Therefore, the binding pose of resveratrol has features that uniquely resemble the structures of EGCg and EMD 57033 when bound to cCTnC. Resveratrol, EMD 57033, and EGCg all share their binding sites with the natural binding partner of cCTnC, cTnI34–71 (figure 6), and as a result, may have a common mode of action. The troponin-dependant Ca2+-sensitizing ability of EMD 57033 has been suggested to involve a competition between EMD 57033 and cTnI34–71 for cCTnC (90). The perturbation of the cTnI34–71-cCTnC interaction may lead to an increase in the affinity of cTnI128–147 for cTnC and thus a decrease in contraction inhibition.
Fig. 6. Structure comparisons.
Superimposition between cCTnC•resveratrol (light blue) and a. cCTnC (red; pdb: 3ctn), b. cCTnC•EMD 57033 (pink; pdb: 1ih0), c. cCTnC•EGCg (magenta; pdb: 2kdh), and d. cCTnC•cTnI34–71 (orange; pdb: 1j1d). The carbon atoms for resveratrol are colored in yellow, and the oxygen atoms are colored in red. For the other structures, ligand carbon atoms are colored in blue, oxygen atoms in red, nitrogen atoms in dark blue, and sulfur atoms in yellow. Ca2+ ions are shown as black spheres.
It was determined that resveratrol bound to cTnC and cCTnC with micromolar affinity. This relatively low affinity of resveratrol for cTnC was anticipated, since too high an affinity would lead to a marked increase in Ca2+-sensitivity. This dramatic increase in Ca2+-sensitivity over the long-term could lead to negative effects, such as hypertrophic cardiomyopathy. On the other hand, it may be useful to optimize the resveratrol-cCTnC interaction for the development of drugs to treat acute heart failure. One method for the analysis of whether a small molecule represents a good lead molecule is to determine its ligand efficiency (LE) (91–93). LE is described by the ratio of free energy of binding over the number of heavy atoms in a compound, and is based on the premise that as a drug is optimized, it often increases in molecular weight; a trend fraught with problems including a decrease in bioavailability through insolubility and membrane permeability (94).
The free energy (ΔG) of binding is:
and the ligand efficiency (binding energy per non-hydrogen atom) (LE) is:
For resveratrol binding to cCTnC; a KD of 240 µM has a binding energy of 4.97 kcal/mol. Resveratrol has 17 non-hydrogen atoms (14 carbon atoms and 3 oxygen atoms), so its ligand efficiency is 0.29 kcal/mol. This ligand efficiency corresponds to a compound with 33 non-hydrogen atoms (approximately 2× the size of resveratrol: 450 MW) with a binding constant of 0.1 µM. The reasonably good ligand efficiency of resveratrol, suggests that the substitution or addition of a few atoms that enhance its affinity for cCTnC may lead toward novel therapies for the treatment of heart failure.
Conclusion
Resveratrol is a natural product found in wine that modulates the Ca2+-sensitivity of myofilaments (31). In this study, the structure of resveratrol in complex with the cardiac regulatory protein troponin C was determined by NMR spectroscopy. Consistent with the small molecules EGCg and EMD 57033, resveratrol targeted the C-domain of troponin C. The binding of resveratrol is primarily stabilized by hydrophobic contacts such as methyl-arene and arene-arene interactions. In addition to providing clues into the cardioprotective nature of resveratrol, the structure highlights several functional groups that could be modified to optimize the binding efficacy of resveratrol. Recently, the polyphenol, propyl gallate, has also been identified to act as a Ca2+-sensitizer as well (95); which alongside the functional and structural data for EGCg and resveratrol, points towards a common mechanism by which these natural compounds target the thin filament to protect against heart failure.
