Abstract
Small heterodimer partner (SHP, NR0B2) is a unique member of the nuclear receptor (NR) superfamily that contains the dimerization and ligand-binding domain found in other family members, but lacks the conserved DNA binding domain. The ability of SHP to bind directly to multiple NRs is crucial for its physiological function as a transcriptional inhibitor of gene expression. A wide variety of interacting partners for SHP have been identified, indicating the potential for SHP to regulate an array of genes in different biological pathways. In this review, we summarize studies concerning the structure and target genes of SHP and discuss recent progress in understanding the function of SHP in bile acid, cholesterol, triglyceride, glucose, and drug metabolism. In addition, we review the regulatory role of SHP in microRNA (miRNA) regulation, liver fibrosis and cancer progression. The fact that SHP controls a complex set of genes in multiple metabolic pathways suggests the intriguing possibility of developing new therapeutics for metabolic diseases, including fatty liver, dyslipidemia and obesity, by regulating SHP with small molecules. To achieve this goal, more progress regarding SHP ligands and protein structure will be required. Besides its metabolic regulatory function, studies by us and other groups provide strong evidence that SHP plays a critical role in the development of cancer, particularly liver and breast cancer. An increased understanding of the fundamental mechanisms by which SHP regulates the development of cancers will be critical in applying knowledge of SHP in diagnostic, therapeutic or preventive strategies for specific cancers.
Keywords: nuclear receptors, small heterodimer partner, metabolic regulation, metabolic syndrome, cancer
1. Introduction
Nuclear receptors (NRs) are a unique family of transcription factors (TFs) that exert critical roles in almost all aspects of mammalian development, metabolism and physiology [1]. Many NRs directly activate or repress their target genes by binding to the hormone response elements (HREs) in promoters or enhancer regions via DNA-binding domains (DBDs). In addition, NRs can bind specific activating molecules through ligand-binding domains (LBDs), interact with other coactivators and corepressors to mediate transcriptional regulation. Dysfunction of nuclear receptor signaling leads to a wide spectra of proliferative, reproductive, and metabolic diseases, including obesity, diabetes, and cancers.
Small heterodimer partner (SHP, NR0B2) is a unique NR which is distinct from conventional NRs in both structure and function [2]. Due to its lack of identified endogenous ligands, SHP belongs to the “orphan” subfamily. SHP executes its regulatory function through protein-protein interactions with other NRs and TFs. In this review, we have attempted to survey all currently available published literatures concerning the structure and target genes of SHP, and discuss recent progress in understanding the function of SHP in regulating bile acid synthesis, cholesterol, lipid and glucose metabolism, as well as its role in microRNA regulation and cancer development.
2. SHP gene structure and genetic variations in humans
SHP was originally cloned in 1996, based on its interaction in yeast two-hybrid assays with several conventional and orphan members of the receptor superfamily, including the constitutive androstane receptor, retinoid receptors, thyroid hormone receptor, and orphan receptor MB67 [2]. The genomic structure of SHP consists of two exons interrupted by a single intron spanning approximately 1.8 kilobases in humans and 1.2 kilobases in mouse. Genomic Southern blot analysis and fluorescence in situ hybridization of human metaphase chromosomes indicated that the SHP gene is located at human chromosome 1p36.1 [3]. Mouse and rat SHP reside in chromosome 4 and 5, respectively.
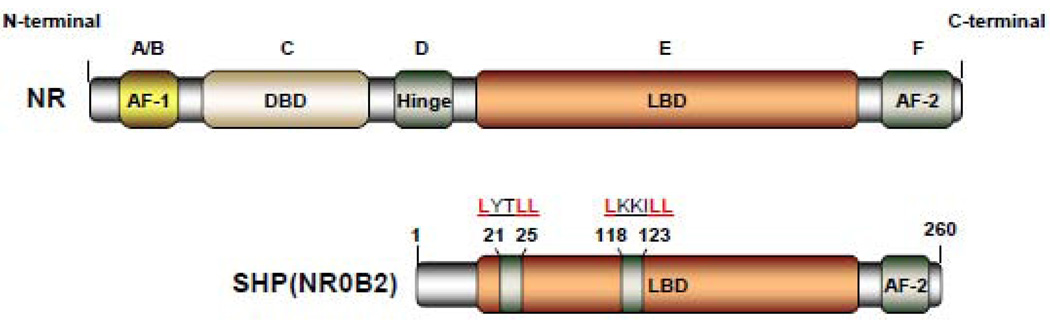
SHP is an orphan member of the nuclear hormone receptor superfamily that contains the dimerization and ligand-binding domain (LBD) found in other family members, but lacks the conserved DNA binding domain (DBD) [2]. The ability of SHP to bind directly to a variety of NRs is crucial for its physiological function as a transcriptional inhibitor of gene expression. SHP binds to the AF-2 domain (the C-terminal transcription activation domain located within the LBD of ligand-regulated and constitutive active NRs) through two functional LXXLL-related motifs (also called NR-boxes) which are located in the putative N-terminal helix 1 of the LBD and in the C-terminal region of helix 5 (Fig. 1) [4].
Figure 1. Domain structure of the orphan nuclear receptor SHP.
Classical nuclear receptor (NR) contains five major functional domains: the N-terminal ligand-independent transactivation domain (A/B domain), the DNA binding domain (DBD or C domain), hinge region (D domain), the C-terminal ligand-binding domain (LBD or E domain), and the ligand-dependent transactivation domain (AF2 or F domain). Compared to the classical NRs, SHP contains the dimerization and LBD domain, but lacks the conserved DBD. SHP represses the transcriptional activities of its targets gene by utilizing two functional LXXLL-related motifs (also called NR-boxes) which are located in the putative N-terminal helix 1 of the LBD and in the C-terminal region of helix 5.
SHP gene is expressed and detected in a variety of tissues. Expression profiling of the 49 known NR mRNAs in 39 tissues from two strains of mice, 12931/SvJ and C57/BL6, showed that SHP is predominantly expressed in the gallbladder and liver, and at lower levels in the brainstem, cerebellum, adrenal, pancreas, stomach, duodenum, jejunum, ileum, colon, kidney, ovary, testis, and heart [5]. In humans, SHP mRNA is detected in the liver, heart, pancreas, kidney, spleen, small intestine, adrenal gland and stomach [3, 6, 7]. Genetic variations including deletions, insertions, repetitive elements, single nucleotide polymorphisms (SNP), and large chromosomal rearrangements can cause diseases by affecting important biological processes. To date, the published data demonstrate numerous mutations in the human SHP gene with a variety of physiological consequences (Table 1). Loss of function mutations in the SHP gene were first reported in Japanese subjects with obesity and diabetes [8]. These mutations included two frameshift mutations, H53fsdel10 (deletion of 10 bases starting at codon 53 for His) and L98fsdel9insAC (deletion of 9 bases and insertion of a dinucleotide AC at codon 98 for Leu), one nonsense mutation R34X (amino acid replacement of Arg codon 34 by terminator), and three missense mutations, R213C, R216H, and A195S (replacements of Arg codons 213 and 216 by Cys and His codons, respectively, and Ala codon 195 by Ser). The H53fsdel10, L98fsdel9insAC, and R34X mutations, found only in subjects with mildly or moderately obesity at onset of diabetes, were truncations that would completely inactivate the putative ligand-binding function and also delete C-terminal sequences associated with transcriptional repression. However, the R216H allele was present in 2 out of 190 control subjects, suggesting that it may be a polymorphism that is not pathogenic. These findings suggest that genetic variations in the SHP gene may contribute to increased body weight and mutations in the SHP gene are likely associated with insulin resistance and mild obesity. In addition to playing a possible role in obesity, SHP may also act as a candidate gene for human lipodystrophy syndromes. In order to investigate possible disease mutations of SHP in association with human lipodystrophy syndromes, another investigator sequenced SHP in 15 subjects with lipodystrophy syndromes who had no mutations in known lipodystrophy genes and 74 clinically normal Caucasian subjects. This led to the identification of four polymorphisms: SNP [−394] C>T in the promoter, a micro-deletion polymorphism [−195] delCTGA in the promoter, a missense SNP 541 G>C in exon 1 (which changed the amino acid sequence G171A), and SNP 903C>T in exon 2 [9]. However, no rare SHP coding sequence variants were found exclusive to patients with lipodystrophy, suggesting that SHP mutations are not commonly seen in patients with lipodystrophy who have no mutations in known disease genes. In 2003, two novel missense mutations, R34G and R36C, and two common polymorphisms, G171A and - 195CTGAdel were identified when the coding regions and 562 bases of the SHP promoter were sequenced in 329 subjects with severe early-onset obesity in the U.K. [10]. The 171A allele was associated with a higher BMI, waist circumference, and children carrying the G171A variant had higher 30-min insulin responses to a glucose load. The results suggest that genetic variation in the SHP locus may influence birth weight and have effects on BMI, possibly through effects on insulin secretion. However, another study investigated SHP in 1927 U.K. subjects, examining relationships with type 2 diabetes, obesity, and birth weight, and found that genetic variation in SHP was unlikely to be common in the predisposition to diabetes, obesity, or increased birth weight in U.K. Caucasians [11]. Moreover, another five novel SHP variants, including three missense variants (c.100C>G [p.R34G], c.278G>A [p.G93D], and c.415C>A [p.P139H]) and two silent variants (c.65C>T [p.Y22Y] and c.339G>A [p.P113P]) were identified in 2004 when the entire coding region of SHP was analyzed in a cohort of 750 Danish men with early-onset obesity [12]. The 34G, 93D, and 139H alleles were rare variants, which were found only among obese subjects. Taken together, these results suggest that further large-scale population studies are necessary to assess the clinical impact of these rare variants on obesity risk among European subjects. Because mutations in SHP may be associated with insulin resistance due to both later obesity and fatty liver in Japanese subjects, the frequencies of SHP mutations in 805 Japanese patients with adult-onset type 2 diabetes (T2DM), 752 non-diabetic controls, and 93 patients with non-alcoholic steatohepatitis (NASH) were examined to evaluate the influence of SHP mutations on the risk of later development of type 2 diabetes in 2008 [13]. A total of 15 different mutations in 44 subjects, including 6 novel mutations (c.160C>T [p.Arg54Cys], c.314T>G [p.Val105Gly], c.618G>A [p.Trp206X], c.112C>T [p.Arg38Cys], c.134G>C [p.Arg45Pro], c.532G>A [p.Asp178Asn]) were identified in this study. Mutations with reduced activity were found in 2.4% of the diabetic group and in 0.8% of the control group, suggesting that SHP mutations associated with mild obesity during childhood increased the susceptibility to type 2 diabetes later in life for Japanese subjects. Recently, we identified two novel missense SHP mutations (p.R38H, p.K170N) in both normal and CADASIL-like (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy) patients and found that the K170 residue played a critical role in controlling SHP ubiquitination and acetylation which are associated with the protein stability and repressive function of SHP [14].
Table 1.
Summary of the mutations identified in the human SHP gene (NR0B2).
Mutation nomenclature was numbered based on GenBank cDNA NM_021969.2 and protein NM_068804.1 sequences.
In open reading frame (ORF) of the coding region, nucleotide +1 corresponds to the A of the ATG start codon.
In intron region, nucleotide +1 corresponds to the 1st nucleotide of intron.
In 3’UTR region, nucleotide +1 corresponds to the 1st nucleotide downstream of the stop codon.
In 5’UTR and promoter region, nucleotide −1 corresponds to the 1st nucleotide upstream of the start codon.
early-onset obesity,
high birth weight,
diabetes,
fatty liver,
decreased insulin sensitivity (Nishigori et al., 2001)
3. Inducers of SHP gene expression
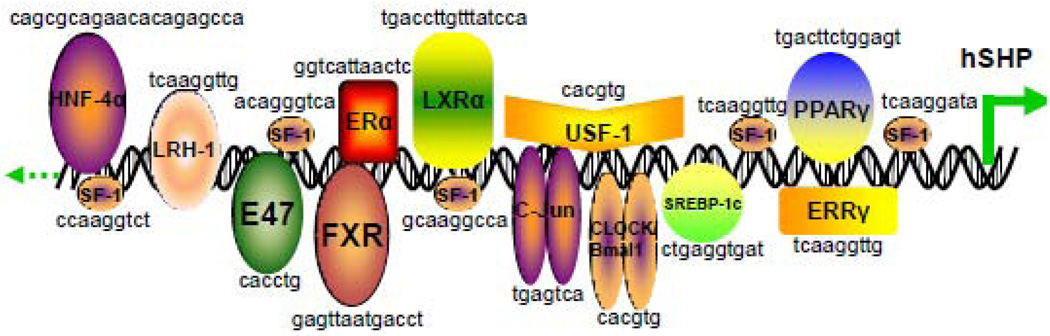
Many NRs and TFs have been reported to target the SHP promoter and regulate SHP gene expression, including steroidogenic factor-1 (SF-1), liver receptor homologue-1 (LRH-1), farsenoid X receptor (FXR), c-jun, hepatocyte nuclear factor 4α (HNF4α), estrogen receptor-related receptor γ (ERRγ), E2A gene products (E47, E12 and E2/5), liver X receptor α (LXRα), estrogen receptor α (ERα), sterol regulatory element binding protein 1c (SREBP-1c), adaptor protein (AP1), pregnane X receptor (PXR), the core circadian component CLOCK-BMAL1, peroxisome proliferator-activated receptor γ (PPARγ), and upstream stimulatory factor-1 (USF-1) (Fig. 2). One of the earliest studies showed that SF-1 and LRH-1, its close relative in liver, potently transactivated the SHP promoter. At least five SF-1 binding sites were identified by DNA footprinting studies of the SHP promoter, and mutagenesis studies demonstrated that each of the three strongest binding sites was required for SF-1 transactivation [15]. Since then, many NRs have been identified to activate the SHP promoter. Two studies showed that binding of bile acids to FXR led to the transcriptional activation of SHP. Elevated SHP protein then inactivated LRH-1 by forming a heterodimeric complex leading to promoter-specific repression of SHP, thereby establishing an elaborate autoregulatory negative feedback loop for SHP and a cascade mediated by nuclear receptors for the maintenance of hepatic cholesterol catabolism [16, 17]. In an attempt to define the molecular mechanism underlying the insulin secretion defect in HNF-1α−/− mice, Shih et al. measured the expression of 50 genes essential for normal beta-cell functions. They found that both the expression of SHP and HNF4α were reduced in adults islets of HNF-1α−/− mice and a putative HNF4α consensus binding sequence was identified in the human and murine SHP promoter [18, 19]. Gupta et al. reported that bile acid-activated JNK pathway played a pivotal role in regulating CYP7A1 levels in primary rat hepatocytes. Overexpression of the wild type c-Jun resulted in increased SHP promoter activity and this effect was suppressed by mutation of a putative AP-1 (c-Jun) element in the SHP promoter [20]. Sanyal et al. reported that the SHP promoter was activated by ERRγ but not its related ERRα and ERRβ isoforms [6]. Kim et al. found that the basic helix-loop-helix (bHLH) transcription factors, namely the E2A proteins (E47, E12 and E2/5), differentially regulated the human and mouse SHP promoters and cooperated with SF-1 for transcriptional activation of the human SHP promoter [21].
