Abstract
A meningococcal group B vaccine containing multiple protein antigens including factor H binding protein (fHbp) and Neisserial heparin binding antigen (NHba) is in clinical development. The ability of antibodies against individual antigens to interact and augment protective immunity is unknown. We assayed human complement-mediated bactericidal activity (SBA) in stored sera from six immunized adults before and after depletion of antibodies to fHbp and/or NHba. All six subjects developed ≥4-fold increases in SBA titer against a test strain with fHbp in the variant 1 group with an amino acid sequence that matched the vaccine antigen (GMT <1:4 baseline, to 1:139 after 3 doses of vaccine). By adsorption 88 to >95 percent of the SBA was directed against fHbp. Four subjects developed ≥4-fold increases in SBA titer against a test strain with a heterologous fHbp variant 2 antigen and a homologous NHba amino acid sequence that matched the vaccine antigen (GMT <1:4 baseline, to 1:45). SBA was directed primarily against NHba in one subject, against fHbp in a second, while depletion of either anti-NHba or anti-fHbp antibody removed the majority of SBA in sera from two subjects. In all four subjects, depletion of both anti-fHbp and anti-NHba antibodies removed more SBA than depletion of either antibody individually. Mixing a mouse non-bactericidal anti-fHbp variant 1 antiserum with a mouse anti-NHba antiserum also augmented the anti-NHba SBA titer against this test strain. For meningococcal vaccines that target relatively sparsely-exposed antigens such fHbp or NHba, non-bactericidal antibodies against individual antigens can cooperate and elicit SBA.
Keywords: Neisseria meningitidis, vaccine, fHbp, NHba, GNA 2132, complement
I. Introduction
Meningococcal vaccines for the prevention of disease caused by group B strains of Neisseria meningitidis are currently in clinical trials. One of the vaccines contains recombinant factor H binding proteins (fHbp) from two antigenic variants [1]. Another contains an outer membrane vesicle vaccine (OMV) [2] combined with three recombinant proteins: NadA, and two fusion proteins, fHbp in the variant 1 (v.1) group fused with GNA2091 (GNA2091-fHbp), and Neisserial heparin binding antigen (NHba) [3] fused with GNA1030 (NHba-GNA1030) [4]. NHba was previously referred to as GNA2132 [5, 6]. In mice immunized with the three recombinant proteins, the principal antigenic targets of serum bactericidal antibody (SBA) were fHbp, NHba, and NadA [4]. The combination of the three proteins also elicited higher SBA titers than any of the individual proteins alone [4].
In humans a vaccine with the three recombinant proteins (no OMV) also elicited SBA responses [7] but the contributions of the antibodies elicited by the individual antigens toward the observed SBA were unclear. Investigating the functional contributions of individual antibody populations in a mixture of serum antibodies can be difficult because of the potential positive or negative interactions of antibodies binding to multiple antigenic targets on the bacterial surface [8–10]. The purpose of the present study was to define the role of antibodies to two of the antigens, fHbp and NHba, in eliciting serum bactericidal activity in immunized humans. Genes encoding these two antigens are present in nearly all disease-causing meningococcal isolates [1, 11–14].
2. Materials and Methods
2.1. Human serum samples
Stored serum samples were available from six adults immunized intramuscularly with an investigational meningococcal vaccine that contained recombinant GNA2091-fHbp, NHba-GNA1030, and NadA [4]. The samples were selected from 36 sera previously investigated from adult participants of phase I studies primarily designed to evaluate vaccine safety and tolerability [7]. The participants were given 3 doses of the vaccine spaced one month apart. Each dose contained 50 µg of each of the three proteins adsorbed with aluminum hydroxide. Blood samples were obtained immediately prior to the first immunization and one month after the third immunization. All subjects provided informed written consent. For the present study, we selected serum samples based on availability of sufficient volumes for the adsorption studies described below. Use of the serum samples for this study was approved by the institutional review board of Children’s Hospital & Research Center, Oakland.
2.2. Mouse serum samples
Stored sera were available from previous experiments in female CD-1 mice (Charles River) immunized with a recombinant NHba vaccine (gene from NZ98/254) [6], or a fHbp ID 1 vaccine. The amino acid sequences of the respective vaccine antigens matched those of the NHba and fHbp components of the vaccine given to the humans [4]. For the mouse study, three injections were given, each spaced 3 weeks apart. The fHbp dose was 20 µg, which was adsorbed with aluminum hydroxide, and the NHba dose was 15 µg, which was given with Freund’s complete adjuvant for dose 1 and incomplete adjuvant for doses 2 and 3. Control mice received the respective adjuvants without the vaccine antigens. Blood samples were obtained 3 weeks after the third dose of vaccine. Sera from 3 to 5 mice from each immunization group were pooled for use in the present studies. The animal immunization experiments were conducted under a protocol approved by the Institutional Animal Care and Use Committee of Children’s Hospital Oakland Research Institute.
2.3. Neisseria meningitidis strains (Table 1)
Table 1.
Group B N. meningitidis strains used to measure bactericidal activity
| Factor H binding protein (fHbp) |
Neisserial Heparin Binding Antigen (NHba) |
NadA gene6 |
||||||
|---|---|---|---|---|---|---|---|---|
| Strain | Reference | Clonal Complex1 |
ID number2 |
Variant Group3 |
Expression4 | Percent amino acid identity5 |
Expression4 | |
| H44/76-SL | Welsch et al [7, 24] | 32 | 1 | 1 | ++ | 91 | +/− | - |
| M4407 | Giuliani et al [15] | 41/44 | 19 | 2 | + | 100 | ++ | - |
| S3446 | Welsch et al [6] | 41/44 | 19 | 2 | + | 98 | ++ | - |
Clonal complex as defined by multilocus sequence typing [27].
fHbp ID number based on amino acid sequence variant as designated on the website, http://pubmlst.org/neisseria/fHbp/. fHbp ID 1 in strain H44/76 matched the amino acid sequence of the fHbp ID 1 antigen in the vaccine; the amino acid sequence identity between fHbp ID 1 in the vaccine and ID 19 in strains M4407 and S3446 was 72%.
fHbp variant group as classified by Masignani et al [25].