Supplementary Material
Acknowledgements
We would like to thank Dr. G. Moyna for making the source code for Jsurf available and for helpful instructions on its use, Drs. Monica Li and Marta Oleszczuk for insightful discussions on drug- troponin C interactions, and Dr. Pascal Mercier and Olivier Julien for helpful advice on structure calculation. The authors would also like to thank David Corson and Melissa Crane for protein expression and purification; and Robert Boyko for spectrometer maintenance and in-house software development. We would like to thank the Canadian National High Field NMR Centre (NANUC) for their assistance and use of the facilities.
Funding:
This work is supported by grants from the Canadian Institutes of Health Research (FRN 37760), the National Institutes of Health (R01 HL-085234), the Heart and Stroke Foundation of Canada to B.D.S., and the Alberta Heritage Foundation for Medical Research to I.M.R. NANUC is funded by the Canadian Institutes of Health Research, the Natural Science and Engineering Research Council of Canada, and the University of Alberta.
Abbreviations
- NMR spectroscopy
Nuclear Magnetic Resonance Spectroscopy
- ROESY
Rotating frame Overhauser effect spectroscopy
- NOESY
Nuclear Overhauser effect spectroscopy
- HSQC
Heteronuclear single quantum coherence
- HMQC
Heteronuclear multiple quantum coherence
- NOE
nuclear Overhauser effect
- TnC
troponin C
- cTnC
cardiac TnC
- cCTnC
C-domain of cTnC
- cNTnC
N-domain of cTnC
- TnI
troponin I
- cTnI
cardiac TnI
- TnT
troponin T
- cTnT
cardiac TnT
- EGCg
epigallocatechin gallate
- TFP
trifluoperazine
- CSP
Chemical shift perturbation
- TCEP
Tris(2-carboxyethyl) phosphine
- ppm
parts per million
- SSE
sum of squared error
Footnotes
Supporting Information Available: Data illustrating the oxidation rate of resveratrol; the resveratrol ROE intensities versus mixing time; the binding of resveratrol to cTnC; the ligand dependant chemical shift perturbations of cCTnC; the intramolecular NOEs of resveratrol in complex with cCTnC; and the structural statistics for the ensemble of cCTnC•resveratrol structures, are all available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Sia SK, Li MX, Spyracopoulos L, Gagne SM, Liu W, Putkey JA, Sykes BD. Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain. J. Biol. Chem. 1997;272:18216–18221. doi: 10.1074/jbc.272.29.18216. [DOI] [PubMed] [Google Scholar]
- 2.Takeda S, Yamashita A, Maeda K, Maeda Y. Structure of the core domain of human cardiac troponin in the Ca(2+)-saturated form. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- 3.Spyracopoulos L, Li MX, Sia SK, Gagne SM, Chandra M, Solaro RJ, Sykes BD. Calcium-induced structural transition in the regulatory domain of human cardiac troponin C. Biochemistry. 1997;36:12138–12146. doi: 10.1021/bi971223d. [DOI] [PubMed] [Google Scholar]
- 4.Li MX, Spyracopoulos L, Sykes BD. Binding of cardiac troponin-I147-163 induces a structural opening in human cardiac troponin-C. Biochemistry. 1999;38:8289–8298. doi: 10.1021/bi9901679. [DOI] [PubMed] [Google Scholar]
- 5.Tripet B, Van Eyk JE, Hodges RS. Mapping of a second actin-tropomyosin and a second troponin C binding site within the C terminus of troponin I, and their importance in the Ca2+-dependent regulation of muscle contraction. J. Mol. Biol. 1997;271:728–750. doi: 10.1006/jmbi.1997.1200. [DOI] [PubMed] [Google Scholar]