Figure 2. Schematic overview of nuclear receptors (NRs) and transcription receptors (TFs) regulating the promoter of the human SHP gene.
Location of the binding sites of NRs and TFs and the consensus sequences are indicated in the human SHP gene promoter. NRs that target SHP promoter include hepatocyte nuclear factor 4a (HNF4α), steroidogenic factor-1 (SF-1), liver receptor homolog-1 (LRH-1), farnesoid X receptor (FXR), estrogen receptor a (ERα), liver X receptor α (LXRα), upstream stimulatory factor-1 (USF-1), estrogen receptor related receptor γ (ERRγ), and peroxisome proliferator-activated receptor γ (PPARγ). TFs that induce SHP expression include E47, c-Jun, sterol regulatory element binding protein -1c (SREBP-1c) and circadian locomotor output cycles kaput (CLOCK)/brain and muscle aryl hydrocarbon receptor nuclear translocator-like 1(Bmal1).
One interesting study revealed a fundamental difference in the regulation of CYP7A1 in rodent and human hepatocytes and demonstrated that SHP was regulated directly by LXRα through a DNA response element that overlapped with the bile acid response element. This study also provided evidence that different species employed distinct molecular strategies to regulate cholesterol homeostasis [22]. Lai et al. showed that estrogens directly induced the expression of SHP in mouse and rat liver and in human HepG2 cells [23]. SHP promoter contained one ERα binding site which overlapped with the known FXR binding site, and the combination of ethynylestradiol plus FXR agonists did not produce an additive induction of SHP expression in mice. One study reported differential regulation of the human and mouse SHP promoters by SREBP-1, which regulates the expression of many genes involved in cholesterol and fatty acid synthesis [24]. Human SHP promoter activation by SREBP-1 was mostly mediated by SRE1 (−186 to −195 bp) in the promoter region, which was not conserved with the mouse SHP promoter. In addition, potential PXR response elements (PXREs) were identified in the SHP promoter, and PXR formed a heterodimer with RXR to activate SHP in HepG2 human hepatoma cells [25]. In 2007, a circadian expression pattern of SHP mRNA was found in mouse liver, which was coordinately regulated by a core circadian clock component CLOCK-BMAL1 and LRH-1 [26]. CLOCK-BMAL1 bound to the E-box (CACGTG) which resulted in stimulation of SHP promoter activity. Furthermore, CLOCK-BMAL1 synergistically enhanced LRH-1-mediated SHP promoter transactivation. The SHP promoter also contains a functional PPAR response element (PPRE), and the PPARγ ligand rosiglitazone activated the binding of PPARγ/RXRα heterodimer to the PPRE and increased SHP expression in primary rat hepatocytes [27]. More recently, HGF and its family member, macrophage-stimulating factor (MSP), were shown to activate the SHP gene promoter and induce SHP mRNA and protein levels. The effect of HGF and MSP on SHP gene expression was demonstrated to be mediated via activation of the AMP-activated protein kinase (AMPK) signaling pathway [28]. Further studies showed that USF-1 bound to E-box-1 in the SHP promoter, and HGF increased USF-1 DNA binding on the SHP promoter via AMPK and DNA-dependent protein kinase-mediated pathways.
To date, many studies have shown that the FXR ligand GW4064 [16, 17], androsterone [29], bile acids (BA) and chenodeoxycholic acid (CDCA) [30, 31] were potent inducers of SHP gene expression. Interestingly, the plant sterol guggulsterone (GS) acted as an antagonist for the farnesoid X receptor (FXR) and decreased expression of bile acid-activated genes in HepG2 cells. However, guggulipid treatment in Fisher rats resulted in significant SHP expression activation, which identified guggulsterone as a novel class of FXR ligands characterized by antagonist activity in coactivator association assays but with the ability to enhance the action of agonists in vivo [32]. The PPARγ coactivator-1α (PGC-1α) has been reported to mediate ligand-dependent activation of FXR and transcription of the SHP gene [33]. In addition, histone methylation, similar to acetylation, was reported to regulate transcriptional activation of SHP. The synthetic BA, 6-Ethyl CDCA (6-ECDCA) was bound to FXR and activated FXR to interact with Protein Arginine Methyl-Transferase type I (PRMT1), which induced recruitment of the H4 methylation and up-regulated SHP mRNA [34]. Whitby et al. demonstrated that the small molecule GSK8470 acted as a high-affinity ligand for LRH-1 and SF-1 by using fluorescence resonance energy transfer (FRET) based biochemical assays, and GSK8470 increased SHP expression in liver cells [35].
SHP plays important roles in negatively regulating the conversion of cholesterol to bile acids and in regulating the expression of genes with roles in bile acid transport, lipid metabolism, and gluconeogenesis. In addition, SHP induces apoptosis in liver and cancer cells. Thus, identifying SHP ligands may lead to new treatments for cancer, obesity, and non-alcohol-related fatty liver diseases. In 2007, 6-[3-(1-adamantyl)-4-hydroxyphenyl]-2-naphthalenecarboxylic acid (CD437/AHPN) and 4-[3-(1-adamantyl)-4-hydroxyphenyl]-3-chlorocinnamic acid (3-Cl-AHPC) were identified as exogenous SHP ligands and their induction of apoptosis was reported to be mediated by their binding to the SHP protein [36]. Further studies demonstrated that AHPN/3-Cl-AHPC and their analogues bound specifically to SHP, and these binding events promoted the interaction of SHP with a corepressor complex containing the repressor mSin3A, NCoR, histone deacetylase 4, and heat shock protein 90 [37]. Loss of SHP or Sin3A expression, while blocking 3-Cl-AHPC-mediated apoptosis, had little effect on 3-Cl-AHPC inhibition of cellular proliferation. Furthermore, 3-Cl-AHPC induction of c-Fos and c-Jun expression as well as NF-kappaB activation were dependent on SHP protein levels [38]. Induction of apoptosis by SHP required the presence of the terminal CO2H group on the AHPN or AHPC scaffold, and optimal apoptotic activity also required both the 3’-(1-Ad) and 4’-OH groups [39]. Recent studies have also shown that AHPN and 3-Cl-AHPC induced SHP mRNA expression in Huh7 human liver cancer cells via increasing LRH-1 protein stability and enhancing the recruitment of LRH-1 to the SHP promoter [40]. We also found that in vivo administration of AHPN and 3-Cl-AHPC to the mice increased SHP expression in liver.
4. SHP in transcriptional regulation
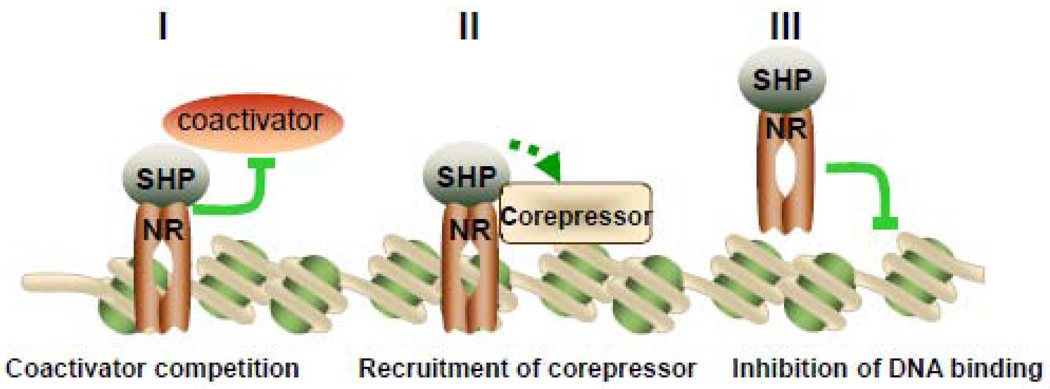
SHP predominantly functions as a transcriptional repressor of gene expression by directly binding to a variety of NRs, including LRH-1, HNF4, ERRs, LXRs, PPARs, GR, ERs, TRβ, RARα, FXR, PXR, CAR, AR, NGFI-B (Nur77) and RXRs which are the common heterodimerization partner for many NRs (Fig. 3). To date, at least three distinct repression mechanisms appear likely to explain the inhibitory function of SHP on the transcription of NR target genes. The NR-binding mode of SHP implies that interfering with the AF-2 domain of NRs through two functional LXXLL-related motifs, resulting in direct competition with coactivator binding, is associated with the inhibitory mechanism of SHP. This mode of inhibition appears pronounced in the case of SHP inhibition on ERs, RXR, LRH-1, HNF4, AR, LXRs, ERRs, GRs and Nur77 induced transcription [4, 6, 17, 41–46]. Two possibilities have been considered in this respect: 1) SHP and AF-1/2 coactivators may compete for a common site; or 2) binding of SHP to the receptor may induce conformational changes leading to the dissociation of AF-1/2 coactivators from the receptors. Interestingly, the competition with coactivator by SHP is not limited to NR targets. A few other TFs have been reported to interact with SHP. One example is that SHP acted as a corepressor for basic helix-loop-helix transcription factor BETA2/NeuroD by competing with coactivator p300 for binding to BETA2/NeuroD [47]. Another example is that SHP interacted with Foxo1 and led to the repression of Foxo1-mediated G6Pase transcription by competing with a coactivator cAMP response element-binding protein [48]. In addition, SHP mediates inhibition of transforming growth factor-beta (TGF-β)-induced gene expression via a direct repression of Smad3 transactivation by competing with its coactivator p300 [49].
Figure 3. Model of three distinct transcriptional repression mechanisms for SHP.
SHP represses nuclear receptor (NR) or transcription factor (TF) mediated transactivation by competition for coactivator binding to NR (I), recruitment of SHP-associated corepressors (II), and inhibition of DNA binding (III). SHP can utilize these three inhibitory steps alternatively or sequentially in a cell type and target gene specific manner.
Although coactivator competition on the AF-2 domain might appear to be important for inhibition, SHP has been suspected to act as a direct transcriptional repressor by recruiting conventional corepressors as SHP contains a strong transcriptional repression domain at its C terminus. The requirement of both these two modes has been found in SHP inhibition on many NRs such as HNF4, ERs and LRH-1 [4, 42, 50]. SHP might utilize these two distinct inhibitory steps in a cell type and target gene dependent manner. The mouse E1A-like inhibitor of differentiation 1 (EID1) acting as a candidate coinhibitor for SHP has been isolated in screens for SHP corepressors [51]. Histone acetyltransferases and histones might act as targets for EID1 action and SHP inhibition of transcription might involve EID1 antagonism of CBP/p300-dependent coactivator functions. Site-specific modification of nucleosomal histones plays a central role in the formation of transcriptionally active and inactive chromatin structures. Components of histone deacetylases (HDACs)/corepressor complexes are other important coinhibitors for SHP. For example, one study showed that mSin3A and a Swi/Snf complex containing Brm as a central ATPase were recruited to the CYP7A1 promoter and SHP was associated with the mSin3A-Swi/Snf complex by direct interaction with Brm and mSin3A through its repression domain to inhibit CYP7A1 gene expression [52]. SHP has also been shown to be associated with unmodified and lysine 9-methylated histone-3 and to functionally interact with HDAC1 and the G9a methyltransferase, which led to histone deacetylation, followed by H3-K9 methylation and stable association of SHP itself with chromatin [53]. Another study showed G9a was recruited to and H3K9 was methylated at the CYP7A1 promoter in a SHP-dependent manner. This study established a critical role for G9a methyltransferase, histone deacetylases, and the Swi/Snf-Brm complex in the SHP-mediated inhibition of hepatic bile acid synthesis via coordinated chromatin modification at target genes [54]. An recent interesting study showed SHP recruited SIRT1, a class III histone deacetylase on LRH1 target gene promoters and SIRT1 deacetylated template-dependent histone H3 and H4 to inhibit transcription of LRH1 target genes [55]. The finding that an HDAC inhibitor affected intrinsic SHP repressive activity provided further evidence for the involvement of HDACs in SHP mediated repression of transcription [56].
Moreover, SHP binding to some NRs resulting in the dissociation of the SHP-NRs complex from certain promoters is the third possible mode of repression by SHP. For example, SHP has been shown to inhibit DNA binding and transcriptional activation by repressing RAR-RXR heterodimers, RAR-PXR heterodimers, agonist dependent ERα dimerization and HNF4α homodimerization [2, 57–59]. Interestingly, SHP also interacts with some TFs and inhibits DNA binding and transactivation. One example is the SHP mediated suppression of agonist-activated arylhydrocarbon receptor (AHR)/AHR nuclear translocator (ARNT) activity by inhibiting AHR/ARNT-DNA binding [60]. HNF-3, JunD and C/EBPα were also reported to be repressed by SHP via inhibition of DNA binding [61–63].
Most studies report that SHP acts as a repressor of gene transcription. In contrast, SHP has been found to activate the nuclear factor-kappa B (NF-κB) in resting macrophage cells treated with oxidized low density lipoprotein (oxLDL) [64]. Moreover, SHP was also reported to up-regulate the transcriptional activity of PPAR by directly binding the DBD/hinge region of PPAR [7]. The authors suggested that the following mechanisms might be involved: 1) SHP might directly inhibit the binding of the PPARγ-RXRα complex to PPRE; 2) SHP might compete with the corepressors for binding to PPARγ; or 3) SHP may possess its own activating function at its C terminus.
5. SHP in microRNA regulation
MicroRNAs (miRNAs, miRs) are recently discovered small 21–23 nts noncoding RNAs that regulate gene expression primarily by translational repression of target mRNAs after binding their 3’-untranslated regions (3’-UTRs) by partial base pairing [65]. They are generally produced from much larger Pol II RNA transcripts [66–69]. MiRNAs participate in the regulation of many cellular processes and changes of their expression are frequently observed in human diseases including cancers [70–72]. Despite intensive studies on their physiological and pathological functions [73], the molecular mechanisms of how miRNA gene transcription is regulated by TFs and NRs remain largely unknown. Recent studies demonstrate that SHP plays an important role in regulating the transcription of several miRNAs. We first showed that the 3'-coding region of pri-miR-433 served as the promoter region of pri-miR-127 [66, 74], and SHP inhibited ERRγ transactivation on the miR-433 and miR-127 promoters, resulted in repression of these two miRNAs [74]. This study revealed a novel mechanism by which the coupled miR-433 and miR-127 genes were regulated by nuclear receptors in a compact genomic space. Further multiple sequence alignments showed that the precursors of miR-433 and of miR-127 exhibited 95% and 100% similarity, respectively, in human, chimpanzee, horse, dog, monkey, rat, cow, and mouse, indicating that the miR-433/127 loci might have evolved from a common gene of origin [75]. Another study from our lab revealed a novel cascade "dual inhibitory" mechanism governing miR-206 gene transcription by SHP: SHP inhibition of ERRγ led to decreased YY1 expression and the de-repression of YY1 on AP (c-Jun and c-Fos) activity, ultimately leading to the activation of miR-206, which elucidates a cascade regulatory mechanism governing miRNAs gene transcription by NRs [67]. A more recent study showed that FXR induced expression of SHP, which in turn resulted in repression of p53 occupancy at the miR-34a gene promoter, leading to the positive regulation of the NAD-dependent deacetylase Sirtuin 1 (SIRT1) in the liver [76]. This study demonstrated a role of the FXR/SHP pathway in controlling SIRT1 levels via miR-34a inhibition and that elevated miR-34a levels in obese mice contribute to decreased SIRT1 levels. The authors proposed that manipulation of this regulatory network might be useful for treating metabolic disease and cancer.