Expression of fHbp and NHba was measured by infrared Western blot [17] (See text).
As compared with the sequence encoded by the gene from strain NZ98/254 [6].
The absence of the NadA gene was confirmed by polymerase chain reaction [14].
Three group B test strains were used to measure serum bactericidal antibody activity. Strain H44/76-SL (B:15:P1.7,16; sequence type 32 [ST-32] [7]), referred to hereafter as H44/76, expressed a fHbp in the variant 1 group that matched the vaccine fHbp antigen (fHbp ID 1), and a NHba with 91% amino acid identity to the vaccine antigen. The other two strains, M4407 (B:NT:P1.15) [15] and S3446 (B:19,14:P1.23,14) [6, 16], were from clonal complex 41/44. Both had fHbp in the variant 2 group (fHbp ID 19), which was a mismatch with the variant 1 vaccine antigen, and expressed NHba with 98 to 100% amino acid identity to the vaccine antigen. All three strains lacked NadA genes. By infrared Western blot [17], strain H44/76 reacted with anti-fHbp mAb JAR 5, which is specific for fHbp in the variant 1 group [18], and was a relatively low expresser of the NHba protein. Strains M4407 and S3446 reacted with anti-fHbp mAb JAR 31, which is specific for fHbp in the variant 2 and 3 groups [18], and had two- to four-fold higher expression of NHba compared with strain H44/76 (Table 1).
2.4. Preparation of immunoadsorbents
Recombinant NHba and fHbp were provided by Novartis Vaccines (Siena, Italy). The recombinant NHba used for coupling with the immunoadsorbent had been prepared from E. coli BL21(DE3), which expressed a NHba gene isolated from group B N. meningitidis strain NZ98/254. The same gene encoded the NHba component of the NHba-GNA1030 fusion protein in the vaccine [4]. The recombinant fHbp (ID 1) was engineered to contain two amino acid substitutions (E218A/E239A) to prevent binding to complement fH, as previously described [19, 20]. The immunoadsorbents were prepared by covalently coupling the individual antigens to cyanogen bromide-activated Sepharose 4B (Sigma, St. Louis, MO) [21]. As a negative control for possible non-specific depletion of antibodies during adsorption, we used the same method to couple recombinant human albumin (rHA, Sigma, St. Louis, MO) to cyanogen bromide-activated Sepharose 4B. After overnight incubation, the coupling reactions were quelled with 0.1M Tris pH 8.0. The immunoadsorbents were washed extensively, blocked and stored in 1% rHA in phosphate-buffered saline (PBS) and sodium azide until use. Prior to use, the immunoadsorbents were extensively washed with PBS to remove all traces of sodium azide.
2.5. Depletion of anti-fHbp and anti-NHba antibodies
Serum samples were diluted 1:5 in PBS and aliquots were added to rHA-, NHba-, or fHbp-conjugated immunoadsorbents. After tumbling the slurries overnight at 4 degrees C, the slurries were loaded onto columns (Bio-Rad, Hercules, CA), and washed with PBS. The flow-through and washes were collected, filtered through a 0.2 µm filter, and concentrated to the original starting volumes of the undiluted serum samples using 10,000 MWCO protein concentrators (Corning, Lowell, MA).
2.6. Anti-fHbp and anti-NHba antibody activity
Serum anti-fHbp and anti-NHba antibody concentrations were measured by ELISA using recombinant fHbp (fHbp ID 1 without the two amino acid substitutions described above), or recombinant NHba as the target antigens. To favor detection of both high and low avidity antibody, the dilutions of sera added to wells of a microtiter plate were incubated overnight at 4°C. After washing, bound antibody was detected with alkaline phosphatase-conjugated goat anti-human IgG, Fcγ-specific (Jackson ImmunoResearch Laboratories, West Grove, PA). Antibody concentrations were assigned in arbitrary units per ml as compared with a reference serum sample from an immunized adult. The reference serum was assigned a value of 1000 units per milliliter (U/ml) for both anti-fHbp and anti-NHba antibodies. Percent depletion of anti-fHbp or anti-NHba antibody for each subject was calculated using the formula: 100 × [1-(antibody concentration of adsorbed sample/concentration of unadsorbed sample)].
2.7. Serum bactericidal antibody activity
All sera were heated for 30 minutes at 56°C to inactivate endogenous complement. SBA was measured as previously described using mid-log phase bacteria grown in Mueller Hinton broth supplemented with 0.25% glucose and 0.02 mM cytidine 5’-monophospho-N-acetylneuraminic acid (Sigma, St. Louis, MO) [7]. The complement was serum from a healthy adult who lacked endogenous bactericidal activity and had normal hemolytic complement activity. To ensure that IgG antibodies in the complement source did not block or augment bactericidal activity in the test sera, we depleted IgG from the complement source using a protein G column (HiTrap Protein G HP, GE Health Sciences, Piscataway, NJ), which was performed as previously described [22].
2.8. Statistical analyses
Statistical calculations were performed using Prism for Mac version 5.0a (GraphPad Software, La Jolla, CA). ELISA or SBA titers below the lower limit of detection were assigned a value of one-half of the lower limit of detection for that assay. Comparisons between the SBA titers of pre- and post-immunization samples, and of samples adsorbed with different immunoadsorbents, were performed using paired t-tests on the log10-transformed values. Titers below the minimum detected were assigned half of the minimum detected titer (i.e, 1:2 for a titer <1:4).