- 6.Li MX, Wang X, Sykes BD. Structural based insights into the role of troponin in cardiac muscle pathophysiology. J. Muscle Res. Cell Motil. 2004;25:559–579. doi: 10.1007/s10974-004-5879-2. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi T, Jin L, de Tombe PP. Cardiac thin filament regulation. Pflugers Archiv-European Journal of Physiology. 2008;457:37–46. doi: 10.1007/s00424-008-0511-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Endoh M. Mechanism of action of Ca2+ sensitizers--update 2001. Cardiovasc. Drugs Ther. 2001;15:397–403. doi: 10.1023/a:1013385305567. [DOI] [PubMed] [Google Scholar]
- 9.Kleerekoper Q, Liu W, Choi D, Putkey JA. Identification of binding sites for bepridil and trifluoperazine on cardiac troponin C. J. Biol. Chem. 1998;273:8153–8160. doi: 10.1074/jbc.273.14.8153. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Love ML, Putkey JA, Cohen C. Bepridil opens the regulatory N-terminal lobe of cardiac troponin C. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5140–5145. doi: 10.1073/pnas.090098997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haikala H, Kaivola J, Nissinen E, Wall P, Levijoki J, Linden IB. Cardiac Troponin-C as a Target Protein for a Novel Calcium Sensitizing Drug, Levosimendan. J. Mol. Cell. Cardiol. 1995;27:1859–1866. doi: 10.1016/0022-2828(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 12.Haikala H, Nissinen E, Etemadzadeh E, Levijoki J, Linden IB. Troponin C-Mediated Calcium Sensitization Induced by Levosimendan Does Not Impair Relaxation. J. Cardiovasc. Pharmacol. 1995;25:794–801. doi: 10.1097/00005344-199505000-00016. [DOI] [PubMed] [Google Scholar]
- 13.Haikala H, Nissinen E, Etemadzadeh E, Linden IB, Pohto P. Levosimendan Increases Calcium Sensitivity without Enhancing Myosin Atpase Activity and Impairing Relaxation. J. Mol. Cell. Cardiol. 1992;24:S97-S97. [Google Scholar]
- 14.Wang X, Li MX, Spyracopoulos L, Beier N, Chandra M, Solaro RJ, Sykes BD. Structure of the C-domain of human cardiac troponin C in complex with the Ca2+ sensitizing drug EMD 57033. J. Biol. Chem. 2001;276:25456–25466. doi: 10.1074/jbc.M102418200. [DOI] [PubMed] [Google Scholar]
- 15.Pan BS, Johnson RG. Interaction of cardiotonic thiadiazinone derivatives with cardiac troponin C. J. Biol. Chem. 1996;271:817–823. [PubMed] [Google Scholar]
- 16.Kleerekoper Q, Putkey JA. Drug binding to cardiac troponin C. J. Biol. Chem. 1999;274:23932–23939. doi: 10.1074/jbc.274.34.23932. [DOI] [PubMed] [Google Scholar]
- 17.Liou YM, Kuo SC, Hsieh SR. Differential effects of a green tea-derived polyphenol (−)-epigallocatechin-3-gallate on the acidosis-induced decrease in the Ca(2+) sensitivity of cardiac and skeletal muscle. Pflugers Arch. 2008;456:787–800. doi: 10.1007/s00424-008-0456-y. [DOI] [PubMed] [Google Scholar]
- 18.Tadano N, Du CK, Yumoto F, Morimoto S, Ohta M, Xie MF, Nagata K, Zhan DY, Lu QW, Miwa Y, Takahashi-Yanaga F, Tanokura M, Ohtsuki I, Sasaguri T. Biological actions of green tea catechins on cardiac troponin C. Br. J. Pharmacol. 2010;161:1034–1043. doi: 10.1111/j.1476-5381.2010.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson IM, Li MX, Sykes BD. Solution Structure of Human Cardiac Troponin C in Complex with the Green Tea Polyphenol, (−)-Epigallocatechin 3-Gallate. J. Biol. Chem. 2009;284:23012–23023. doi: 10.1074/jbc.M109.021352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renaud S, Delorgeril M. Wine, Alcohol, Platelets, and the French Paradox for Coronary Heart-Disease. Lancet. 1992;339:1523–1526. doi: 10.1016/0140-6736(92)91277-f. [DOI] [PubMed] [Google Scholar]