5. SHP in metabolism
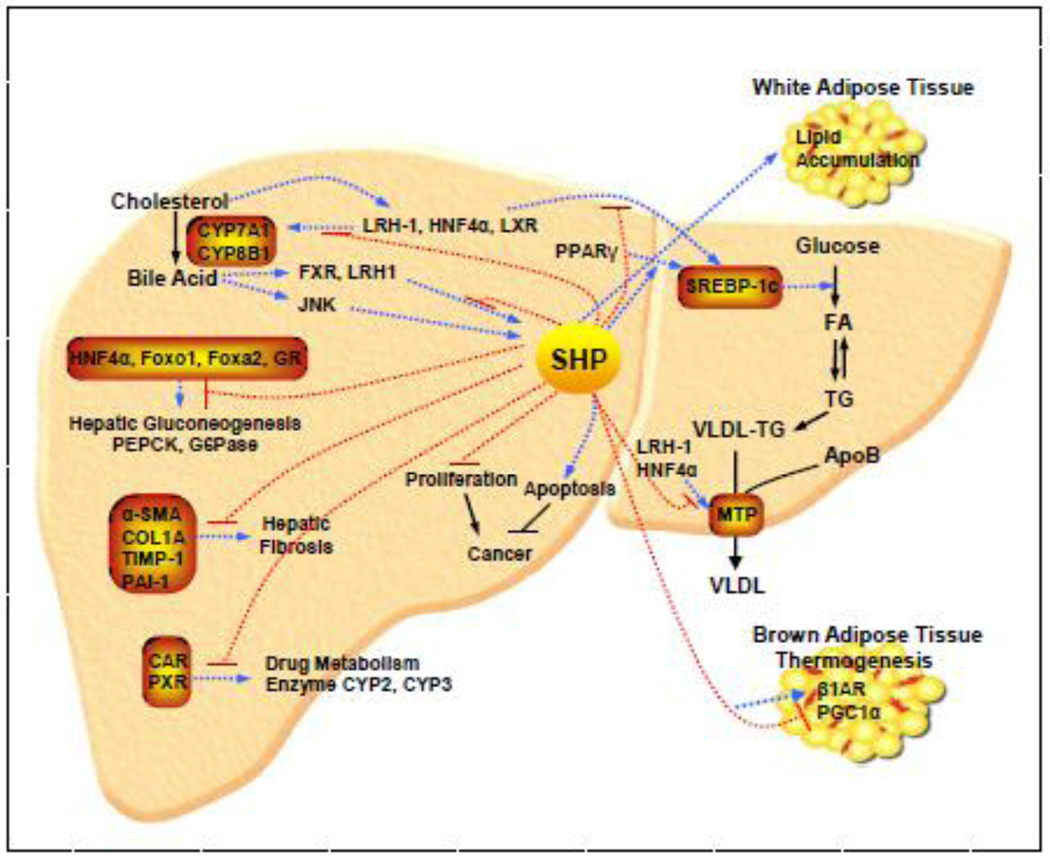
Numerous independent in vitro studies have identified a number of interaction partners for SHP, indicating the potential for SHP to regulate a wide array of genes in various biological pathways (Fig. 4). In line with this, an in vivo study performing global gene expression profiling combined with chromatin immunoprecipitation assays in transgenic mice constitutively expressing SHP in the liver demonstrates that SHP is a pleiotropic regulator, influencing multiple target genes involved in diverse biological processes in a direct or indirect manner, including regulation of metabolic pathways, stress and inflammatory response, detoxification and cell cycle control [77].
Figure 4. Integrated view of SHP as a central transcriptional coregulator in diverse physiological pathways in the liver.
Activation pathways are shown as broken blue arrows and inhibitory pathways are shown as broken red lines. FA, fatty acid; TG, triglyceride; VLDL, very low density lipoprotein; CYP7A1, cytochrome P450 enzyme cholesterol 7α-hydroxylase; G6P, glucose-6-phosphatase; HNF-4α, hepatocyte nuclear factor 4 α; JNK, c-jun N-terminal kinase; LRH-1, liver receptor homolog-1; LXR, liver X receptor; MTP, microsomal triglyceride transfer protein; PEPCK, phosphoenolpyruvate carboxykinase; PPARγ, peroxisome proliferator activated receptor γ; SHP, small heterodimer partner; SREBP-1c, sterol regulatory element binding protein-1c; WAT, white adipose tissue; and BAT, brown adipose tissue.
(i) SHP in feedback regulation of bile acid synthesis
Bile acids are the end products of the cholesterol catabolic pathway and their synthesis in the liver requires the coordinated actions of many enzymes. SHP was shown to regulate genes involved in bile acid synthesis (CYP7A1, CYP8B1, and CYP7B1), conjugation (BAT), and transport (BSEP, MDR2, NTCP, SLC10A2, ABCB11, ABCB4, ABCA1, CETP, and SCARB1) [16, 17, 45, 77–82]. At least two pathways of bile acid biosynthesis have been described. The first and rate-limiting enzyme is cytochrome P450 enzyme cholesterol 7α-hydroxylase (CYP7A1) in the endoplasmic reticulum initiated neutral pathway which is regulated by FXR and LXR [16, 83, 84]. The alternative acidic pathway is initiated in the mitochondria by the enzyme sterol 27-hydroxylase (CYP27A1), followed by distinct oxysterol 7α-hydroxylase (CYP7B) [85, 86]. The feedback inhibition of CYP7A1 expression is regulated by multiple bile acid-activated signaling pathways, including the FXR/SHP nuclear receptor cascade [16, 17], cellular kinases [20, 87–89], and the fibroblast growth factor 15/19 (FGF15/19) [90, 91]. SHP has been implicated as a key downstream regulator in all of these inhibitory pathways.
The function of CYP7A1 in bile acid biosynthesis has been studied extensively. Bile acids, steroid hormones, inflammatory cytokines, insulin, and growth factors have been found to inhibit CYP7A1 transcription through the promoter region. Two bile acid response elements (BARE) that are putative nuclear receptor binding sites were identified in the proximal promoter of rat CYP7A1 [92]. BARE-I is located at −73 to −55 of the rat CYP7A1 promoter containing LXR binding site in all species except humans. This has been confirmed by the finding that bile acid synthesis was increased in wild-type mice but not in LXRα null mice fed a high cholesterol diet [83]. In contrast, LXRα did not bind the human CYP7A1 promoter and the hCYP7A1 promoter was not induced by feeding cholesterol to transgenic mice harboring the gene [93–95]. A subsequent study has shown that activation of LXRα repressed CYP7A1 expression through induction of SHP in primary cultures of human hepatocytes [22]. These findings indicate that there are fundamental differences in the regulation of CYP7A1 and cholesterol homeostasis in rodents and humans. While rodents appear to deal with excess cholesterol by stimulating its conversion to bile acids for excretion from the body via stimulating CYP7A1 expression, humans may have evolved an alternate strategy against excessive cholesterol, in which the absorption of cholesterol in the intestine is reduced through decreased bile acid production via repression of CYP7A1 expression. BARE-II contains a HNF4α binding site located in a region from −149 to −118 of the rat CYP7A1 promoter, which has an 18 nucleotide sequence that is completely conserved in humans [96]. In the CYP7A1 and CYP8B1 promoters, the HNF4α binding site overlaps with a binding site for LRH-1. In the FXR/SHP pathway, bile acids bind to the bile acid receptor FXR leading to the induction of SHP, which in turn inactivates LRH-1 and HNF4α, results in repression of CYP7A1 and SHP [16, 17]. In this process, induced SHP inhibits transcription of CYP7A1 by coordinately recruiting the chromatin modifying cofactors, mSin3A/HDAC corepressors, a class III histone deacetylase SIRT1, G9a methyltransferase, and Swi/Snf–Brm chromatin remodeling complex to the CYP7A1 promoter [52, 54, 55, 97]. However, two recent studies using liver specific conditional knockouts of LRH-1 in mice suggest that LRH-1 may not be essential for bile acid inhibition of CYP7A1. Mataki et al. reported that while the expression of CYP8B1 was almost abolished in mouse liver lacking LRH-1, the expression of CYP7A1 remained unchanged [98]. Lee et al. generated conditional knockouts of LRH-1 in either hepatocytes or intestinal epithelium and demonstrated that LRH-1 deficiency in hepatocytes had no significant effect on either basal CYP7A1 expression or its repression by FXR [99]. However, basal CYP8B1 mRNA levels were significantly decreased and this decrease was accompanied by a decreased concentration of taurocholic acid and increased levels of tauromuricholic acids in the bile acid pool. These results indicate that LRH-1 is required for FXR inhibition of CYP8B1 gene transcription, but may not be involved in FXR inhibition of CYP7A1, which also suggests different roles for CYP7A1 and CYP8B1 in regulating the bile acid pool size and composition.
While this SHP-dependent regulatory cascade accounts for the inhibitory effects of bile acid synthesis, studies from two different groups suggest that other pathways are also involved. One study reported that SHP-deficient mice showed abnormal accumulation and increased synthesis of bile acids due to derepression of the rate-limiting CYP7A1 and CYP8B1 hydroxylase enzymes in the biosynthetic pathway. Dietary bile acids induced liver damage and restored feedback regulation. Reduction of the bile acid pool with cholestyramine enhanced CYP7A1 and CYP8B1 expression. The authors proposed that three negative regulatory pathways controlled bile acid synthesis, one was mediated by SHP, and two were SHP independent and invoked by liver damage and changes in bile acid pool size [87]. Another study describe of mice lacking the SHP gene showed that they exhibited mild defects in bile acid homeostasis and failed to repress CYP7A1 expression in response to a specific agonist for the bile acid receptor FXR. However, this repression was retained in the SHP−/− mice fed bile acids, demonstrating the existence of compensatory repression pathways for bile acid signaling. Further studies suggested that both the c-Jun N-terminal kinase JNK and PXR pathways might be involved in CYP repression by bile acids in SHP null animals [88]. In addition, bile acids have been shown to inhibit CYP7A1 by activating PKB (AKT) kinases [89]. It should be noticed that bile acids also activate the steroid and xenobiotic receptor PXR, which induces human cytochrome P4503A4 (CYP3A4) in drug metabolism and CYP7A1 in bile acid synthesis in the liver. Rifampicin, a human PXR agonist, inhibits bile acid synthesis and has been used to treat cholestatic diseases. However, rifampicin reduced CYP7A1 and SHP mRNA expression suggesting that SHP was not involved in the PXR inhibition of CYP7A1. Further investigation showed that activation of PXR by rifampicin promoted the interaction of PXR with HNF4α and blocked PGC-1α activation by HNF4α, resulting in inhibition of CYP7A1 gene transcription [100].
Bile acids repressed CYP8B1 transcription by reducing the transactivation of HNF4α by SHP contributes to the repression of bile acid production [101]. In contrast to the increased sensitivity predicted from the loss of negative feedback regulation, the SHP−/− mice were relatively resistant to the hepatotoxicity of a diet containing 0.5% cholic acid and the more severe effects of a diet containing both 0.5% cholic acid and 2% cholesterol in long term treatments. This was associated with the decreased hepatic accumulation of cholesterol and triglycerides in SHP−/− mice. CYP8B1 was strongly re-expressed in the SHP−/− mice, but not in wild type mice, fed either diet containing bile acid. This contrasts to the strong repression of CYP8B1 observed with short term bile acid feeding, as well as the effects of long term feeding on other bile acid biosynthetic enzymes such as CYP7A1. The induction of CYP8B1 could contribute to the decreased toxicity of chronic bile acid treatment by increasing the hydrophilicity of the bile acid pool. These results identified an unexpected role for SHP in hepatotoxicity and suggested new approaches to modulating effects of chronically elevated bile acids in cholestasis [102].
Bile acid-activated FXR was demonstrated recently to induce expression of FGF15/19 (FGF15 is the mouse homolog of human FGF19) in the small intestine and secreted FGF15 repressed CYP7A1 transcription in liver through a mechanism involving the membrane FGF receptor 4 (FGFR4) and SHP [91]. The role of this intestinal FXR/FGF15 pathway has been further confirmed in intestine-specific FXR knockout mice [103]. In this study, the FXR agonist GW4064 repressed CYP7A1 in liver-specific FXR knockout mice but not in intestine-specific FXR knockout mice, indicating that the activation of intestinal FXR but not liver FXR is required for bile acid inhibition of CYP7A1 gene expression. In contrast, FXR-mediated repression of CYP8B1 was more preferentially dependent on the liver FXR/SHP pathway and less dependent on the intestine FXR/FGF19/FGFR4 pathway indicating differences in bile acid feedback repression of CYP7A1 and CYP8B1.
While the induction of SHP gene expression by increased bile acids is well established, whether SHP protein activity is also modulated remains largely unknown. A recent study showed that SHP was a rapidly degraded protein via the ubiquitin-proteasomal pathway and that bile acids or bile acid-induced FGF19 increased stability of hepatic SHP by inhibiting proteasomal degradation in an extracellular signal-regulated kinase (ERK)-dependent manner. SHP was ubiquitinated at Lys122 and Lys123, and mutation of these sites altered its stability and gene repression activity. Furthermore, SHP was phosphorylated at Ser26 by ERK during bile acid treatment, and mutation of this site dramatically decreased the stability of SHP. Moreover, the stability of SHP was abnormally elevated in ob/ob mice and diet-induced obese mice. The authors proposed the important role for regulation of SHP stability in bile acid signaling under normal conditions, and that abnormal stabilization of SHP might be associated with obesity and diabetes [104].
(ii) SHP in lipid metabolism and obesity
Cellular cholesterol and lipid homeostasis is tightly controlled by a complex network of transcriptional programs regulating their production and clearance, which depends on a large number of nuclear receptor family members. For instance, increased activity of PPARγ has been directly associated with steatosis [105]. To date, multiple lines of evidence demonstrate that SHP regulates several metabolic pathways involved in fatty liver and obesity by acting as a transcriptional repressor of other nuclear receptors [106, 107]. One untypical example is that SHP gene augmented PPARγ transactivation [7] and the acute PPARγ overexpression resulted in marked hepatic lipid accumulation [105]. SREBP-1c is another important factor in regulating the lipogenic program in liver and is also a target of SHP. It has been shown that down-regulation of SREBP-1c expression by SHP was mediated via FXR activation by either natural or synthetic FXR agonists, followed by the down-regulation of LXRα and LXRβ, and consequently repressing other transcriptional factors to stimulate SREBP-1c transcription [106]. In agreement with these observations, in transgenic mice constitutively expressing SHP in the liver, SHP was shown to affect genes involved in bile acid conjugation, transport and lipogenic pathways. Constitutive SHP expression led to the depletion of hepatic bile acid pool and accumulation of triglycerides in the liver, which may account for the direct repression of downstream target genes and the bile acid sensor FXR, and an indirect activation of the major regulators of lipogenic genes such as PPARγ and SREBP-1c via SHP [77]. Furthermore, the mRNA levels of other genes involved in fatty acid and triglyceride biosynthesis, such as the fatty acid synthase (FAS), ATP citrate lyase (ACL), acetyl-CoA carboxy-lase (ACC-1) and stearoyl-CoA reductase (SCD1), were also increased significantly in SHP transgenic mice [77]. However, another study showed LRH-1 stimulated FAS transcription via LXR, and this response was blocked by increased SHP and that FAS mRNA was overexpressed in SHP−/− mice [107].