3. Results
3.1. Adequacy of depletion of anti-NHba or anti-fHbp antibodies
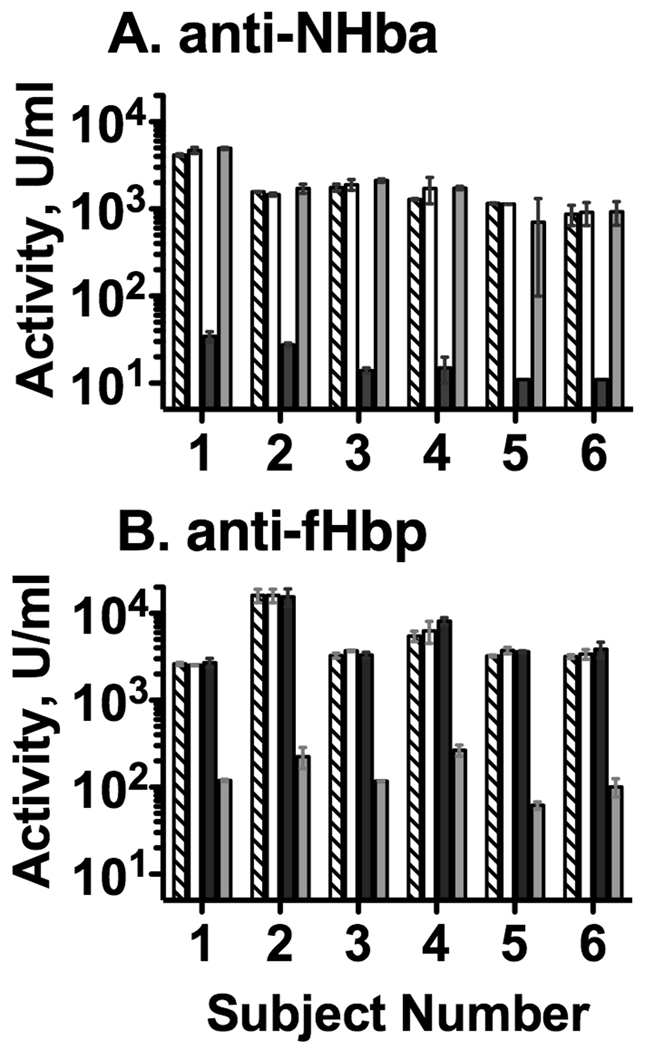
The percent depletion of antibodies by treatment of the sera with the respective immunoadsorbents was determined by ELISA (Figure 1). As an example, the post-immunization serum anti-NHba antibody concentration of Subject 1 decreased from 4175 U/ml in the unadsorbed sample to 35 U/ml after adsorption using the NHba-coupled immunoadsorbent (>99% decrease comparing the diagonal-hatched and the black bars, Panel A). There was no corresponding decrease in anti-NHba antibody concentrations after adsorption using the fHbp- (gray bars) or human recombinant albumin-coupled immunoadsorbents (white bars). Similarly, adsorption of post-immunization serum from subject 1 using the fHbp-coupled immunoadsorbent decreased the anti-fHbp antibody concentration from 2629 U/ml in unadsorbed serum to 120 U/ml after adsorption (>95% decrease comparing the diagonal-hatched and the gray bars, Figure 1, Panel B). There was no corresponding decrease in anti-fHbp antibody concentrations after adsorption with the NHba- (black bars) or rHA-coupled immunoadsorbents (white bars). These data indicated that the depletion of the antibodies was specific for each of the respective immunoadsorbents. For all six post-immunization sera, the adsorption method depleted >98% of the serum anti-NHba activity (Figure 1, Panel A), and >95% of the anti-fHbp antibody activity (Panel B).
Figure 1.
Adequacy of adsorption of post-immunization serum anti-NHba (Panel A) and anti-fHbp antibodies (Panel B) as measured by ELISA. The Y-axis shows the mean antibody concentrations and ranges of duplicate independent dilutions in arbitrary units (U) per ml (See methods). On some bars, the ranges are not evident because the differences between the respective replicate titers were small. Diagonal hatched bars, unadsorbed sera; open bars, sera adsorbed with irrelevant antigen (recombinant human albumin); black bars, adsorbed with NHba; gray bars, adsorbed with fHbp. The antibody concentrations in unadsorbed sera obtained before vaccination were <100 U/ml.