- 21.Langcake P, Pryce RJ. Production of Resveratrol by Vitis-Vinifera and Other Members of Vitaceae as a Response to Infection or Injury. Physiol. Plant Pathol. 1976;9:77–86. [Google Scholar]
- 22.Siemann EH, Creasy LL. Concentration of the Phytoalexin Resveratrol in Wine. Am. J. Enol. Vitic. 1992;43:49–52. [Google Scholar]
- 23.Das DK, Mukherjee S, Ray D. Resveratrol and red wine, healthy heart and longevity. Heart Failure Reviews. 2010;15:467–477. doi: 10.1007/s10741-010-9163-9. [DOI] [PubMed] [Google Scholar]
- 24.Lekli I, Ray D, Das DK. Longevity nutrients resveratrol, wines and grapes. Genes and Nutrition. 2010;5:55–60. doi: 10.1007/s12263-009-0145-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nichols JA, Katiyar SK. Skin photoprotection by natural polyphenols: anti-inflammatory, antioxidant and DNA repair mechanisms. Archives of Dermatological Research. 2010;302:71–83. doi: 10.1007/s00403-009-1001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun AY, Wang Q, Simonyi A, Sun GY. Resveratrol as a Therapeutic Agent for Neurodegenerative Diseases. Molecular Neurobiology. 2010;41:375–383. doi: 10.1007/s12035-010-8111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jang MS, Cai EN, Udeani GO, Slowing KV, Thomas CF, Beecher CWW, Fong HHS, Farnsworth NR, Kinghorn AD, Mehta RG, Moon RC, Pezzuto JM. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- 28.Holme AL, Pervaiz S. Resveratrol in cell fate decisions. J. Bioenerg. Biomembr. 2007;39:59–63. doi: 10.1007/s10863-006-9053-y. [DOI] [PubMed] [Google Scholar]
- 29.Das DK, Maulik N. Red wine and heart: A cardioprotective journey from grape to resveratrol. Alcoholism-Clinical and Experimental Research. 2006;30:84a-84a. [Google Scholar]
- 30.Ray PS, Maulik G, Cordis GA, Bertelli AAE, Bertelli A, Das DK. The red wine antioxidant resveratrol protects isolated rat hearts from ischemia reperfusion injury. Free Radical Biology and Medicine. 1999;27:160–169. doi: 10.1016/s0891-5849(99)00063-5. [DOI] [PubMed] [Google Scholar]
- 31.Liew R, Stagg MA, MacLeod KT, Collins P. The red wine polyphenol, resveratrol, exerts acute direct actions on guinea-pig ventricular myocytes. Eur. J. Pharmacol. 2005;519:1–8. doi: 10.1016/j.ejphar.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 32.Bertini I, Fragai M, Giachetti A, Luchinat C, Maletta M, Parigi G, Yeo KJ. Combining in silico tools and NMR data to validate protein-ligand structural models: Application to matrix metalloproteinases. J. Med. Chem. 2005;48:7544–7559. doi: 10.1021/jm050574k. [DOI] [PubMed] [Google Scholar]
- 33.Cioffi M, Hunter CA, Packer MJ, Spitaleri A. Determination of protein-ligand binding modes using complexation-induced changes in H-1 NMR chemical shift. J. Med. Chem. 2008;51:2512–2517. doi: 10.1021/jm701194r. [DOI] [PubMed] [Google Scholar]
- 34.Krishnamoorthy J, Yu VCK, Mok YK. Auto-FACE: An NMR Based Binding Site Mapping Program for Fast Chemical Exchange Protein-Ligand Systems. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0008943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pintacuda G, John M, Su XC, Otting G. NMR structure determination of protein-ligand complexes by lanthanide labeling. Acc. Chem. Res. 2007;40:206–212. doi: 10.1021/ar050087z. [DOI] [PubMed] [Google Scholar]