Microsomal triglyceride transfer protein (MTP) is a dedicated chaperone required for biosynthesis of apolipoprotein B (apoB), and is transcriptional repressed by SHP via interactions with HNF4α and LRH-1 [108, 109]. Our previous study showed that the deletion of SHP in obese leptin-deficient mice (ob/ob), an animal model of severe obesity and insulin resistance, prevented the development of nonalcoholic fatty liver by enhancing lipid secretion (MTP) as well as decreasing de novo hepatic fatty acid synthesis (SREBP-1c and FAS) and uptake (PPARγ). These findings established a major function of SHP in modulating hepatic lipid synthesis and transport [109].
Several studies demonstrated that plasma triglycerides show diurnal variations [110–112]. MTP expression and plasma lipids underwent diurnal regulation and exhibited peaks and nadirs at similar times, indicating that circadian changes in MTP could contribute to plasma lipids circadian rhythmicity [112]. Oiwa and associates reported rhythmic expression of SHP in the mouse liver. They demonstrated that clock genes, namely circadian locomotor output cycles kaput (CLOCK) and brain and muscle aryl hydrocarbon receptor nuclear translocator-like 1 (Bmal1), bound to the E box elements present in the SHP promoter and synergistically activated SHP transcription with LRH-1, which was in turn suppressed by SHP itself [26]. However, little was known about the mechanisms controlling diurnal changes in plasma lipids and MTP. To address this issue, Pan et al. examined the role of the CLOCK gene in the diurnal regulation of MTP, and found that CLOCK up-regulation of SHP led to a consequent suppression of MTP expression resulting in MTP diurnal changes [113]. This study points to the importance of both CLOCK and SHP in the circadian regulation of MTP and plasma triglycerides, and further indicates that disruptions in circadian rhythms might induce hyperlipidemia. Mice with the clock gene knocked out are prone to developing metabolic syndrome. These findings have important applications in understanding why some night shift workers are at increased risk for developing the metabolic syndrome [114–116].
To further characterize the effect of loss of SHP in protecting against dyslipidemia, Hartman et al crossed the SHP−/− mice with the LDLR−/− mice and found that the elevated levels of VLDL and LDL cholesterol induced by a lipid rich western diet in LDLR−/− mice were greatly reduced by SHP-deficiency in LDLR−/−SHP−/− mice [117]. In addition, hepatic inflammatory marker genes including TNFα, VCAM and ICAM-1 were induced by the same diet in LDLR−/− mice but their induction was completely blocked in LDLR−/−SHP−/− mice. The authors confirmed that the loss of SHP expression specifically in hepatocytes was responsible for protecting against dyslipidemia by using liver-specific SHP-deleted mice.
Obesity is the result of an imbalance between energy intake and expenditure. Energy intake is mainly dependent on food ingestion and energy expenditure depends on several factors, including exercise and heat production, or so-called adaptive thermogenesis. Brown adipose tissue (BAT) is the major site for adrenergic mediated adaptive thermogenesis involving the uncoupling protein-1 (UCP1), whereas white adipose tissue (WAT) is mostly implicated in the regulation of lipid storage and catabolism [118, 119]. Previous study showed that basal gene expression of UCP1 and PGC-1α was increased in BAT of SHP−/− mice and this was associated with increased oxygen consumption, heat production and decreased obesity, indicating that SHP may be a negative regulator of energy utilization [120]. To further investigate the effects of SHP activation in adipose tissues, we generated transgenic mice over-expressing SHP in WAT and BAT, and unexpectedly found the enhanced whole body energy metabolism and up-regulation of β1AR and PGC-1α in BAT of the young mice [121]. On the other hand, SHP activation in WAT had a dominant role in obesity which exacerbated the obese phenotype associated with the high-fat diet. Developing small molecule therapeutics that antagonize the effect of SHP may provide a new approach for treating obesity.
(iii) SHP in liver fibrosis
Hepatic fibrosis, the precursor to cirrhosis, represents the wound healing response to liver injury from a wide variety of etiologies, and includes components of both increased and altered deposition of extracellular matrix (ECM) and wound contraction. Remarkable progress has been made in identifying the cellular sources of ECM and the pathways regulating the regression of fibrosis [122]. Following hepatic injury, hepatic stellate cells (HSCs), the major source of ECM in the liver, become activated and undergo a progressive process of trans-differentiation from a resting, fat-storing phenotype toward a myofibro blast-like phenotype, characterized by increased expression of fibroblastic cell markers such as α-smooth muscle actin (α-SMA) [123, 124].
Several studies demonstrated that SHP has a preventative role in liver fibrosis. One early study showed that exposure of HSCs to FXR ligands caused a 3-fold increase of SHP, reduced alpha1 (I) collagen and TGF-β1 by approximately 60%–70% and abrogated alpha1(I) collagen mRNA up-regulation induced by thrombin and TGF-β1. SHP bound JunD and inhibited DNA binding of adaptor protein (AP)-1 induced by thrombin. Based on these findings, the authors proposed that FXR ligands might represent a novel therapeutic option to treat liver fibrosis [62]. In addition, 6-ethyl chenodeoxycholic acid (6-ECDCA or INT-747), a semisynthetic derivative of chenodeoxycholic acid (CDCA), modulated tissue metalloproteinase inhibitor (TIMP)-1 and matrix metalloprotease (MMP)-2 expression and activity in HSCs and in the liver of rats rendered cirrhotic by a 4-week treatment with CCl(4). Further studies showed that activation/overexpression of FXR caused a SHP-dependent inhibition of JunD binding to its consensus element in the TIMP-1 promoter. Consequently, inhibition of TIMP-1 expression by SHP enhanced the sensitivity of HSCs to proapoptogenic stimuli, demonstrating that a FXR-SHP regulatory cascade promoted the development of a quiescent phenotype and increased apoptosis of HSCs [125].
However, one study investigated the impact of genetic FXR ablation in four different mouse models of hepatic fibrosis, including CCl(4), 3,5-diethoxycarbonyl-1,4-dihydrocollidine feeding, common bile duct ligation (BDL), or Schistosoma mansoni-infection. The authors found that FXR-deficiency had no effect in CCl(4) treated and Schistosoma infected mice, but significantly decreased liver fibrosis of the biliary type (common bile duct ligation, 3,5-diethoxycarbonyl-1,4-dihydrocollidine feeding). Moreover, FXR protein was undetectable in mouse and human HSCs and portal myofibroblasts (MFBs). This study suggested that the loss of FXR significantly reduced fibrosis of the biliary type, but had no impact on non-cholestatic liver fibrosis. Since there is no FXR expression in HSCs and MFBs during liver fibrosis, the authors questioned the justification of using FXR ligands as therapeutics for hepatic fibrosis [126].
Loss of SHP resulted in increased sensitivity to liver damage induced by bile duct ligation (BDL) further confirmed the protective effect of SHP in fibrosis. SHP−/− mice showed increased sensitivity in this model of acute obstructive cholestasis, with greater numbers of bile infarcts and higher mortality than wild-type C57BL/6 mice. At 3 hours, CYP7A1 expression still remained elevated in SHP−/− with respect to wild-type mice, and the hepatic and serum bile acid levels and total hepatobiliary bile acid pool were significantly increased. The increased sensitivity of mice lacking SHP contrasts with the decreased sensitivity of mice lacking FXR to BDL, which has been associated with decreased intraductal pressure and fewer bile infarcts [127].
Plasminogen activator inhibitor type I (PAI-1) is a marker of the fibrinolytic system and serves as a possible predictor for hepatic metabolic syndromes. Recently, Fenofibrate, a peroxisome proliferator-activated receptor alpha (PPARα) agonist, has been shown to increase SHP gene expression in cultured liver cells and in normal and diabetic mouse liver by activating the adenosine monophosphate-activated protein kinase (AMPK) signaling pathway in a PPARα-independent manner. Furthermore, administration of TGF-β or a methionine-deficient and choline-deficient (MCD) diet to induce progressive fibrosing steatohepatitis in C57BL/6 mice was reversed by fenofibrate via AMPK-mediated induction of SHP. This was accompanied by a dramatic decrease in PAI-1 messenger RNA (mRNA) and protein expression along with other fibrotic marker genes, which was not observed in SHP−/− treated with fenofibrate [128]. These findings raise the possibility that synthetic ligands that activate SHP might have antifibrotic effect in some types of liver diseases.
(iv) SHP in glucose metabolism
The maintenance of glucose homeostasis depends on the coordination and integration of several physiological systems to balance glucose production in the liver and glucose use in peripheral tissues. This process is normally controlled by the balanced secretion and action of insulin on one side and glucagon, epinephrine, cortisol, and growth hormone on the other [129]. A variety of transcription factors and cofactors such as PGC-1α [130] are known to regulate transcription of glucose-6-phosphatase (G6Pase) and phosphoenolpyruvate carboxykinase (PEPCK), which encodes a rate-determining enzyme in hepatic gluconeogenesis. Specific functions of SHP in gluconeogenesis, particularly in the liver, were suspected based on the abundance of mRNA in this tissue and its interaction with NRs implicated in liver gluconeogenesis. The glucocorticoid receptor (GR) plays an important role in regulating blood glucose levels in mammals, and has been identified as a downstream target for SHP inhibition. The fact that SHP antagonizes the GR coactivator PGC-1α coactivation leading to inhibition of PEPCK expression implies a physiologically relevant role for SHP in modulating hepatic glucocorticoid action [44]. In addition, it has been shown that SHP is involved in the repression of G6Pase and PEPCK gene expression via inhibition of the forkhead transcription factors hepatocyte nuclear factor-3 (HNF3) [61] and HNF6 [131]. Another study showed that SHP repressed C/EBPalpha (CCAAT/enhancer-binding protein alpha) driven transcription of PEPCK through a direct interaction with the C/EBPalpha protein, resulting in the inhibition of PEPCK gene transcription [63]. This observation provides further evidence that SHP has a major function in regulating hepatic gluconeogenesis. Other important targets of SHP in glucose metabolism are the forkhead transcription factor FOXO1, the basic helix-loop-helix transcriptional factor BETA2/NeuroD, and the aryl hydrocarbon receptor (AHR)/nuclear translocator (ARNT) [47, 48, 60].
The observation that bile acids inhibit the expression of gluconeogenic genes, including G6Pase, PEPCK, and fructose 1, 6-bis phosphatase (FBP1) in a SHP-dependent manner and the absence of this repression in both FXR−/− and SHP−/− mice indicate that FXR-SHP nuclear receptor cascade also regulates glucose metabolism [48, 132]. Reporter assays demonstrated that over-expression SHP and chenodeoxicholic acid treatment down-regulated PGC-1 promoter activity via a member of the forkhead transcription factors, Foxo1, Foxo3a, and Foxo4 [133]. The above findings reveal that bile acids inhibit hepatic gluconeogenesis in a SHP-dependent manner. In addition, hyperglycemia, insulin resistance, and insulin response to glucose, as well as dyslipidemia, were significantly ameliorated in a type 2 diabetes animal model by treatment with the bile acid binding resin colestimide provides further evidence for a link between bile acids and glucose metabolism [134]. These observations suggest that the bile acid metabolism pathway could be a novel target in treating obesity, insulin resistance, and type 2 diabetes.
AMP-activated protein kinase (AMPK) is a serine/threonine kinase that regulates hepatic glucose and lipid homeostasis by affecting a diverse set of target genes associated with these metabolic pathways. Metformin, an antidiabetic drug widely used for the treatment of type 2 diabetes, and sodium arsenite which was previously reported to exhibit insulin-mimetic effects on glucose homeostasis have been reported to inhibit hepatic gluconeogenesis through SHP mediated inhibition of PEPCK and G6Pase gene expression in an AMPK-dependent manner [135, 136]. These studies provide a novel molecular mechanism of SHP-mediated regulation of hepatic glucose homeostasis and indicate that SHP may be one of the primary targets of AMPK. More recently, hepatocyte growth factor (HGF), a pleiotropic growth factor, was identified as a novel inducer of SHP mediated suppression of gluconeogenesis via activation of the AMPK signaling pathway [28]. HGF and its family member, macrophage-stimulating factor (MSP), increased upstream stimulatory factor-1 (USF-1) DNA binding on the SHP promoter to activate SHP gene transcription, resulting in suppression of PEPCK and G6Pase gene expression. This study provides a novel signaling pathway through HGF/AMPK/USF-1/SHP to inhibit hepatic gluconeogenesis.
Liver glucokinase (LGK), an important hepatic glucokinase, plays an essential role in sensing and maintaining proper blood glucose levels. One recent study showed that SHP decreased LGK gene expression by inhibiting the transcriptional activity of LXRα and PPARγ via interacting directly with their common heterodimer partner RXRα to mediate suppression of gluconeogenesis [137].
(v) SHP in drug metabolism
A vast majority of drugs are metabolized in liver, where prodrugs are frequently converted to active metabolites or drugs are inactivated by specific cytochrome P450 (CYP) enzymes. The levels of cytochrome P450 enzymes control the rate at which many drugs are metabolized. It has become clear that these drug metabolizing enzymes and drug efflux transporters are directly under the control of tissue-specific NRs such as CAR and PXR, which coordinately regulate expression of detoxifying enzymes and transporters [138]. Originally, SHP had been reported to bind to and inhibit CAR, a nuclear receptor involved in the induction of the CYP2 and CYP3 genes in the metabolism of xenobiotics [139]. Further studies showed the induction of CYP2B gene expression by phenobarbital was mediated by the translocation of CAR from the cytoplasm to the nucleus, and coexpression of SHP inhibited the transactivation of the CYP2B gene mediated by CAR via either inhibiting recruitment of other coactivators through the p160 coactivator GRIP1 or actively recruiting corepressors directly to the CAR/RXR/phenobarbital responsive unit (PBRU) complex in the CYP2B gene [140].