3.2. Serum bactericidal activity against a strain with fHbp v.1
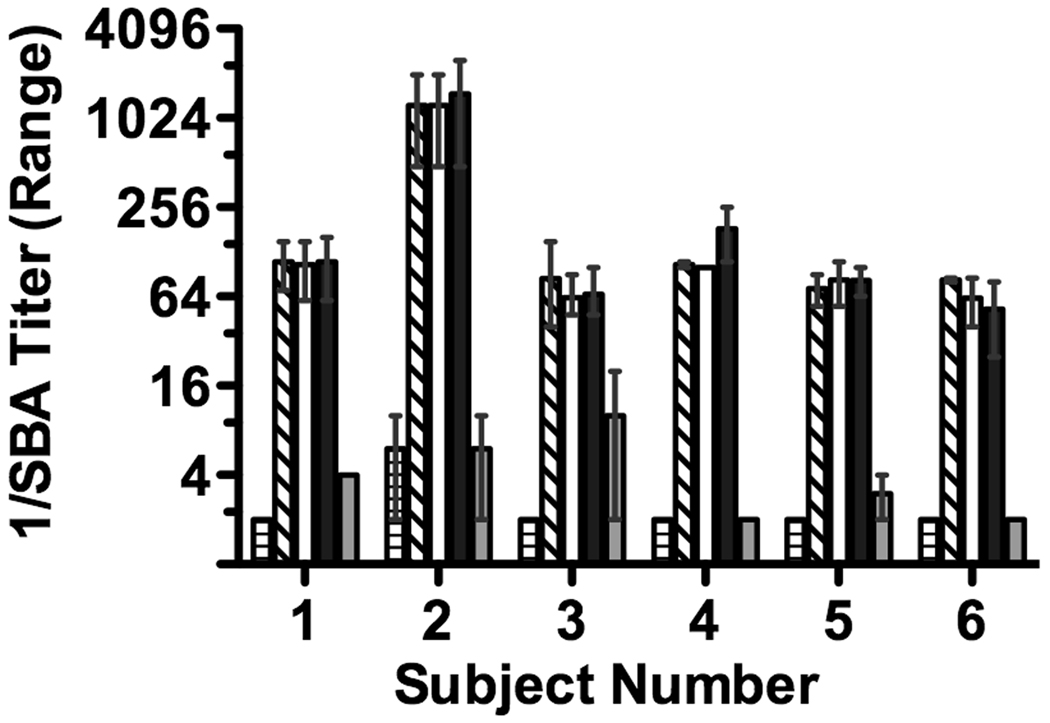
The reciprocal serum bactericidal GMT of the six subjects when measured against strain H44/76 increased from <4 in pre-immunization sera to 139 in post-immunization samples (p<0.0001); all six subjects had greater than 8-fold increases in titer after immunization. There were no significant changes in the respective post-immunization SBA titers after depletion of anti-NHba antibodies (Figure 2, black bars). After depletion of anti-fHbp antibodies, the SBA titers decreased by >95% in five of the subjects; and by 88% (1:85 to 1:10) in the sixth subject (subject 3). Thus, for all six subjects the serum bactericidal activity elicited by the vaccine against strain H44/76 was primarily directed against fHbp.
Figure 2.
Human complement-mediated serum bactericidal antibody titer against group B strain H44/76, which expressed fHbp in the variant 1 group that matched the amino acid sequence of the vaccine antigen. The titers and error bars represent the respective mean and range of values measured in two to three independent assays. Pre-immune serum, bar with horizontal hatching; symbols for bars indicating respective post-immunization unadsorbed and adsorbed sera are described in legend to Figure 1.
3.3. Serum bactericidal activity against strains with fHbp v.2
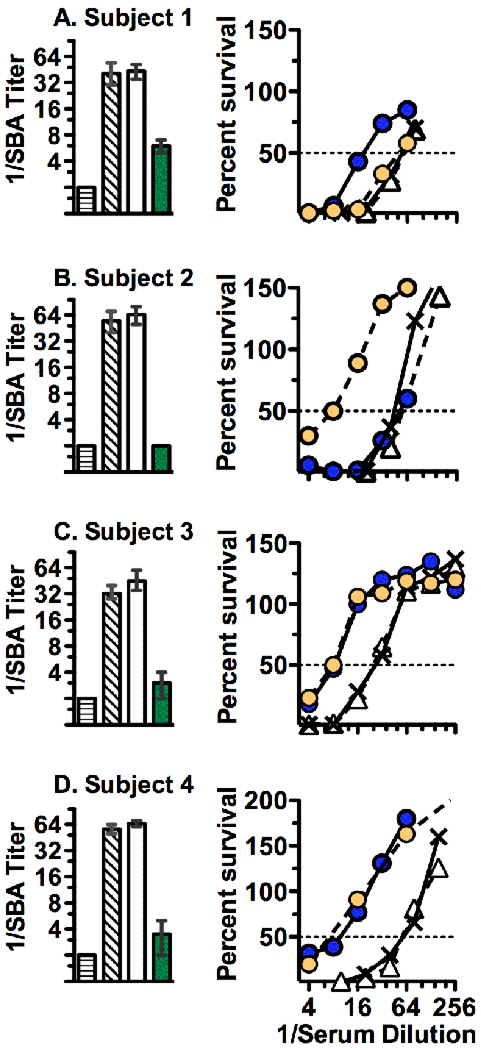
In response to vaccination, four of the six subjects showed 4-fold or greater increases in serum bactericidal titer against strain M4407, which expressed fHbp ID 19 in the variant 2 group and was heterologous to the variant 1 antigen in the vaccine. In these four subjects, the reciprocal GMT increased from <4 in pre-immunization serum to 45 in post-immunization serum samples (p=0.0003). After depletion of antibodies to both fHbp and NHba (Figure 3, left panels), the reciprocal SBA titers decreased from 40 (diagonal-hatched bar) to 6 (green bar) for subject 1 (85% decrease), from 32 to ≤4 for subject 3 (>87% decrease) and from 56 to ≤4 for subjects 2 and 4 (>93% decrease). Non-specific loss of bactericidal activity attributable to the adsorption process was not observed since SBA titers did not decrease in samples that underwent adsorption twice with the rHA-coupled negative control immunoadsorbent (Figure 3, white bars compared with the reciprocal SBA titers of the unadsorbed post-immunization sera represented by the diagonal hatched bars). Collectively the results indicated that the antibodies responsible for SBA were largely directed at fHbp and/or NHba and that there was minimal, if any, functional contribution of antibodies to the GNA2091 or GNA1030 components of the respective fHbp- or NHba- fusion proteins toward SBA against this strain.
Figure 3.