- 36.Hoffman RMB, Sykes BD. Structure of the Inhibitor W7 Bound to the Regulatory Domain of Cardiac Troponin C. Biochemistry. 2009;48:5541–5552. doi: 10.1021/bi9001826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chandra M, Dong WJ, Pan BS, Cheung HC, Solaro RJ. Effects of protein kinase A phosphorylation on signaling between cardiac troponin I and the N-terminal domain of cardiac troponin C. Biochemistry. 1997;36:13305–13311. doi: 10.1021/bi9710129. [DOI] [PubMed] [Google Scholar]
- 38.Li MX, Gagne SM, Tsuda S, Kay CM, Smillie LB, Sykes BD. Calcium-Binding to the Regulatory N-Domain of Skeletal-Muscle Troponin-C Occurs in a Stepwise Manner. Biochemistry. 1995;34:8330–8340. doi: 10.1021/bi00026a014. [DOI] [PubMed] [Google Scholar]
- 39.Lykkesfeldt J. Determination of ascorbic acid and dehydroascorbic acid in biological samples by high-performance liquid chromatography using subtraction methods: Reliable reduction with tris[2-carboxyethyl] phosphine hydrochloride. Anal. Biochem. 2000;282:89–93. doi: 10.1006/abio.2000.4592. [DOI] [PubMed] [Google Scholar]
- 40.Gemmecker G, Olejniczak ET, Fesik SW. An Improved Method for Selectively Observing Protons Attached to C-12 in the Presence of H-1-C-13 Spin Pairs. J. Magn. Reson. 1992;96:199–204. [Google Scholar]
- 41.Ikura M, Bax A. Isotope-Filtered 2d Nmr of a Protein Peptide Complex - Study of a Skeletal-Muscle Myosin Light Chain Kinase Fragment Bound to Calmodulin. J. Am. Chem. Soc. 1992;114:2433–2440. [Google Scholar]
- 42.Stuart AC, Borzilleri KA, Withka JM, Palmer AG. Compensating for variations in H-1-C-13 scalar coupling constants in isotope-filtered NMR experiments. J. Am. Chem. Soc. 1999;121:5346–5347. [Google Scholar]
- 43.Lee W, Revington MJ, Arrowsmith C, Kay LE. A Pulsed-Field Gradient Isotope-Filtered 3d C-13 Hmqc-Noesy Experiment for Extracting Intermolecular Noe Contacts in Molecular-Complexes. FEBS Lett. 1994;350:87–90. doi: 10.1016/0014-5793(94)00740-3. [DOI] [PubMed] [Google Scholar]
- 44.Robertson IM, Spyracopoulos L, Sykes BD. The Evaluation of Isotope Editing and Filtering for Protein-Ligand Interaction Elucidation by Nmr. Biophysics and the Challenges of Emerging Threats. 2009:101–119. [Google Scholar]
- 45.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 46.Johnson BA, Blevins RA. Nmr View - a Computer-Program for the Visualization and Analysis of Nmr Data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 47.Lin X, Krudy GA, Howarth J, Brito RMM, Rosevear PR, Putkey JA. Assignment and Calcium-Dependence of Methionyl Epsilon-C and Epsilon-H Resonances in Cardiac Troponin-C. Biochemistry. 1994;33:14434–14442. doi: 10.1021/bi00252a009. [DOI] [PubMed] [Google Scholar]
- 48.Schuttelkopf AW, van Aalten DMF. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 49.Kleywegt GJ, Zou JY, Kjeldgaard M, Jones TA, Around O. In: International Tables for Crystallography, Vol. F. Crystallography of Biological Macromolecules. Rossmann MG, Arnold E, editors. The Netherlands: Dordrecht: Kluwer Academic Publishers; 2001. pp. 353–356.pp. 366–367. [Google Scholar]