Like CAR, PXR was initially reported to induce CYP3A gene transcription following xenobiotic induction by phenobarbital or rifampicin [141]. Upon activation, PXR controlled elimination of these compounds by inducing CYP3A4 and Na(+)-independent organic anion transporter 2 (Oatp2) expression [141–143]. More recently, it has been shown that PXR is a broad-specificity sensor that recognizes a wide variety of synthetic drugs as well as endogenous compounds such as bile acid precursors [59]. The bile acid mediated induction of SHP resulted in SHP interacting with both murine and human PXR in a ligand-dependent manner to provoke a repression of PXR-mediated CYP3A induction. This study revealed an elaborate regulatory cascade, tightly controlled by SHP, for regulating both the maintenance of bile acid production and detoxification in the liver [59]. A better understanding of these mechanisms may aid the development of better agents to decrease serum bile acids in chronic cholestatic liver diseases such as primary biliary cirrhosis and sclerosing cholangitis [144, 145].
6. SHP in cancer
Nuclear receptors have been implicated in regulating the biology of a wide variety of cancers, including the initiation and progression of hepatocellular carcinoma (HCC). In fact, inhibition of SHP in vivo causes regression of hepatoma [146]. Because of their important physiologic roles in cancers, nuclear receptors and the critical genes that they regulate are emerging targets for molecular diagnostic tests and cancer therapeutics. Accumulating evidence points to the involvement of SHP in the development of some cancers, particularly liver cancer and breast cancer.
(i) SHP in liver cancer
Our recent studies demonstrated that SHP has potent tumor suppressive activity by inhibiting cellular growth and activating apoptosis [40, 146–148]. Aberrant hypermethylation of promoter regions in cytosine-guanine dinucleotides (CpG) islands has been shown to be associated with transcriptional silencing of tumor-suppressor genes in many cancers. We evaluated the methylation profile and tumor-suppressive function of SHP in the development of HCC. The expression of SHP was markedly diminished in human HCC pathologic specimens and cell lines by epigenetic silencing due to SHP promoter hypermethylation. In vitro methylation decreased SHP promoter transactivation and nuclear receptor LRH-1 binding which was reversed by demethylation. Overexpression of SHP also inhibited human HCC foci formation, arrested HCC tumor growth in xenografted nude mice, and increased the sensitivity of HCC cells to apoptotic stimuli. Further analysis of a total of 19 normal human liver and 57 HCC specimens showed that down-regulation of SHP gene expression may be a common denominator in HCC [147].
Study of SHP−/− mice provided additional evidence that SHP has a biological tumor suppressor function by negatively regulating cellular growth. SHP−/− mice 12 to 15 months of age developed spontaneous hepatocellular carcinomas, which were found to be strongly associated with enhanced hepatocyte proliferation and increased cyclin D1 expression. In contrast, overexpressing SHP in hepatocytes of SHP-transgenic mice reversed this effect. Embryonic fibroblasts lacking SHP showed enhanced proliferation and produced increased cyclin D1 messenger RNA and protein, and SHP was shown to be a direct negative regulator of cyclin D1 gene transcription. In agreement with this, the immortal SHP−/− fibroblasts displayed characteristics of malignant transformed cells and formed tumors in nude mice [146].
In addition to suppressing the transcription of a critical cell proliferating gene, SHP can also activate cell death. Apoptosis is a distinct form of programmed cell death that is best defined morphologically by nuclear and cell fragmentation. Apoptosis plays a major role in liver disease, and altered regulation of apoptosis has been shown to be associated with the pathogenesis of HCC and dysplasia in the liver [149]. Our recent studies showed that at least two mechanisms are involved in the activation of apoptosis and inhibition of tumor growth by SHP. One mechanism is that inhibition of ERRγ by SHP leads to decreased YY1 expression and the de-repression of YY1 on AP1 activity, which in turn results in the activation of miR-206 and inhibition of notch3 signaling [67, 148]. Expression of miR-206 markedly induced apoptotic cell death and blocked the anti-apoptotic activity of Notch3. In addition, ectopic expression of miR-206 inhibited HeLa cell migration and focus formation. Therefore, we identified miR-206 as a pro-apoptotic activator of cell death, which was associated with its inhibition of notch3 signaling and tumor formation [148].
More recently, we reported a novel cytoplasmic function of SHP through its regulation of mitochondrial induced apoptosis. SHP acted as a pivotal cell death receptor that targets mitochondria by binding Bcl-2, disrupting the Bcl-2/Bid interaction, and inducing cytochrome c release. The apoptosis inducer AHPN acted by increasing SHP gene expression and promoting the translocation of SHP from the nucleus to the mitochondria. Furthermore, induction of apoptosis by activation of SHP inhibited peritoneal pancreatic tumor growth, suggesting this may be a mechanism functioning in a variety of cancers [40].
(ii) SHP in breast cancer
Breast cancer is the most common diagnosed cancer of women in the Western world and second only to lung cancer in mortality rate. Estrogen is a classical etiological factor for breast cancer and exerts its biological function via ERα and ERβ in a ligand-dependent manner. Approximately 70% of breast cancers are ER positive, and adjuvant anti-estrogen therapy plays an important role in treatment. Previous studies have reported links between SHP and estrogen signaling, such as the direct interaction of SHP with ERs [151], inhibition ER-mediated transcriptional activation by SHP [41] and ER-dependent transcriptional regulation of SHP in a feedback loop [23]. In postmenopausal women with breast cancer the source of estrogen driving tumor growth is produced locally within the tumor and in the surrounding adipose tissue and is due to activity of aromatase CYP19 [152]. In further support of this hypothesis, overexpression of aromatase in adipose tissue contributes to development and progression of breast cancer. Clinical trials have shown that aromatase inhibitors are superior to tamoxifen as a first line treatment of advanced disease in postmenopausal women [153]. The fact that SHP enhances the transcriptional activity of PPARγ in adipose tissue via an atypical interaction with the DNA binding domain and hinge region of PPARγ [7] and ligands for PPARγ are very effective inhibitors of aromatase expression in adipose stromal cells [154] suggested that SHP might exert an inhibitory effect on aromatase expression. Indeed, SHP was shown to inhibit aromatase expression and estrogen production in breast preadipocytes by preventing LRH-1 transactivation on aromatase promoter II [155]. These studies suggest that SHP inhibits estrogen action at multiple levels and induction of SHP expression or activity in breast would be relatively specific for inhibiting estrogen signaling in breast cancer. Recently, the FXR agonist GW4064 was found to up-regulate SHP expression, inhibit aromatase activation and induce breast carcinoma cell line MCF-7 and MDA-MB-468 apoptosis [156]. It is, therefore, tempting to speculate that this FXR-SHP-LRH-1 pathway is functionally active in breast cancer and may represent a therapeutic approach to repress proliferation and to induce apoptosis in breast cancer treatment.
(iii) SHP in other cancers
Data mining of a microarray database http://www.genecards.org showed that SHP was down-regulated in many cancers, including adrenal cortex, cerebellum, kidney, skin, and thyroid, suggesting that SHP may function as a common tumor suppressor. A more recent study showed that SHP decreased glioma-associated oncogene homologue (Gli) target gene expression by repressing the transcriptional activity of Gli [150]. Gli was originally identified as an amplified gene in gliomas, and acted as a terminal effector of the Hedgehog signaling pathway in cell proliferation and anti-apoptosis tumorigenesis. SHP was shown to inhibit transcriptional activity and nuclear localization of Gli1 via direct protein interaction. These findings provide further evidence for a possible link between anti-tumor functions of SHP and the role of SHP as an inhibitory regulator of Hedgehog/Gli signaling pathway.
7. Conclusion and perspective
Since its discovery in 1996, SHP has been shown to interact with a wide variety of transcription factors, including nuclear receptors and nonnuclear receptor transcription factors, which regulate the expression of many target genes in a tissue-specific manner. This wide variety of SHP-interacting partners is indicative of its regulatory role in many cellular and physiological pathways, thereby establishing its role as a critical transcriptional coregulator in metabolic processes. The fact that SHP regulates bile acid synthesis, lipid, cholesterol, and glucose metabolism provide a potential means to develop SHP targeted therapeutics for at least several metabolic diseases. Of particular interest is the observation that the loss of SHP function prevents dyslipidemia, fatty liver and insulin resistance caused by LDLR- or leptin-deficiency. These observations raise the possibility of developing synthetic SHP antagonists to selectively inhibit SHP activity in hepatocytes and adipocytes as therapeutics for fatty liver, hypercholesterolemia and obesity. Moreover, SHP plays a preventive role in liver fibrosis providing a rational to develop SHP agonists as antifibrotic therapeutics. Since many of the target genes and functions of SHP have been derived from in vitro systems, additional in vivo studies are needed. In this context, it is also important to keep in mind that possible species differences might exist with regard to SHP expression, regulation and function. Regarding the pharmacological modulation of SHP activity, the protein contains a conserved ligand-binding domain suggesting the possibility that its activity may be regulated by natural or synthetic ligands. Although no array of ligands has been identified to date, several ligands have been identified [37]. An important topic of future studies will be to define the structure of the ligand binding domain of SHP to aid the identification and synthesis of activators and inhibitors of SHP.
The new finding about the cytoplasmic function of SHP through its regulation of mitochondrial induced apoptosis, which is mediated by its subcellular localization, indicates that the subcellular localization of SHP protein has a different biological function than its original nuclear receptor role. In view of these different functions, studies of posttranslational modifications of the SHP protein such as acetylation, methylation, and phosphorylation are essential topics of future studies. Although a role for SHP stability has been identified in bile acid signaling pathways, it will be interesting to see if SHP protein stability is critical in regulating other pathways such as lipid and glucose metabolism.
Besides its metabolic regulatory functions, studies by our and other groups provide strong evidence that SHP plays a critical role in the development of cancer, particularly in liver and breast cancer. Although this is a new area of investigation, an increased understanding of the fundamental mechanisms by which SHP regulates cancer progression, especially its intriguing role in regulating miRNA expression, is of great importance in applying this knowledge to new diagnostics, therapeutics and prevention strategies for cancer. In this context, identification of SHP regulated miRNA oncogenes or anti-oncogenes is of particular interest.
Table 2.
Interaction of SHP with other nuclear receptors and transcription factors, or coregulators
| Classification | Interacting protein |
Methods | Target gene |
References |
|---|---|---|---|---|
| NR | AR | Y2H and GST pull down: rSHP/hAR | Gobinet et al., 2001 | |
| CAR | Y2H and GST pull down: mSHP/mCAR | Seol et al., 1996 | ||
| Y2H and GST pull down: mSHP/mCAR, mSHPΔ128–139/mCAR | Park et al., 2004 | |||
| GST pull down: mSHP/mCAR | ↓CYP2B | Bae et al., 2004 | ||
| DAX-1 | Co-IP: hSHP/hDAX-1 | Iyer et al., 2006 | ||
| ERα | Y2H and GST pull down: rSHP/hERα | Johansson et al., 1999 | ||
| Y2H, M2H, and GST pull down: SHP/hERα | Johansson et al., 2000 | |||
| Y2H and GST pull down: rSHP/ERα | Goodwin et al., 2000 | |||
| GST pull down: mSHP/hERα | Klinge et al., 2002 | |||
| Co-IP: hSHP/hERα | Iyer et al., 2006 | |||
| ERβ | Y2H and GST pull down: rSHP/hERβ | Johansson et al., 1999; | ||
| Y2H, M2H, and GST pull down: SHP/hERβ | Johansson et al., 2000 | |||
| ERRα | Y2H, and M2H: hSHP/ERRα, mSHP/ERRα | Sanyal et al., 2002 | ||
| ERRβ | Y2H, and M2H: hSHP/ERRβ, mSHP/ERRβ | Sanyal et al., 2002 | ||
| ERRγ | Y2H: and M2H: hSHP/ERRγ, mSHP/ERRγ | Sanyal et al., 2002 | ||