Bactericidal activity against group B strain M4407. Panels A-D show data for subjects 1–4, respectively. Left panels, mean bactericidal titers of unadsorbed pre-immunization sera (horizontal hatched bars) and the respective post-immunization sera: unadsorbed (bars with diagonal hatching), adsorbed two times with the negative control rHA-coupled immunoadsorbent (open bars) and adsorbed with both anti-NHba and anti-fHbp antibodies (green bars). The error bars represent the respective ranges of values measured in two to three independent assays. On some bars, the ranges are not evident because the respective ranges were small. Right panels, percent survival after incubation of the test strain for 60 minutes with human complement and dilutions of post-immunization test sera before (X with solid line) or after depletion of antibodies to fHbp (yellow circles, dashed lines), NHba (blue circles, solid lines), or recombinant human albumin (negative control, open triangles, dashed lines). Data shown are from a representative experiment, which were confirmed in at least two or three independent experiments.
To dissect the individual contributions of the anti-fHbp and anti-NHba antibodies to SBA, we measured the percent survival of strain M4407 when incubated with human complement and different dilutions of unabsorbed and adsorbed post-immunization sera depleted of antibodies to fHbp or NHba (Figure 3, right panels). For subject 1 (Panel A), approximately half of the bactericidal activity was removed after depletion of anti-NHba antibodies (blue circles), while none of the bactericidal activity was removed by depletion of anti-fHbp antibodies (yellow circles). The reverse was observed in the post-immunization serum from subject 2 (Panel B); >80% of the activity was removed after depletion of anti-fHbp antibodies, but no activity was removed after depletion of anti-NHba antibodies. In subjects 3 and 4, depletion of either anti-fHbp or anti-NHba antibodies removed 50% to 75% or more of the SBA (Panels C and D, respectively). Note that depletion of both antibodies in sera from all four subjects removed more bactericidal activity than depletion of either antibody individually. In contrast, there was no loss of activity after adsorption twice with the negative control rHA-couple immunoadsorbent. These data indicated that the loss of SBA after depletion of both antibodies was specific and that there was cooperative bactericidal activity between antibodies to fHbp and NHba for all four subjects. Similar cooperative anti-fHbp and anti-NHba bactericidal activity was found against a second group B strain, S3446, that also expressed a fHbp in the variant 2 group (ID 19), lacked a NadA gene, and was a relatively high expresser of NHba with 98% amino acid sequence identity to the vaccine antigen (Table 1). Against the second strain, SBA in subject 1 was mainly directed against NHba while that of subject 2 was against fHbp (Supplemental Figure 1, Panels A and B, respectively). The respective cooperative contributions of anti-NHba and anti-fHbp antibodies of subjects 3 and 4 toward bactericidal activity against S3446 were similar to that observed against strain M4407 but not as pronounced, in part, because the differences between the respective pre- and post-immunization titers against strain S3446 were smaller than against strain M4407. In subjects 3 and 4, depletion of both anti-fHbp and anti-NHba antibodies decreased SBA titers to 1:4 (Supplemental Figure 2), which was consistent with cooperative activity between both antibody populations.
3.4. Mouse antibodies to fHbp and NHba elicit cooperative complement-mediated bactericidal activity against strain M4407
Based on antibody depletion of sera from immunized humans, both anti-fHbp variant 1 and anti-NHba antibodies exhibited cooperative activity to elicit complement-mediated SBA against the two strains with fHbp v.2. We therefore investigated whether there was similar cooperative bactericidal activity against strain M4407 by anti-fHbp variant 1 and anti-NHba antibodies in sera from immunized mice. For these experiments we had serum pools available from mice immunized in previous studies with recombinant fHbp variant 1 or NHba vaccines [6, 23]. The amino acid sequences of the vaccine antigens matched those of the respective fHbp variant 1 and NHba components of the fusion proteins used to immunize humans.
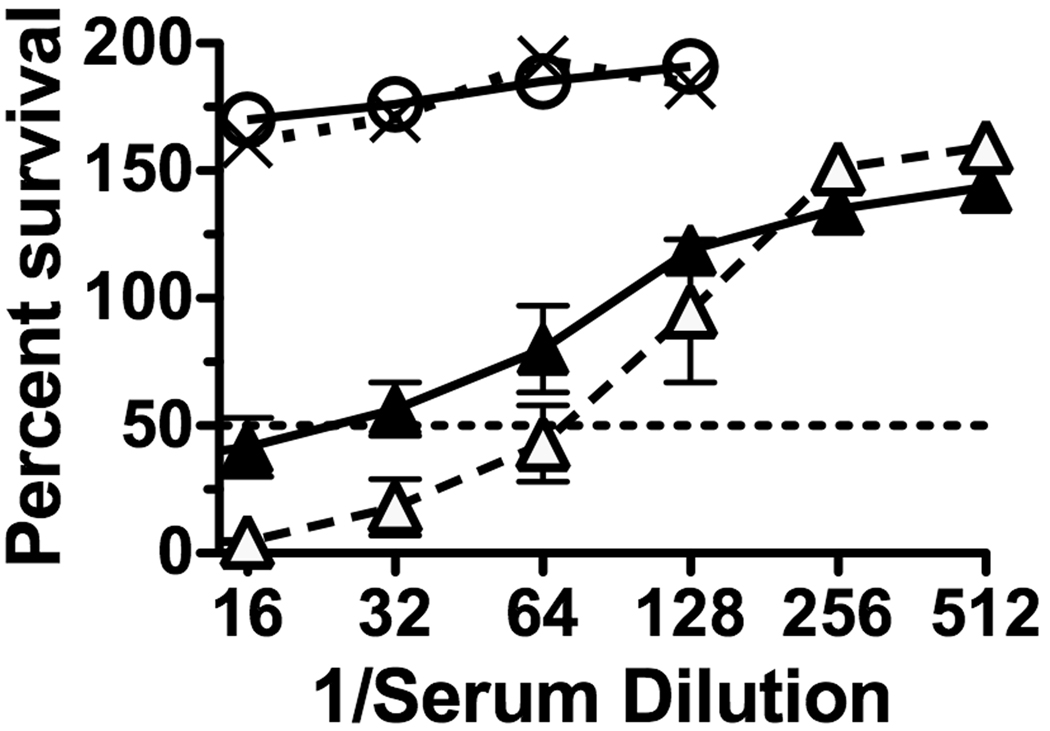
Figure 4 shows the percent survival of strain M4407 when incubated for 1 hour with human complement and different dilutions of 1:1 mixtures of a mouse anti-NHba serum pool and a non-bactericidal mouse anti-fHbp variant 1 antiserum pool (open triangles, dashed line) or, as a negative control, a non-bactericidal serum pool from mice given the aluminum hydroxide adjuvant alone (filled triangles, solid line). The reciprocal SBA titer was ~20 when the anti-NHba antiserum was mixed with the negative control mouse serum pool, which increased to ~70 when the anti-NHba serum pool was mixed with the non-bactericidal anti-fHbp variant 1 antiserum pool.