- 50.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 51.Frisch MJ, T GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Vreven T, Jr, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, Gaussian 03 ed. Wallingford CT: Gaussian, Inc; 2004. [Google Scholar]
- 52.Hoffman RMB, Li MX, Sykes BD. The bindin of W7, an inhibitor of striated muscle contraction, to cardiac troponin C. Biochemistry. 2005;44:15750–15759. doi: 10.1021/bi051583y. [DOI] [PubMed] [Google Scholar]
- 53.McCoy MA, Wyss DF. Spatial localization of ligand binding sites from electron current density surfaces calculated from NMR chemical shift perturbations. J. Am. Chem. Soc. 2002;124:11758–11763. doi: 10.1021/ja026166c. [DOI] [PubMed] [Google Scholar]
- 54.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 55.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 56.Linge JP, Williams MA, Spronk CAEM, Bonvin AMJJ, Nilges M. Refinement of protein structures in explicit solvent. Proteins-Structure Function and Bioinformatics. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 57.Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 58.Jayatilake GS, Jayasuriya H, Lee ES, Koonchanok NM, Geahlen RL, Ashendel CL, Mclaughlin JL, Chang CJ. Kinase Inhibitors from Polygonum-Cuspidatum. J. Nat. Prod. 1993;56:1805–1810. doi: 10.1021/np50100a021. [DOI] [PubMed] [Google Scholar]
- 59.Koh D, Park KH, Jung J, Yang H, Mok KH, Lim Y. Complete assignment of the H-1 and C-13 NMR spectra of resveratrol derivatives. Magn. Reson. Chem. 2001;39:768–770. [Google Scholar]
- 60.Commodari F, Khiat A, Ibrahimi S, Brizius AR, Kalkstein N. Comparison of the phytoestrogen trans-resveratrol (3,4 ',5-trihydroxystilbene) structures from x-ray diffraction and solution NMR. Magn. Reson. Chem. 2005;43:567–572. doi: 10.1002/mrc.1583. [DOI] [PubMed] [Google Scholar]
- 61.Bonechi C, Martini S, Magnani A, Rossi C. Stacking interaction study of trans-resveratrol (trans-3,5,4 '-trihydroxystilbene) in solution by nuclear magnetic resonance and Fourier transform infrared spectroscopy. Magn. Reson. Chem. 2008;46:625–629. doi: 10.1002/mrc.2217. [DOI] [PubMed] [Google Scholar]
- 62.Karplus M. Contact Electron-Spin Coupling of Nuclear Magnetic Moments. J. Chem. Phys. 1959;30:11–15. [Google Scholar]
- 63.Trela BC, Waterhouse AL. Resveratrol: Isomeric molar absorptivities and stability. J. Agric. Food Chem. 1996;44:1253–1257. [Google Scholar]
- 64.Caruso F, Tanski J, Villegas-Estrada A, Rossi M. Structural basis for antioxidant activity of trans-resveratrol: Ab initio calculations and crystal and molecular structure. J. Agric. Food Chem. 2004;52:7279–7285. doi: 10.1021/jf048794e. [DOI] [PubMed] [Google Scholar]
- 65.Yin Q, Shi YM, Liu HM, Li CB, Zhang WQ. (E)-3,5,4 '-Trimethoxystilbene. Acta Crystallographica Section E-Structure Reports Online. 2002;58:O1180–O1181. [Google Scholar]
- 66.Robertson IM, Pineda-Sanabria S, Sykes BD. Approaches to protein-ligand structure determination by NMR spectroscopy: applications in drug binding to the cardiac regulatory protein troponin C. Biophysics and Structure to Counter Threats and Challenges. (In Press) [Google Scholar]
- 67.Baryshnikova OK, Robertson IM, Mercier P, Sykes BD. Dilated cardiomyopathy G159D mutation in cardiac troponin C weakens the anchoring interaction with troponin I. Biochemistry. 2008;47:10950–10960. doi: 10.1021/bi801165c. [DOI] [PubMed] [Google Scholar]