| Y2H: rSHP/rERRγ | Razzaque et al., 2004 | |||
| Y2H and GST pull down: mSHP/ERRγ, mSHPΔ128–139/ERRγ | Park et al., 2004 | |||
| GST pull down: hSHP/mERRγ | ↓YY1 | Zhou et al., 2010 | ||
| FXR | M2H and GST pull down: hSHP/mFXR, mSHP/mFXR | ↓PCK1 | Lu et al., 2000 | |
| GR | Co-IP: rSHP/hGR | Borgius et al., 2002 | ||
| HNF4α | M2H and GST pull down: mSHP/rHNF4α | Lee et al., 2000 | ||
| M2H and GST pull down: hSHP/rHNF4α, hSHP/hHNF4α | Boulias et al., 2004 | |||
| GST pull down: hSHP/hHNF4α | ↓AGT ↓FBP1 |
Shimamoto et al., 2004 | ||
| M2H: SHP/HNF4α | ↓MTP | Hirokane, et al., 2004 | ||
| GST pull down: hSHP/hHNF4α | ↓ApoCIII ↓APoB |
Zhou et al., 2010 | ||
| LRH-1 | Y2H: rSHP/mLRH-1 LBD M2H: mSHP/hLRH-1 LBD |
Lee et al., 2000 | ||
| M2H and GST pull down: mSHP/m LRH-1 | ↓SHP | Lu et al., 2000 | ||
| M2H: SHP/hLRH-1, SHP/mLRH-1 GST pull down: SHP/hLRH-1 |
↓CYP7A1 | Goodwin et al., 2000 | ||
| M2H and GST pull down: hSHP/hLRH-1 | Lee and Moore, 2002 | |||
| Y2H: hSHP/hLRH-1 M2H: mSHP/hLRH-1 |
Brendel et al., 2002 | |||
| M2H: mSHP/mLRH-1 | Boulias et al., 2004 | |||
| Y2H and GST pull down: mSHP/ LRH1, mSHPΔ128–139/LRH1 | Park et al., 2004 | |||
| SHP/LRH-1 | ↓MTP | Huang et al., 2007 | ||
| SHP/LRH-1 | ↓cyclinD1 | Zhang et al., 2008 | ||
| GST pull down: hSHP/mLRH-1 | ↓SHP | Zhou et al., 2010 | ||
| LXRα | GST pull down: mSHP/hLXRα | Brendel et al., 2002 | ||
| LXRβ | GST pull down: mSHP/mLXRβ | Brendel et al., 2002 | ||
| Nuf77 | M2H and Co-IP: SHP/Nuf77 | ↓CYP17 | Yeo et al., 2005 | |
| PPARα | Y2H: rSHP/r PPARα LBD GST pull down: rSHP/rPPARα |
Kassam et al., 2001 | ||
| PPARγ | M2H and GST pull down: hSHP/hPPARα | Nishizawa et al., 2002 | ||
| PXR | GST pull down: mSHP/mPXR, mSHP/hPXR | ↓CYP3A | Ourlin et al., 2003 | |
| RARα | Y2H: mSHP/RARαLBD M2H: hSHP/hRARαLBD |
Seol et al., 1996 | ||
| Y2H: mSHPΔ148/mRARαLBD | Seol et al., 1997 | |||
| RXRα | Y2H, M2H, and GST pull down: mSHP/hRXRα | Seol et al., 1997 | ||
| M2H and GST pull down: mSHP/mRXRα | Goodwin et al., 2000 | |||
| SF-1 | M2H: hSHP/mSF-1LBD GST pull down: mSHP/mSF-1 | Lu et al., 2000 | ||
| SHP | Co-IP: SHP/SHP | Iyer et al., 2007 | ||
| TRβ | Y2H: mSHP/TRβ LBD Y2H: hSHP/hTRβ LBD |
Seol et al., 1996; Seol et al., 1997 | ||
| TF (Non-NR) | ARNT | GST pull down: SHP/ARNT | ↓CYP1A1 ↓UGT1A6 |
Klinge et al., 2001 |
| BETA2/NeuroD | Y2H and GST pull down: SHP/BETA2 | ↓βGK ↓p21 |
Kim et al., 2004 | |
| C-jun | Co-IP: rSHP/rC-jun | Fiorucci et al., 2004 | ||
| Jun D | Co-IP: rSHP/rJun D | Fiorucci et al., 2004 | ||
| C/EBPα | M2H and GST pull down: SHP/N-terminal domain of C/EBPα Co-IP: SHP/C/EBPα |
↓PEPCK | Park et al., 2007 | |
| Foxo1 | GST pull down: SHP N1 or NT/Foxo1 CT Co-IP: SHP/Foxo1 |
↓G6Pase | Yamagata et al., 2004 | |
| HNF3 | GST pull down and Co-IP: SHP/HNF3α, SHP/HNF3β, SHP/HNF3γ | ↓G6Pase ↓CYP7A1 |
Kim et al., 2004 | |
| NF-κB | Y2H, GST pull down and Co-IP: SHP/p65 | Kim et al., 2001 | ||
| Smad3 | Y2H, GST pull down and Co-Ip: SHP/Smad3 | ↓TGFβ ↓p21 ↓smad7 ↓PAI-1 |
Suh et al., 2006 | |
| Bcl2 | Co-IP: mSHP/hBcl2 | Zhang et al., 2010 | ||
| Gli | GST pull down and Co-IP: SHP/Gli | ↓PTCH1 ↓BCl2 ↓BClxL |
Kim et al., 2010 | |
| P53 | GST pull down and Co-IP: SHP/P53 | ↓miR34a | Lee et al., 2010 | |
| Co-regulator | ||||
| Co-repressor | BAF155 | GST pull down: mSHP/BAF155 | ↓CYP7A1 | Kemper et al., 2004 |
| BAF57 | GST pull down: mSHP/BAF57 | ↓CYP7A1 | Kemper et al., 2004 | |
| Brm | GST pull down and Co-IP: mSHP/Brm | ↓CYP7A1 | Kemper et al., 2004 | |
| Co-IP: mSHP/Brm | ↓CYP7A1 ↓SHP |
Miao et al., 2009 | ||
| EID1 | Y2H, M2H, GST pull down and Co-IP: rSHP/mEID1 | Bavner et al., 2002 | ||
| Y2H and GST pull down: mSHP/EID1 | Park et al., 2004 | |||
| GST pull down: hSHP/mEID1 | Zhou et al., 2010 | |||
| G9a | GST pull down and Co-IP: SHP/G9a | Boulias et al., 2004 | ||
| GST pull down and Co-IP: SHP/G9a | ↓CYP7A1 ↓CYP8B1 |
Fang et al., 2007 | ||
| HDAC1 | GST pull down: SHP/HDAc1 | ↓CYP2B | Bae et al., 2004 | |
| GST pull down and Co-IP: SHP/HDAC1 | Boulias et al., 2004 | |||
| GST pull down and Co-IP: SHP/HDAC1 | Gobinet et al., 2005 | |||
| HDAC3 | GST pull down: SHP/HDAC3 | Gobinet et al., 2005 | ||
| HDAC5 | GST pull down: SHP/HDAC5 | Gobinet et al., 2005 | ||
| HDAC6 | GST pull down: SHP/HDAC6 | Gobinet et al., 2005 | ||
| mSin3A | GST pull down and Co-IP: mSHP/mSin3A | ↓CYP7A1 | Kemper et al., 2004 | |
| GST pull down: SHP/mSin3A | ↓CYP2B | Bae et al., 2004 | ||
| SMRT | GST pull down: SHP/SMRT | Bae et al., 2004 | ||
| GST pull down: SHP/SMRT | ↓CYP2B | Bae et al., 2004 | ||
| SIRT1 | M2H, GST pull down and Co-IP: SHP/SIRT1 | ↓CYP7A1 ↓SHP | Chanda et al., 2010 | |
| Co-activator | CBP/p300 | Co-IP: SHP/CBP | Yeo et al., 2005 | |
| M2H and GST pull down: SHP/p300 | ↓TGFβ ↓p21 ↓smad7 ↓PAI-1 |
Suh et al., 2006 | ||
| SRC-1 | GST pull down: SHP/SRC-D (759–1141), SHP/SRC-E (1101–1441) | Kim et al., 2001 |
Abbreviations: NR, Nuclear receptor; TF, Transcription factor; Co-IP, coimmunoprecipitation assay; M2H, mammalian two hybrid system; Y2H, yeast two-hybrid system; h, human; m, mouse; r, rat.
Table 3.
SHP target genes and their function in physiology and disease
| Tissue | Target gene | Function | Reference |
|---|---|---|---|
| Cholesterol and Bile acid metabolism | |||
| Liver | ↓CYP7A1 | Bile acid synthesis | Goodwin et al., 2000 Lu et al., 2000 Boulias et al., 2005 |
| ↓CYP8B1 | Bile acid synthesis | Yang et al., 2002 Boulias et al., 2005 | |
| ↓CYP7B1 | Bile acid synthesis | Boulias et al., 2005 | |
| ↓BAT | Bile acid conjugation | Boulias et al., 2005 | |
| ↓BSEP | Bile acid transport | Boulias et al., 2005 | |
| ↓MDR2 | Bile acid transport | Boulias et al., 2005 | |
| ↓NTCP | Bile acid transport/uptake | Denson et al., 2001 Boulias et al., 2005 | |
| ↓SLC10A2 | Bile acid transport | Neimark et al., 2004 | |
| ↓ABCB11 | Bile acid export | Boulias et al., 2005 | |
| ↓ABCB4 | Bile acid export | Boulias et al., 2005 | |
| ↓ABCA1 | Cholesterol efflux | Brendel et al., 2002 Boulias et al., 2005 | |
| ↓APOA1 | Reverse cholesterol transport | Delerive et al., 2004 | |
| ↓CETP | Cholesteryl easter transfer | Luo et al., 2001 | |
| ↓SCARb1 | Cholesterol uptake | Boulias et al., 2005 | |
| Lipid metabolism and Obesity | |||
| Liver | ↑PPARγ | Lipogenesis | Nishizawa et al., 2002 Boulias et al., 2005 Huang et al., 2007 |
| ↑SREBP1C | Lipogenesis | Boulias et al., 2005 Huang et al., 2007 | |
| ↑CD36 | Long chain fatty acids uptake | Boulias et al., 2005 | |
| ↑FAS | Fatty acid and triglyceride biosynthesis | Boulias et al., 2005 Huang et al., 2007 | |
| ↑ACL | Fatty acid and triglyceride biosynthesis | Boulias et al., 2005 | |
| ↑ACC-1 | Fatty acid and triglyceride biosynthesis | Boulias et al., 2005 | |
| ↑SCD-1 | Fatty acid and triglyceride biosynthesis | Boulias et al., 2005 | |
| ↓MTP | Plasma lipoprotein secretion | Hirokane et al., 2004 Huang et al., 2007 Pan et al., 2010 | |
| Brown adipose tissue | ↓UCP1 | Mitochondrial thermogenesis and biogenesis | Wang et al., 2005 |
| ↓PGC1α | Thermogenesis | Wang et al., 2005 | |
| ↓Dio2 | Thermogenesis | Wang et al., 2005 Tabbi-Anneni., 2010 | |
| ↑HSL | Lipolysis | Tabbi-Anneni., 2010 | |
| ↑LPL | Thermogenesis | Tabbi-Anneni., 2010 | |
| ↑β1AR | Mitochondrial thermogenesis and biogenesis | Tabbi-Anneni., 2010 | |
| White adipose tissue | ↓SREBP1 ↓LPL |
Lipid accumulation | Tabbi-Anneni., 2010 |
| Liver fibrosis | |||
| Liver | ↓alpha1 (I) collagen | Fibrogenesis | Fiorucci et al., 2004 |
| ↓A-SMA | |||
| ↓TIMP-1 | Regulation of metalloproteinase activity | Fiorucci et al., 2005 | |
| ↓PAI-1 | Fibrogenesis | Chanda et al., 2009 | |
| Glucose metabolism | |||
| Liver | ↓PEPCK | Rate limiting gluconeogenic enzyme | Borgius et al.,2002 Park et al., 2007 Chanda et al., 2007 |
| ↓G6Pase | Regulate blood glucose levels | Kim et al., 2004 Lee et al., 2008 Chanda et al., 2009 | |
| ↓FBP1 | Gluconeogenesis | Yamagata et al., 2004 | |
| ↓LGK | Hepatic glucokinase | Kim et al., 2009 | |
| Drug metabolism | |||
| Liver | ↓CYP2B | Drug and hormone metabolism | Bae et al., 2004 |
| ↓CYB3A | Drug and hormone metabolism | Ourlin et al., 2003 | |
| Cancer | |||
| Liver | ↓CyclinD1 | Cell cycle | Zhang et al., 2008 |
| ↓Bcl2 | Apoptosis | Zhang et al., 2010 | |
| Breast | ↓CYP19 | Estrogen production | Kovacic et al., 2004 |
| Other | ↓Gli | Hedgehog signaling pathway | Kim et al., 2010 |
↓: inhibition; ↑: induction.
Acknowledgements
YZ is supported by T32CA093247 Multidisciplinary Cancer Research Training Program (MCRTP). CHH is supported by NIH CA63640. This work is in part supported by NIDDK R01 080440 to L.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
No conflicts of interest exist for the authors.
References
- 1.Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov. 2004;3:950–964. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- 2.Seol W, Choi HS, Moore DD. An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science. 1996;272:1336–1339. doi: 10.1126/science.272.5266.1336. [DOI] [PubMed] [Google Scholar]
- 3.Lee HK, Lee YK, Park SH, Kim YS, Lee JW, Kwon HB, Soh J, Moore DD, Choi HS. Structure and expression of the orphan nuclear receptor SHP gene. J Biol Chem. 1998;273:14398–14402. doi: 10.1074/jbc.273.23.14398. [DOI] [PubMed] [Google Scholar]
- 4.Johansson L, Bavner A, Thomsen JS, Farnegardh M, Gustafsson JA, Treuter E. The orphan nuclear receptor SHP utilizes conserved LXXLL-related motifs for interactions with ligand-activated estrogen receptors. Mol Cell Biol. 2000;20:1124–1133. doi: 10.1128/mcb.20.4.1124-1133.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanyal S, Kim JY, Kim HJ, Takeda J, Lee YK, Moore DD, Choi HS. Differential regulation of the orphan nuclear receptor small heterodimer partner (SHP) gene promoter by orphan nuclear receptor ERR isoforms. J Biol Chem. 2002;277:1739–1748. doi: 10.1074/jbc.M106140200. [DOI] [PubMed] [Google Scholar]
- 7.Nishizawa H, Yamagata K, Shimomura I, Takahashi M, Kuriyama H, Kishida K, Hotta K, Nagaretani H, Maeda N, Matsuda M, Kihara S, Nakamura T, Nishigori H, Tomura H, Moore DD, Takeda J, Funahashi T, Matsuzawa Y. Small heterodimer partner, an orphan nuclear receptor, augments peroxisome proliferator-activated receptor gamma transactivation. J Biol Chem. 2002;277:1586–1592. doi: 10.1074/jbc.M104301200. [DOI] [PubMed] [Google Scholar]
- 8.Nishigori H, Tomura H, Tonooka N, Kanamori M, Yamada S, Sho K, Inoue I, Kikuchi N, Onigata K, Kojima I, Kohama T, Yamagata K, Yang Q, Matsuzawa Y, Miki T, Seino S, Kim MY, Choi HS, Lee YK, Moore DD, Takeda J. Mutations in the small heterodimer partner gene are associated with mild obesity in Japanese subjects. Proc Natl Acad Sci U S A. 2001;98:575–580. doi: 10.1073/pnas.021544398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao H, Hegele RA. Identification of polymorphisms in the human SHP1 gene. J Hum Genet. 2002;47:445–447. doi: 10.1007/s100380200062. [DOI] [PubMed] [Google Scholar]
- 10.Hung CC, Farooqi IS, Ong K, Luan J, Keogh JM, Pembrey M, Yeo GS, Dunger D, Wareham NJ, S OR. Contribution of variants in the small heterodimer partner gene to birthweight, adiposity, and insulin levels: mutational analysis and association studies in multiple populations. Diabetes. 2003;52:1288–1291. doi: 10.2337/diabetes.52.5.1288. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell SM, Weedon MN, Owen KR, Shields B, Wilkins-Wall B, Walker M, McCarthy MI, Frayling TM, Hattersley AT. Genetic variation in the small heterodimer partner gene and young-onset type 2 diabetes, obesity, and birth weight in U.K. subjects. Diabetes. 2003;52:1276–1279. doi: 10.2337/diabetes.52.5.1276. [DOI] [PubMed] [Google Scholar]
- 12.Echwald SM, Andersen KL, Sorensen TI, Larsen LH, Andersen T, Tonooka N, Tomura H, Takeda J, Pedersen O. Mutation analysis of NR0B2 among 1545 Danish men identifies a novel c.278G>A (p.G93D) variant with reduced functional activity. Hum Mutat. 2004;24:381–387. doi: 10.1002/humu.20090. [DOI] [PubMed] [Google Scholar]
- 13.Enya M, Horikawa Y, Kuroda E, Yonemaru K, Tonooka N, Tomura H, Oda N, Yokoi N, Yamagata K, Shihara N, Iizuka K, Saibara T, Seino S, Takeda J. Mutations in the small heterodimer partner gene increase morbidity risk in Japanese type 2 diabetes patients. Hum Mutat. 2008;29:E271–E277. doi: 10.1002/humu.20865. [DOI] [PubMed] [Google Scholar]
- 14.Zhou T, Zhang Y, Macchiarulo A, Yang Z, Cellanetti M, Coto E, Xu P, Pellicciari R, Wang L. Novel polymorphisms of nuclear receptor shp associated with functional and structural changes. J Biol Chem. 2010 doi: 10.1074/jbc.M110.133280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YK, Parker KL, Choi HS, Moore DD. Activation of the promoter of the orphan receptor SHP by orphan receptors that bind DNA as monomers. J Biol Chem. 1999;274:20869–20873. doi: 10.1074/jbc.274.30.20869. [DOI] [PubMed] [Google Scholar]
- 16.Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 17.Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Willson TM, Kliewer SA. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 18.Shih DQ, Bussen M, Sehayek E, Ananthanarayanan M, Shneider BL, Suchy FJ, Shefer S, Bollileni JS, Gonzalez FJ, Breslow JL, Stoffel M. Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat Genet. 2001;27:375–382. doi: 10.1038/86871. [DOI] [PubMed] [Google Scholar]