Figure 4.
Percent survival of group B strain M4407 when incubated for 60 mins with human complement and dilutions of serum pools from immunized mice. Open circles with solid line, serum pool from mice immunized with recombinant fHbp variant 1 vaccine; X’s with dotted lines, serum pool from negative control mice immunized with aluminum hydroxide adjuvant alone; filled triangles with solid line, 1:1 mixture of the serum pool from mice immunized with the recombinant NHba vaccine and the pool from negative control mice immunized with aluminum hydroxide; open triangles with dashed line, 1:1 mixture of the serum pool from mice immunized with recombinant NHba vaccine and the pool from mice immunized with the recombinant fHbp variant 1 vaccine. The error bars represent the range at each dilution for independently diluted serum samples. The respective results were confirmed in a second independent experiment.
4. Discussion
In a previous study, adults immunized with the meningococcal recombinant protein vaccine developed high serum bactericidal antibody responses against group B strain H44/76 [7]. In the present study we confirmed the high bactericidal titers against this strain in all six sera retested. By adsorption, we found that nearly all of the SBA against this strain was directed at fHbp. This result was expected. In immunized mice, three of the antigens in the vaccine, fHbp, NHba and NadA, were responsible for SBA [4]. The H44/76 test strain was a relatively low expresser of NHba [15] and lacked a gene for NadA, but was a high expresser of the fHbp v.1 antigen [24] with an amino acid sequence that matched the fHbp ID 1 vaccine antigen.
To investigate the bactericidal role of anti-NHba antibodies, we selected a second group B strain, M4407, which had been used in a previous study to measure anti-NHba bactericidal activity [15]. This strain expressed a heterologous fHbp in the variant 2 group, was a relatively high expresser of NHba with 100% amino acid identity to the antigen used in the vaccine, and lacked a gene for NadA. Four of the six subjects showed four-fold or greater increases in SBA titer against strain M4407. For three of the four subjects (subjects 1, 3, and 4), depletion of anti-NHba antibodies individually removed the majority of the SBA, which was consistent with susceptibility of this strain to anti-NHba antibody activity. Depletion of anti-fHbp antibodies alone however, removed the majority of the SBA in the fourth serum (subject 2), and also removed the majority of SBA in two of the three sera in which depletion of anti-NHba antibodies had decreased SBA. Similar respective results were observed against a second group B strain, S3446, with fHbp in variant group 2 (supplemental data).
The loss of SBA against strains M4407 or S3446 after depletion of anti-fHbp antibodies elicited by the fHbp v.1 vaccine antigen was unexpected since in previous studies there was minimal cross-reactive SBA of mouse anti-fHbp variant 1 antibodies against strains with fHbp in the variant 2 group [14, 23, 25]. The present results from studies of sera from immunized adults indicated that antibodies elicited by the fHbp v.1 vaccine antigen can be sufficiently cross-reactive with fHbp v.2 to cooperate with antibodies to NHba to elicit human complement-mediated bactericidal activity. Because human antisera to the individual fHbp v.1 and NHba vaccines were unavailable, we tested the ability of mouse antisera produced against the individual antigens to cooperate and augment SBA. The results confirmed the conclusions from the human serum studies: mouse antibodies to fHbp v.1, which individually had no bactericidal activity, augmented human complement-mediated anti-NHba bactericidal activity against strain M4407 (Figure 4).
Our results differed from those recently reported by Giuliani et al. who investigated the antigenic targets of bactericidal activity against strain M4407 in sera from adults immunized with the same three recombinant proteins given together or in combination with an outer membrane vesicle vaccine [15]. They found that inhibition of serum anti-fHbp antibody did not decrease bactericidal titers against strain M4407, whereas inhibition of serum anti-NHba antibodies removed most of the activity. One possible explanation for the discordant data is that we used an immunoadsorbent coupled to fHbp to deplete serum anti-fHbp antibodies whereas Giuliani et al. inhibited anti-fHbp antibody activity by the addition of 500 µg/ml of the GNA2091-fHbp fusion protein to the test sera. The addition of the fusion protein to the bactericidal reaction mixture would be expected to inhibit both anti-fHbp antibodies and bind the human complement inhibitor, factor H. Decreased fH complement inhibition would be expected to increase susceptibility to anti-NHba bactericidal activity. We also used human serum that had been depleted of IgG as a complement source [22] whereas Giuliani et al. used a normal human serum that lacked intrinsic SBA, which was not IgG-depleted. Non-bactericidal IgG antibodies in the human complement source could have cooperated with vaccine-induced anti-NHba antibodies to augment anti-NHba bactericidal activity in the fHbp-inhibited sera, and obviated the need for anti-fHbp cooperative activity.