- 68.Gasmi-Seabrook GM, Howarth JW, Finley N, Abusamhadneh E, Gaponenko V, Brito RM, Solaro RJ, Rosevear PR. Solution structures of the C-terminal domain of cardiac troponin C free and bound to the N-terminal domain of cardiac troponin I. Biochemistry. 1999;38:8313–8322. doi: 10.1021/bi9902642. [DOI] [PubMed] [Google Scholar]
- 69.Lindhout DA, Sykes BD. Structure and dynamics of the C-domain of human cardiac troponin C in complex with the inhibitory region of human cardiac troponin I. J. Biol. Chem. 2003;278:27024–27034. doi: 10.1074/jbc.M302497200. [DOI] [PubMed] [Google Scholar]
- 70.Ferrer JL, Jez JM, Bowman ME, Dixon RA, Noel JP. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat. Struct. Biol. 1999;6:775–784. doi: 10.1038/11553. [DOI] [PubMed] [Google Scholar]
- 71.Klabunde T, Petrassi HM, Oza VB, Raman P, Kelly JW, Sacchettini JC. Rational design of potent human transthyretin amyloid disease inhibitors. Nat. Struct. Biol. 2000;7:312–321. doi: 10.1038/74082. [DOI] [PubMed] [Google Scholar]
- 72.Austin MB, Bowman ME, Ferrer JL, Schroder J, Noel JP. An aldol switch discovered in stilbene synthases mediates cyclization specificity of type III polyketide synthases. Chem. Biol. 2004;11:1179–1194. doi: 10.1016/j.chembiol.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 73.Buryanovskyy L, Fu Y, Boyd M, Ma YL, Hsieh TC, Wu JM, Zhang ZT. Crystal structure of quinone reductase 2 in complex with resveratrol. Biochemistry. 2004;43:11417–11426. doi: 10.1021/bi049162o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shomura Y, Torayama I, Suh DY, Xiang T, Kita A, Sankawa U, Miki K. Crystal structure of stilbene synthase from Arachis hypogaea. Proteins-Structure Function and Bioinformatics. 2005;60:803–806. doi: 10.1002/prot.20584. [DOI] [PubMed] [Google Scholar]
- 75.Gledhill JR, Montgomery MG, Leslie AGW, Walker JE. Mechanism of inhibition of bovine F-1-ATPase by resveratrol and related polyphenols. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13632–13637. doi: 10.1073/pnas.0706290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davies DR, Mamat B, Magnusson OT, Christensen J, Haraldsson MH, Mishra R, Pease B, Hansen E, Singh J, Zembower D, Kim H, Kiselyov AS, Burgin AB, Gurney ME, Stewart LJ. Discovery of Leukotriene A4 Hydrolase Inhibitors Using Metabolomics Biased Fragment Crystallography (vol 52, pg 4694, 2009) J. Med. Chem. 2010;53:2330–2331. doi: 10.1021/jm900259h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li MX, Robertson IM, Sykes BD. Interaction of cardiac troponin with cardiotonic drugs: a structural perspective. Biochem. Biophys. Res. Commun. 2008;369:88–99. doi: 10.1016/j.bbrc.2007.12.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sorsa T, Pollesello P, Solaro RJ. The contractile apparatus as a target for drugs against heart failure: Interaction of levosimendan, a calcium sensitiser, with cardiac troponin c. Mol. Cell. Biochem. 2004;266:87–107. doi: 10.1023/b:mcbi.0000049141.37823.19. [DOI] [PubMed] [Google Scholar]
- 79.Mirza M, Marston S, Willott R, Ashley C, Mogensen J, McKenna W, Robinson P, Redwood C, Watkins H. Dilated cardiomyopathy mutations in three thin filament regulatory proteins result in a common functional phenotype. J. Biol. Chem. 2005;280:28498–28506. doi: 10.1074/jbc.M412281200. [DOI] [PubMed] [Google Scholar]