- 19.Shih DQ, Screenan S, Munoz KN, Philipson L, Pontoglio M, Yaniv M, Polonsky KS, Stoffel M. Loss of HNF-1alpha function in mice leads to abnormal expression of genes involved in pancreatic islet development and metabolism. Diabetes. 2001;50:2472–2480. doi: 10.2337/diabetes.50.11.2472. [DOI] [PubMed] [Google Scholar]
- 20.Gupta S, Stravitz RT, Dent P, Hylemon PB. Down-regulation of cholesterol 7alpha-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J Biol Chem. 2001;276:15816–15822. doi: 10.1074/jbc.M010878200. [DOI] [PubMed] [Google Scholar]
- 21.Kim HJ, Kim JY, Park YY, Choi HS. Synergistic activation of the human orphan nuclear receptor SHP gene promoter by basic helix-loop-helix protein E2A and orphan nuclear receptor SF-1. Nucleic Acids Res. 2003;31:6860–6872. doi: 10.1093/nar/gkg906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodwin B, Watson MA, Kim H, Miao J, Kemper JK, Kliewer SA. Differential regulation of rat and human CYP7A1 by the nuclear oxysterol receptor liver X receptor-alpha. Mol Endocrinol. 2003;17:386–394. doi: 10.1210/me.2002-0246. [DOI] [PubMed] [Google Scholar]
- 23.Lai K, Harnish DC, Evans MJ. Estrogen receptor alpha regulates expression of the orphan receptor small heterodimer partner. J Biol Chem. 2003;278:36418–36429. doi: 10.1074/jbc.M303913200. [DOI] [PubMed] [Google Scholar]
- 24.Kim HJ, Kim JY, Park SK, Seo JH, Kim JB, Lee IK, Kim KS, Choi HS. Differential regulation of human and mouse orphan nuclear receptor small heterodimer partner promoter by sterol regulatory element binding protein-1. J Biol Chem. 2004;279:28122–28131. doi: 10.1074/jbc.M313302200. [DOI] [PubMed] [Google Scholar]
- 25.Frank C, Makkonen H, Dunlop TW, Matilainen M, Vaisanen S, Carlberg C. Identification of pregnane X receptor binding sites in the regulatory regions of genes involved in bile acid homeostasis. J Mol Biol. 2005;346:505–519. doi: 10.1016/j.jmb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Oiwa A, Kakizawa T, Miyamoto T, Yamashita K, Jiang W, Takeda T, Suzuki S, Hashizume K. Synergistic regulation of the mouse orphan nuclear receptor SHP gene promoter by CLOCK-BMAL1 and LRH-1. Biochem Biophys Res Commun. 2007;353:895–901. doi: 10.1016/j.bbrc.2006.12.131. [DOI] [PubMed] [Google Scholar]
- 27.Kim HI, Koh YK, Kim TH, Kwon SK, Im SS, Choi HS, Kim KS, Ahn YH. Transcriptional activation of SHP by PPAR-gamma in liver. Biochem Biophys Res Commun. 2007;360:301–306. doi: 10.1016/j.bbrc.2007.05.171. [DOI] [PubMed] [Google Scholar]
- 28.Chanda D, Li T, Song KH, Kim YH, Sim J, Lee CH, Chiang JY, Choi HS. Hepatocyte growth factor family negatively regulates hepatic gluconeogenesis via induction of orphan nuclear receptor small heterodimer partner in primary hepatocytes. J Biol Chem. 2009;284:28510–28521. doi: 10.1074/jbc.M109.022244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S, Lai K, Moy FJ, Bhat A, Hartman HB, Evans MJ. The nuclear hormone receptor farnesoid X receptor (FXR) is activated by androsterone. Endocrinology. 2006;147:4025–4033. doi: 10.1210/en.2005-1485. [DOI] [PubMed] [Google Scholar]
- 30.Ellis E, Axelson M, Abrahamsson A, Eggertsen G, Thorne A, Nowak G, Ericzon BG, Bjorkhem I, Einarsson C. Feedback regulation of bile acid synthesis in primary human hepatocytes: evidence that CDCA is the strongest inhibitor. Hepatology. 2003;38:930–938. doi: 10.1053/jhep.2003.50394. [DOI] [PubMed] [Google Scholar]
- 31.Nishimaki-Mogami T, Une M, Fujino T, Sato Y, Tamehiro N, Kawahara Y, Shudo K, Inoue K. Identification of intermediates in the bile acid synthetic pathway as ligands for the farnesoid X receptor. J Lipid Res. 2004;45:1538–1545. doi: 10.1194/jlr.M400102-JLR200. [DOI] [PubMed] [Google Scholar]
- 32.Cui J, Huang L, Zhao A, Lew JL, Yu J, Sahoo S, Meinke PT, Royo I, Pelaez F, Wright SD. Guggulsterone is a farnesoid X receptor antagonist in coactivator association assays but acts to enhance transcription of bile salt export pump. J Biol Chem. 2003;278:10214–10220. doi: 10.1074/jbc.M209323200. [DOI] [PubMed] [Google Scholar]
- 33.Kanaya E, Shiraki T, Jingami H. The nuclear bile acid receptor FXR is activated by PGC-1alpha in a ligand-dependent manner. Biochem J. 2004;382:913–921. doi: 10.1042/BJ20040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rizzo G, Renga B, Antonelli E, Passeri D, Pellicciari R, Fiorucci S. The methyl transferase PRMT1 functions as co-activator of farnesoid X receptor (FXR)/9-cis retinoid X receptor and regulates transcription of FXR responsive genes. Mol Pharmacol. 2005;68:551–558. doi: 10.1124/mol.105.012104. [DOI] [PubMed] [Google Scholar]
- 35.Whitby RJ, Dixon S, Maloney PR, Delerive P, Goodwin BJ, Parks DJ, Willson TM. Identification of small molecule agonists of the orphan nuclear receptors liver receptor homolog-1 and steroidogenic factor-1. J Med Chem. 2006;49:6652–6655. doi: 10.1021/jm060990k. [DOI] [PubMed] [Google Scholar]
- 36.Dawson MI, Xia Z, Liu G, Ye M, Fontana JA, Farhana L, Patel BB, Arumugarajah S, Bhuiyan M, Zhang XK, Han YH, Stallcup WB, Fukushi J, Mustelin T, Tautz L, Su Y, Harris DL, Waleh N, Hobbs PD, Jong L, Chao WR, Schiff LJ, Sani BP. An adamantyl-substituted retinoid-derived molecule that inhibits cancer cell growth and angiogenesis by inducing apoptosis and binds to small heterodimer partner nuclear receptor: effects of modifying its carboxylate group on apoptosis, proliferation, and protein-tyrosine phosphatase activity. J Med Chem. 2007;50:2622–2639. doi: 10.1021/jm0613323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farhana L, Dawson MI, Leid M, Wang L, Moore DD, Liu G, Xia Z, Fontana JA. Adamantyl-substituted retinoid-related molecules bind small heterodimer partner and modulate the Sin3A repressor. Cancer Res. 2007;67:318–325. doi: 10.1158/0008-5472.CAN-06-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farhana L, Dawson MI, Dannenberg JH, Xu L, Fontana JA. SHP and Sin3A expression are essential for adamantyl-substituted retinoid-related molecule-mediated nuclear factor-kappaB activation, c-Fos/c-Jun expression, and cellular apoptosis. Mol Cancer Ther. 2009;8:1625–1635. doi: 10.1158/1535-7163.MCT-08-0964. [DOI] [PubMed] [Google Scholar]
- 39.Dawson MI, Xia Z, Jiang T, Ye M, Fontana JA, Farhana L, Patel B, Xue LP, Bhuiyan M, Pellicciari R, Macchiarulo A, Nuti R, Zhang XK, Han YH, Tautz L, Hobbs PD, Jong L, Waleh N, Chao WR, Feng GS, Pang Y, Su Y. Adamantyl-substituted retinoid-derived molecules that interact with the orphan nuclear receptor small heterodimer partner: effects of replacing the 1-adamantyl or hydroxyl group on inhibition of cancer cell growth, induction of cancer cell apoptosis, and inhibition of SRC homology 2 domain-containing protein tyrosine phosphatase-2 activity. J Med Chem. 2008;51:5650–5662. doi: 10.1021/jm800456k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Soto J, Park K, Viswanath G, Kuwada S, Abel ED, Wang L. Nuclear receptor SHP, a death receptor that targets mitochondria, induces apoptosis and inhibits tumor growth. Mol Cell Biol. 2010;30:1341–1356. doi: 10.1128/MCB.01076-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johansson L, Thomsen JS, Damdimopoulos AE, Spyrou G, Gustafsson JA, Treuter E. The orphan nuclear receptor SHP inhibits agonist-dependent transcriptional activity of estrogen receptors ERalpha and ERbeta. J Biol Chem. 1999;274:345–353. doi: 10.1074/jbc.274.1.345. [DOI] [PubMed] [Google Scholar]
- 42.Lee YK, Dell H, Dowhan DH, Hadzopoulou-Cladaras M, Moore DD. The orphan nuclear receptor SHP inhibits hepatocyte nuclear factor 4 and retinoid X receptor transactivation: two mechanisms for repression. Mol Cell Biol. 2000;20:187–195. doi: 10.1128/mcb.20.1.187-195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gobinet J, Auzou G, Nicolas JC, Sultan C, Jalaguier S. Characterization of the interaction between androgen receptor and a new transcriptional inhibitor, SHP. Biochemistry. 2001;40:15369–15377. doi: 10.1021/bi011384o. [DOI] [PubMed] [Google Scholar]
- 44.Borgius LJ, Steffensen KR, Gustafsson JA, Treuter E. Glucocorticoid signaling is perturbed by the atypical orphan receptor and corepressor SHP. J Biol Chem. 2002;277:49761–49766. doi: 10.1074/jbc.M205641200. [DOI] [PubMed] [Google Scholar]
- 45.Brendel C, Schoonjans K, Botrugno OA, Treuter E, Auwerx J. The small heterodimer partner interacts with the liver X receptor alpha and represses its transcriptional activity. Mol Endocrinol. 2002;16:2065–2076. doi: 10.1210/me.2001-0194. [DOI] [PubMed] [Google Scholar]
- 46.Yeo MG, Yoo YG, Choi HS, Pak YK, Lee MO. Negative cross-talk between Nur77 and small heterodimer partner and its role in apoptotic cell death of hepatoma cells. Mol Endocrinol. 2005;19:950–963. doi: 10.1210/me.2004-0209. [DOI] [PubMed] [Google Scholar]
- 47.Kim JY, Chu K, Kim HJ, Seong HA, Park KC, Sanyal S, Takeda J, Ha H, Shong M, Tsai MJ, Choi HS. Orphan nuclear receptor small heterodimer partner, a novel corepressor for a basic helix-loop-helix transcription factor BETA2/neuroD. Mol Endocrinol. 2004;18:776–790. doi: 10.1210/me.2003-0311. [DOI] [PubMed] [Google Scholar]
- 48.Yamagata K, Daitoku H, Shimamoto Y, Matsuzaki H, Hirota K, Ishida J, Fukamizu A. Bile acids regulate gluconeogenic gene expression via small heterodimer partner-mediated repression of hepatocyte nuclear factor 4 and Foxo1. J Biol Chem. 2004;279:23158–23165. doi: 10.1074/jbc.M314322200. [DOI] [PubMed] [Google Scholar]
- 49.Suh JH, Huang J, Park YY, Seong HA, Kim D, Shong M, Ha H, Lee IK, Lee K, Wang L, Choi HS. Orphan nuclear receptor small heterodimer partner inhibits transforming growth factor-beta signaling by repressing SMAD3 transactivation. J Biol Chem. 2006;281:39169–39178. doi: 10.1074/jbc.M605947200. [DOI] [PubMed] [Google Scholar]
- 50.Lee YK, Moore DD. Dual mechanisms for repression of the monomeric orphan receptor liver receptor homologous protein-1 by the orphan small heterodimer partner. J Biol Chem. 2002;277:2463–2467. doi: 10.1074/jbc.M105161200. [DOI] [PubMed] [Google Scholar]
- 51.Bavner A, Johansson L, Toresson G, Gustafsson JA, Treuter E. A transcriptional inhibitor targeted by the atypical orphan nuclear receptor SHP. EMBO Rep. 2002;3:478–484. doi: 10.1093/embo-reports/kvf087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kemper JK, Kim H, Miao J, Bhalla S, Bae Y. Role of an mSin3A-Swi/Snf chromatin remodeling complex in the feedback repression of bile acid biosynthesis by SHP. Mol Cell Biol. 2004;24:7707–7719. doi: 10.1128/MCB.24.17.7707-7719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boulias K, Talianidis I. Functional role of G9a-induced histone methylation in small heterodimer partner-mediated transcriptional repression. Nucleic Acids Res. 2004;32:6096–6103. doi: 10.1093/nar/gkh947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fang S, Miao J, Xiang L, Ponugoti B, Treuter E, Kemper JK. Coordinated recruitment of histone methyltransferase G9a and other chromatin-modifying enzymes in SHP-mediated regulation of hepatic bile acid metabolism. Mol Cell Biol. 2007;27:1407–1424. doi: 10.1128/MCB.00944-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chanda D, Xie YB, Choi HS. Transcriptional corepressor SHP recruits SIRT1 histone deacetylase to inhibit LRH-1 transactivation. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gobinet J, Carascossa S, Cavailles V, Vignon F, Nicolas JC, Jalaguier S. SHP represses transcriptional activity via recruitment of histone deacetylases. Biochemistry. 2005;44:6312–6320. doi: 10.1021/bi047308d. [DOI] [PubMed] [Google Scholar]
- 57.Klinge CM, Jernigan SC, Risinger KE. The agonist activity of tamoxifen is inhibited by the short heterodimer partner orphan nuclear receptor in human endometrial cancer cells. Endocrinology. 2002;143:853–867. doi: 10.1210/endo.143.3.8676. [DOI] [PubMed] [Google Scholar]
- 58.Shimamoto Y, Ishida J, Yamagata K, Saito T, Kato H, Matsuoka T, Hirota K, Daitoku H, Nangaku M, Fujii H, Takeda J, Fukamizu A. Inhibitory effect of the small heterodimer partner on hepatocyte nuclear factor-4 mediates bile acid-induced repression of the human angiotensinogen gene. J Biol Chem. 2004;279:7770–7776. doi: 10.1074/jbc.M310577200. [DOI] [PubMed] [Google Scholar]
- 59.Ourlin JC, Lasserre F, Pineau T, Fabre JM, Sa-Cunha A, Maurel P, Vilarem MJ, Pascussi JM. The small heterodimer partner interacts with the pregnane X receptor and represses its transcriptional activity. Mol Endocrinol. 2003;17:1693–1703. doi: 10.1210/me.2002-0383. [DOI] [PubMed] [Google Scholar]
- 60.Klinge CM, Jernigan SC, Risinger KE, Lee JE, Tyulmenkov VV, Falkner KC, Prough RA. Short heterodimer partner (SHP) orphan nuclear receptor inhibits the transcriptional activity of aryl hydrocarbon receptor (AHR)/AHR nuclear translocator (ARNT) Arch Biochem Biophys. 2001;390:64–70. doi: 10.1006/abbi.2001.2366. [DOI] [PubMed] [Google Scholar]
- 61.Kim JY, Kim HJ, Kim KT, Park YY, Seong HA, Park KC, Lee IK, Ha H, Shong M, Park SC, Choi HS. Orphan nuclear receptor small heterodimer partner represses hepatocyte nuclear factor 3/Foxa transactivation via inhibition of its DNA binding. Mol Endocrinol. 2004;18:2880–2894. doi: 10.1210/me.2004-0211. [DOI] [PubMed] [Google Scholar]