One limitation of the present study was the small number of immunized adults (N=6). Therefore, it was not possible to predict vaccine efficacy in immunized adults in general or, importantly, in immunized children who may have a different antibody repertoire than adults. The adsorption and SBA results described, however, were robust and reproducible in multiple independent assays of each serum sample. Furthermore, in all four subjects who developed SBA responses to the test strain with fHbp v.2, depletion of both anti-fHbp and anti-NHba antibodies removed more bactericidal activity than depletion of either antibody individually. These data indicated that the two antibody populations can have cooperative activity even when there is a mismatch between the fHbp variant 1 antigen in the vaccine and the fHbp variant 2 in the test strain.
A second limitation of our study was that because of insufficient volumes of sera, we tested bactericidal activity of the adsorbed sera against only two strains with fHbp in the variant 2 group, and both had identical fHbp amino acid sequences (ID 19). Among invasive group B isolates with fHbp in the variant 2 group, ID 19 is the most common, accounting for 8 to 19% of recent group B disease in the United States, France and the United Kingdom [26]. Therefore, while our data are limited to two fHbp variant 2 strains with ID 19, this sequence variant is prevalent among group B isolates. Additional studies are needed to determine whether vaccine-induced anti-fHbp v.1 antibody can elicit cooperative anti-NHba bactericidal activity against other strains with fHbps in the variant 2 or 3 groups.
The recombinant protein vaccine used for immunization of the adults contained five antigens, fHbp, NHba and NadA, which in mice elicited SBA responses, and GNA2091 and GNA1030, which (when tested individually) did not elicit SBA in mice [4]. Although depletion of antibodies to both fHbp and NHba in post-immunization sera removed >85% of the bactericidal activity against strain M4407, low-levels of residual bactericidal activity was observed in three of the adsorbed samples. The residual titers of two of these adsorbed sera (subjects 3 and 4), however, were at the lower range of detection (titers of 1:3 to 1:4), and were not significantly different than the <1:4 titers in the respective pre-immunization sera. Thus, only one subject (subject 1) had definite residual bactericidal activity (titer of 1:6) after adsorption of both anti-NHba and anti-fHbp antibodies. The antigenic target(s) of this activity were not defined. We can exclude NadA since the test strain did not contain the NadA gene; among the other four vaccine antigens it is possible that serum antibodies to GNA2091 and/or GNA1030, either individually or in combination with the low levels of residual antibodies to fHbp or NHba, contributed to the bactericidal activity since depletion of antibodies to fHbp and NHba in the adsorbed sera was not 100% complete.
In conclusion, our results demonstrated that vaccine-induced mouse or human serum antibodies to fHbp variant 1 can cooperate with anti-NHba antibodies to elicit high SBA titers against some strains with fHbp variant 2. The data support the hypothesis that for antibodies directed at these relatively sparsely-distributed antigens, a critical density of antibodies directed at more than one surface antigen can enhance engagement of C1q and augment classical complement pathway killing [8]. The results underscore a potential advantage of using multiple antigens in a meningococcal vaccine that targets sparsely-exposed antigens.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported, in part, by a grant from Novartis Vaccines and Diagnostics, Siena Italy, and by Public Health Service grants R01 AI 046464 and AI 082263 (to D.M.G.) from the National Institute of Allergy and Infectious Diseases, NIH. The work at Children’s Hospital Oakland Research Institute was performed in a facility funded by Research Facilities Improvement Program grant number C06 RR 016226 from the National Center for Research Resources, NIH. N. meningitidis strain M4407, recombinant fHbp and NHba were generous gifts of Silvana Savino, Marzia Giuliani, and Davide Serruto, Novartis Vaccines and Diagnostics, Siena, Italy. Dan M. Granoff holds a paid consultancy from Novartis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
David Vu and Tracy Wong declare no conflicts.
REFERENCES
- 1.Fletcher LD, Bernfield L, Barniak V, Farley JE, Howell A, Knauf M, et al. Vaccine potential of the Neisseria meningitidis 2086 lipoprotein. Infect Immun. 2004;72:2088–2100. doi: 10.1128/IAI.72.4.2088-2100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oster P, O'Hallahan J, Aaberge I, Tilman S, Ypma E, Martin D. Immunogenicity and safety of a strain-specific MenB OMV vaccine delivered to under 5-year olds in New Zealand. Vaccine. 2007;25:3075–3079. doi: 10.1016/j.vaccine.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 3.Serruto D, Spadafina T, Ciucchi L, Lewis LA, Ram S, Tontini M, et al. Neisseria meningitidis GNA2132, a heparin-binding protein that induces protective immunity in humans. Proc Natl Acad Sci U S A. 2010;107:3770–3775. doi: 10.1073/pnas.0915162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giuliani MM, Adu-Bobie J, Comanducci M, Arico B, Savino S, Santini L, et al. A universal vaccine for serogroup B meningococcus. Proc Natl Acad Sci U S A. 2006;103:10834–10839. doi: 10.1073/pnas.0603940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 6.Welsch JA, Moe GR, Rossi R, Adu-Bobie J, Rappuoli R, Granoff DM. Antibody to genome-derived neisserial antigen 2132, a Neisseria meningitidis candidate vaccine, confers protection against bacteremia in the absence of complement-mediated bactericidal activity. J Infect Dis. 2003;188:1730–1740. doi: 10.1086/379375. [DOI] [PubMed] [Google Scholar]