- 80.Landstrom AP, Parvatiyar MS, Pinto JR, Marquardt ML, Bos JM, Tester DJ, Ornmen SR, Potter JD, Ackerman MJ. Molecular and functional characterization of novel hypertrophic cardiomyopathy susceptibility mutations in TNNC1-encoded troponin C. J. Mol. Cell. Cardiol. 2008;45:281–288. doi: 10.1016/j.yjmcc.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Swindle N, Tikunova SB. Hypertrophic Cardiomyopathy-Linked Mutation D145E Drastically Alters Calcium Binding by the C-Domain of Cardiac Troponin C. Biochemistry. 2010;49:4813–4820. doi: 10.1021/bi100400h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Szczesna D, Guzman G, Miller T, Zhao JJ, Farokhi K, Ellemberger H, Potter JD. The role of the four Ca2+ binding sites of troponin C in the regulation of skeletal muscle contraction. J. Biol. Chem. 1996;271:8381–8386. doi: 10.1074/jbc.271.14.8381. [DOI] [PubMed] [Google Scholar]
- 83.Agianian B, Krzic U, Qiu F, Linke WA, Leonard K, Bullard B. A troponin switch that regulates muscle contraction by stretch instead of calcium. EMBO J. 2004;23:772–779. doi: 10.1038/sj.emboj.7600097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Solaro RJ, Gambassi G, Warshaw DM, Keller MR, Spurgeon HA, Beier N, Lakatta EG. Stereoselective Actions of Thiadiazinones on Canine Cardiac Myocytes and Myofilaments. Circ. Res. 1993;73:981–990. doi: 10.1161/01.res.73.6.981. [DOI] [PubMed] [Google Scholar]
- 85.Biffinger JC, Kim HW, DiMagno SG. The polar hydrophobicity of fluorinated compounds. Chembiochem. 2004;5:622–627. doi: 10.1002/cbic.200300910. [DOI] [PubMed] [Google Scholar]
- 86.Smart BE. Fluorine substituent effects (on bioactivity) J. Fluorine Chem. 2001;109:3–11. [Google Scholar]
- 87.Cao H, Pan XL, Li C, Zhou C, Deng FY, Li TH. Density functional theory calculations for resveratrol. Bioorg. Med. Chem. Lett. 2003;13:1869–1871. doi: 10.1016/s0960-894x(03)00283-x. [DOI] [PubMed] [Google Scholar]
- 88.Del Nero J, De Melo CP. Investigation of the excited states of resveratrol and related molecules. Int. J. Quantum Chem. 2003;95:213–218. [Google Scholar]
- 89.Leopoldini M, Marino T, Russo N, Toscano M. Antioxidant properties of phenolic compounds: H-atom versus electron transfer mechanism. J. Phys. Chem. A. 2004;108:4916–4922. [Google Scholar]
- 90.Li MX, Spyracopoulos L, Beier N, Putkey JA, Sykes BD. Interaction of cardiac troponin C with Ca2+ sensitizer EMD 57033 and cardiac troponin I inhibitory peptide. Biochemistry. 2000;39:8782–8790. doi: 10.1021/bi000473i. [DOI] [PubMed] [Google Scholar]
- 91.Andrews PR, Craik DJ, Martin JL. Functional-Group Contributions to Drug Receptor Interactions. J. Med. Chem. 1984;27:1648–1657. doi: 10.1021/jm00378a021. [DOI] [PubMed] [Google Scholar]
- 92.Hopkins AL, Groom CR, Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discov. Today. 2004;9:430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 93.Kuntz ID, Chen K, Sharp KA, Kollman PA. The maximal affinity of ligands. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9997–10002. doi: 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 95.Tadano N, Morimoto S, Takahashi-Yanaga F, Miwa Y, Ohtsuki I, Sasaguri T. Propyl Gallate, a Strong Antioxidant, Increases the Ca2+ Sensitivity of Cardiac Myofilament. J. Pharmacol. Sci. 2009;109:456–458. doi: 10.1254/jphs.08266sc. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.