- 7.Plested JS, Welsch JA, Granoff DM. Ex vivo model of meningococcal bacteremia using human blood for measuring vaccine-induced serum passive protective activity. Clin Vaccine Immunol. 2009;16:785–791. doi: 10.1128/CVI.00007-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weynants VE, Feron CM, Goraj KK, Bos MP, Denoel PA, Verlant VG, et al. Additive and synergistic bactericidal activity of antibodies directed against minor outer membrane proteins of Neisseria meningitidis. Infect Immun. 2007;75:5434–5442. doi: 10.1128/IAI.00411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenqvist E, Musacchio A, Aase A, Hoiby EA, Namork E, Kolberg J, et al. Functional activities and epitope specificity of human and murine antibodies against the class 4 outer membrane protein (Rmp) of Neisseria meningitidis. Infect Immun. 1999;67:1267–1276. doi: 10.1128/iai.67.3.1267-1276.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munkley A, Tinsley CR, Virji M, Heckels JE. Blocking of bactericidal killing of Neisseria meningitidis by antibodies directed against class 4 outer membrane protein. Microb Pathog. 1991;11:447–452. doi: 10.1016/0882-4010(91)90041-8. [DOI] [PubMed] [Google Scholar]
- 11.Lucidarme J, Comanducci M, Findlow J, Gray SJ, Kaczmarski EB, Guiver M, et al. Characterization of fHbp, nhba (gna2132), nadA, porA, and sequence type in group B meningococcal case isolates collected in England and Wales during January 2008 and potential coverage of an investigational group B meningococcal vaccine. Clin Vaccine Immunol. 2010;17:919–929. doi: 10.1128/CVI.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lucidarme J, Comanducci M, Findlow J, Gray SJ, Kaczmarski EB, Guiver M, et al. Characterization of fHbp, nhba (gna2132), nadA, porA, sequence type (ST), and genomic presence of IS1301 in group B meningococcal ST269 clonal complex isolates from England and Wales. J Clin Microbiol. 2009;47:3577–3585. doi: 10.1128/JCM.00936-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bambini S, Muzzi A, Olcen P, Rappuoli R, Pizza M, Comanducci M. Distribution and genetic variability of three vaccine components in a panel of strains representative of the diversity of serogroup B meningococcus. Vaccine. 2009;27:2794–2803. doi: 10.1016/j.vaccine.2009.02.098. [DOI] [PubMed] [Google Scholar]
- 14.Beernink PT, Welsch JA, Harrison LH, Leipus A, Kaplan SL, Granoff DM. Prevalence of factor H-binding protein variants and NadA among meningococcal group B isolates from the United States: implications for the development of a multicomponent group B vaccine. J Infect Dis. 2007;195:1472–1479. doi: 10.1086/514821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giuliani MM, Biolchi A, Serruto D, Ferlicca F, Vienken K, Oster P, et al. Measuring antigen-specific bactericidal responses to a multicomponent vaccine against serogroup B meningococcus. Vaccine. 2010;28:5023–5030. doi: 10.1016/j.vaccine.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 16.Frasch CE, Mocca LF. Strains of Neisseria meningitidis isolated from patients and their close contacts. Infect Immun. 1982;37:155–159. doi: 10.1128/iai.37.1.155-159.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pajon R, Beernink PT, Harrison LH, Granoff DM. Frequency of factor H-binding protein modular groups and susceptibility to cross-reactive bactericidal activity in invasive meningococcal isolates. Vaccine. 2010;28:2122–2129. doi: 10.1016/j.vaccine.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beernink PT, Welsch JA, Bar-Lev M, Koeberling O, Comanducci M, Granoff DM. Fine antigenic specificity and cooperative bactericidal activity of monoclonal antibodies directed at the meningococcal vaccine candidate factor h-binding protein. Infect Immun. 2008;76:4232–4240. doi: 10.1128/IAI.00367-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beernink PT, Shaughnessy J, Ram S, Granoff DM. Impaired immunogenicity of a meningococcal factor H-binding protein vaccine engineered to eliminate factor H binding. Clin Vaccine Immunol. 2010;17:1074–1078. doi: 10.1128/CVI.00103-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider MC, Prosser BE, Caesar JJ, Kugelberg E, Li S, Zhang Q, et al. Neisseria meningitidis recruits factor H using protein mimicry of host carbohydrates. Nature. 2009;458:890–893. doi: 10.1038/nature07769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koeberling O, Giuntini S, Seubert A, Granoff DM. Meningococcal outer membrane vesicle vaccines derived from mutant strains engineered to express factor H binding proteins from antigenic variant groups 1 and 2. Clin Vaccine Immunol. 2009;16:156–162. doi: 10.1128/CVI.00403-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beernink PT, Caugant DA, Welsch JA, Koeberling O, Granoff DM. Meningococcal factor H-binding protein variants expressed by epidemic capsular group A, W-135, and X strains from Africa. J Infect Dis. 2009;199:1360–1368. doi: 10.1086/597806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beernink PT, Granoff DM. Bactericidal antibody responses induced by meningococcal recombinant chimeric factor H-binding protein vaccines. Infect Immun. 2008;76:2568–2575. doi: 10.1128/IAI.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welsch JA, Ram S, Koeberling O, Granoff DM. Complement-dependent synergistic bactericidal activity of antibodies against factor H-binding protein, a sparsely distributed meningococcal vaccine antigen. J Infect Dis. 2008;197:1053–1061. doi: 10.1086/528994. [DOI] [PubMed] [Google Scholar]
- 25.Masignani V, Comanducci M, Giuliani MM, Bambini S, Adu-Bobie J, Arico B, et al. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J Exp Med. 2003;197:789–799. doi: 10.1084/jem.20021911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy E, Andrew L, Lee KL, Dilts DA, Nunez L, Fink PS, et al. Sequence diversity of the factor H binding protein vaccine candidate in epidemiologically relevant strains of serogroup B Neisseria meningitidis. J Infect Dis. 2009;200:379–389. doi: 10.1086/600141. [DOI] [PubMed] [Google Scholar]
- 27.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.