- 62.Fiorucci S, Antonelli E, Rizzo G, Renga B, Mencarelli A, Riccardi L, Orlandi S, Pellicciari R, Morelli A. The nuclear receptor SHP mediates inhibition of hepatic stellate cells by FXR and protects against liver fibrosis. Gastroenterology. 2004;127:1497–1512. doi: 10.1053/j.gastro.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 63.Park MJ, Kong HJ, Kim HY, Kim HH, Kim JH, Cheong JH. Transcriptional repression of the gluconeogenic gene PEPCK by the orphan nuclear receptor SHP through inhibitory interaction with C/EBPalpha. Biochem J. 2007;402:567–574. doi: 10.1042/BJ20061549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim YS, Han CY, Kim SW, Kim JH, Lee SK, Jung DJ, Park SY, Kang H, Choi HS, Lee JW, Pak YK. The orphan nuclear receptor small heterodimer partner as a novel coregulator of nuclear factor-kappa b in oxidized low density lipoprotein-treated macrophage cell line RAW 264.7. J Biol Chem. 2001;276:33736–33740. doi: 10.1074/jbc.M101977200. [DOI] [PubMed] [Google Scholar]
- 65.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 66.Song G, Wang L. MiR-433 and miR-127 arise from independent overlapping primary transcripts encoded by the miR-433-127 locus. PLoS One. 2008;3:e3574. doi: 10.1371/journal.pone.0003574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Song G, Wang L. Nuclear receptor SHP activates miR-206 expression via a cascade dual inhibitory mechanism. PLoS One. 2009;4:e6880. doi: 10.1371/journal.pone.0006880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. Rna. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, Cairns BR. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 17:620–628. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fabbri M, Croce CM, Calin GA. MicroRNAs. Cancer J. 2008;14:1–6. doi: 10.1097/PPO.0b013e318164145e. [DOI] [PubMed] [Google Scholar]
- 71.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 72.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 73.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 74.Song G, Wang L. Transcriptional mechanism for the paired miR-433 and miR-127 genes by nuclear receptors SHP and ERRgamma. Nucleic Acids Res. 2008;36:5727–5735. doi: 10.1093/nar/gkn567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song G, Wang L. A conserved gene structure and expression regulation of miR-433 and miR-127 in mammals. PLoS One. 2009;4:e7829. doi: 10.1371/journal.pone.0007829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee J, Padhye A, Sharma A, Song G, Miao J, Mo YY, Wang L, Kemper JK. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J Biol Chem. 2010;285:12604–12611. doi: 10.1074/jbc.M109.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boulias K, Katrakili N, Bamberg K, Underhill P, Greenfield A, Talianidis I. Regulation of hepatic metabolic pathways by the orphan nuclear receptor SHP. EMBO J. 2005;24:2624–2633. doi: 10.1038/sj.emboj.7600728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang Y, Zhang M, Eggertsen G, Chiang JY. On the mechanism of bile acid inhibition of rat sterol 12alpha-hydroxylase gene (CYP8B1) transcription: roles of alpha-fetoprotein transcription factor and hepatocyte nuclear factor 4alpha. Biochim Biophys Acta. 2002;1583:63–73. doi: 10.1016/s1388-1981(02)00186-5. [DOI] [PubMed] [Google Scholar]
- 79.Denson LA, Sturm E, Echevarria W, Zimmerman TL, Makishima M, Mangelsdorf DJ, Karpen SJ. The orphan nuclear receptor, shp, mediates bile acid-induced inhibition of the rat bile acid transporter, ntcp. Gastroenterology. 2001;121:140–147. doi: 10.1053/gast.2001.25503. [DOI] [PubMed] [Google Scholar]
- 80.Neimark E, Chen F, Li X, Shneider BL. Bile acid-induced negative feedback regulation of the human ileal bile acid transporter. Hepatology. 2004;40:149–156. doi: 10.1002/hep.20295. [DOI] [PubMed] [Google Scholar]
- 81.Delerive P, Galardi CM, Bisi JE, Nicodeme E, Goodwin B. Identification of liver receptor homolog-1 as a novel regulator of apolipoprotein AI gene transcription. Mol Endocrinol. 2004;18:2378–2387. doi: 10.1210/me.2004-0132. [DOI] [PubMed] [Google Scholar]
- 82.Luo Y, Liang CP, Tall AR. The orphan nuclear receptor LRH-1 potentiates the sterol-mediated induction of the human CETP gene by liver X receptor. J Biol Chem. 2001;276:24767–24773. doi: 10.1074/jbc.M100912200. [DOI] [PubMed] [Google Scholar]
- 83.Peet DJ, Turley SD, Ma W, Janowski BA, Lobaccaro JM, Hammer RE, Mangelsdorf DJ. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- 84.Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 85.Axelson M, Shoda J, Sjovall J, Toll A, Wikvall K. Cholesterol is converted to 7 alpha-hydroxy-3-oxo-4-cholestenoic acid in liver mitochondria. Evidence for a mitochondrial sterol 7 alpha-hydroxylase. J Biol Chem. 1992;267:1701–1704. [PubMed] [Google Scholar]
- 86.Payne DW, Shackleton C, Toms H, Ben-Shlomo I, Kol S, deMoura M, Strauss JF, Adashi EY. A novel nonhepatic hydroxycholesterol 7 alpha-hydroxylase that is markedly stimulated by interleukin-1 beta. Characterization in the immature rat ovary. J Biol Chem. 1995;270:18888–18896. doi: 10.1074/jbc.270.32.18888. [DOI] [PubMed] [Google Scholar]
- 87.Kerr TA, Saeki S, Schneider M, Schaefer K, Berdy S, Redder T, Shan B, Russell DW, Schwarz M. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev Cell. 2002;2:713–720. doi: 10.1016/s1534-5807(02)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang L, Lee YK, Bundman D, Han Y, Thevananther S, Kim CS, Chua SS, Wei P, Heyman RA, Karin M, Moore DD. Redundant pathways for negative feedback regulation of bile acid production. Dev Cell. 2002;2:721–731. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 89.Dent P, Fang Y, Gupta S, Studer E, Mitchell C, Spiegel S, Hylemon PB. Conjugated bile acids promote ERK1/2 and AKT activation via a pertussis toxin-sensitive mechanism in murine and human hepatocytes. Hepatology. 2005;42:1291–1299. doi: 10.1002/hep.20942. [DOI] [PubMed] [Google Scholar]
- 90.Holt JA, Luo G, Billin AN, Bisi J, McNeill YY, Kozarsky KF, Donahee M, Wang DY, Mansfield TA, Kliewer SA, Goodwin B, Jones SA. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 2003;17:1581–1591. doi: 10.1101/gad.1083503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, Gerard RD, Repa JJ, Mangelsdorf DJ, Kliewer SA. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 92.Chiang JY, Stroup D. Identification and characterization of a putative bile acid-responsive element in cholesterol 7 alpha-hydroxylase gene promoter. J Biol Chem. 1994;269:17502–17507. [PubMed] [Google Scholar]
- 93.Chiang JY, Kimmel R, Stroup D. Regulation of cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription by the liver orphan receptor (LXRalpha) Gene. 2001;262:257–265. doi: 10.1016/s0378-1119(00)00518-7. [DOI] [PubMed] [Google Scholar]
- 94.Agellon LB, Drover VA, Cheema SK, Gbaguidi GF, Walsh A. Dietary cholesterol fails to stimulate the human cholesterol 7alpha-hydroxylase gene (CYP7A1) in transgenic mice. J Biol Chem. 2002;277:20131–20134. doi: 10.1074/jbc.C200105200. [DOI] [PubMed] [Google Scholar]
- 95.Chen JY, Levy-Wilson B, Goodart S, Cooper AD. Mice expressing the human CYP7A1 gene in the mouse CYP7A1 knock-out background lack induction of CYP7A1 expression by cholesterol feeding and have increased hypercholesterolemia when fed a high fat diet. J Biol Chem. 2002;277:42588–42595. doi: 10.1074/jbc.M205117200. [DOI] [PubMed] [Google Scholar]
- 96.Stroup D, Crestani M, Chiang JY. Identification of a bile acid response element in the cholesterol 7 alpha-hydroxylase gene CYP7A. Am J Physiol. 1997;273:G508–G517. doi: 10.1152/ajpgi.1997.273.2.G508. [DOI] [PubMed] [Google Scholar]
- 97.Miao J, Fang S, Lee J, Comstock C, Knudsen KE, Kemper JK. Functional specificities of Brm and Brg-1 Swi/Snf ATPases in the feedback regulation of hepatic bile acid biosynthesis. Mol Cell Biol. 2009;29:6170–6181. doi: 10.1128/MCB.00825-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mataki C, Magnier BC, Houten SM, Annicotte JS, Argmann C, Thomas C, Overmars H, Kulik W, Metzger D, Auwerx J, Schoonjans K. Compromised intestinal lipid absorption in mice with a liver-specific deficiency of liver receptor homolog 1. Mol Cell Biol. 2007;27:8330–8339. doi: 10.1128/MCB.00852-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee YK, Schmidt DR, Cummins CL, Choi M, Peng L, Zhang Y, Goodwin B, Hammer RE, Mangelsdorf DJ, Kliewer SA. Liver receptor homolog-1 regulates bile acid homeostasis but is not essential for feedback regulation of bile acid synthesis. Mol Endocrinol. 2008;22:1345–1356. doi: 10.1210/me.2007-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li T, Chiang JY. Mechanism of rifampicin and pregnane X receptor inhibition of human cholesterol 7 alpha-hydroxylase gene transcription. Am J Physiol Gastrointest Liver Physiol. 2005;288:G74–G84. doi: 10.1152/ajpgi.00258.2004. [DOI] [PubMed] [Google Scholar]
- 101.Zhang M, Chiang JY. Transcriptional regulation of the human sterol 12alpha-hydroxylase gene (CYP8B1): roles of heaptocyte nuclear factor 4alpha in mediating bile acid repression. J Biol Chem. 2001;276:41690–41699. doi: 10.1074/jbc.M105117200. [DOI] [PubMed] [Google Scholar]
- 102.Wang L, Han Y, Kim CS, Lee YK, Moore DD. Resistance of SHP-null mice to bile acid-induced liver damage. J Biol Chem. 2003;278:44475–44481. doi: 10.1074/jbc.M305258200. [DOI] [PubMed] [Google Scholar]
- 103.Kim I, Ahn SH, Inagaki T, Choi M, Ito S, Guo GL, Kliewer SA, Gonzalez FJ. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J Lipid Res. 2007;48:2664–2672. doi: 10.1194/jlr.M700330-JLR200. [DOI] [PubMed] [Google Scholar]
- 104.Miao J, Xiao Z, Kanamaluru D, Min G, Yau PM, Veenstra TD, Ellis E, Strom S, Suino-Powell K, Xu HE, Kemper JK. Bile acid signaling pathways increase stability of Small Heterodimer Partner (SHP) by inhibiting ubiquitin-proteasomal degradation. Genes Dev. 2009;23:986–996. doi: 10.1101/gad.1773909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gavrilova O, Haluzik M, Matsusue K, Cutson JJ, Johnson L, Dietz KR, Nicol CJ, Vinson C, Gonzalez FJ, Reitman ML. Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J Biol Chem. 2003;278:34268–34276. doi: 10.1074/jbc.M300043200. [DOI] [PubMed] [Google Scholar]
- 106.Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, Heyman RA, Moore DD, Auwerx J. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Matsukuma KE, Wang L, Bennett MK, Osborne TF. A key role for orphan nuclear receptor liver receptor homologue-1 in activation of fatty acid synthase promoter by liver X receptor. J Biol Chem. 2007;282:20164–20171. doi: 10.1074/jbc.M702895200. [DOI] [PubMed] [Google Scholar]
- 108.Hirokane H, Nakahara M, Tachibana S, Shimizu M, Sato R. Bile acid reduces the secretion of very low density lipoprotein by repressing microsomal triglyceride transfer protein gene expression mediated by hepatocyte nuclear factor-4. J Biol Chem. 2004;279:45685–45692. doi: 10.1074/jbc.M404255200. [DOI] [PubMed] [Google Scholar]
- 109.Huang J, Iqbal J, Saha PK, Liu J, Chan L, Hussain MM, Moore DD, Wang L. Molecular characterization of the role of orphan receptor small heterodimer partner in development of fatty liver. Hepatology. 2007;46:147–157. doi: 10.1002/hep.21632. [DOI] [PubMed] [Google Scholar]
- 110.Seaman GV, Engel R, Swank RL, Hissen W. Circadian periodicity in some physicochemical parameters of circulating blood. Nature. 1965;207:833–835. doi: 10.1038/207833a0. [DOI] [PubMed] [Google Scholar]
- 111.Schlierf G, Dorow E. Diurnal patterns of triglycerides, free fatty acids, blood sugar, and insulin during carbohydrate-induction in man and their modification by nocturnal suppression of lipolysis. J Clin Invest. 1973;52:732–740. doi: 10.1172/JCI107235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pan X, Hussain MM. Diurnal regulation of microsomal triglyceride transfer protein and plasma lipid levels. J Biol Chem. 2007;282:24707–24719. doi: 10.1074/jbc.M701305200. [DOI] [PubMed] [Google Scholar]
- 113.Pan X, Zhang Y, Wang L, Hussain MM. Diurnal Regulation of MTP and Plasma Triglyceride by CLOCK Is Mediated by SHP. Cell Metab. 12:174–186. doi: 10.1016/j.cmet.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Garaulet M, Ordovas JM, Madrid JA. The chronobiology, etiology and pathophysiology of obesity. Int J Obes (Lond) doi: 10.1038/ijo.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pietroiusti A, Neri A, Somma G, Coppeta L, Iavicoli I, Bergamaschi A, Magrini A. Incidence of metabolic syndrome among night-shift healthcare workers. Occup Environ Med. 67:54–57. doi: 10.1136/oem.2009.046797. [DOI] [PubMed] [Google Scholar]
- 116.Biggi N, Consonni D, Galluzzo V, Sogliani M, Costa G. Metabolic syndrome in permanent night workers. Chronobiol Int. 2008;25:443–454. doi: 10.1080/07420520802114193. [DOI] [PubMed] [Google Scholar]
- 117.Hartman HB, Lai K, Evans MJ. Loss of small heterodimer partner expression in the liver protects against dyslipidemia. J Lipid Res. 2009;50:193–203. doi: 10.1194/jlr.M800323-JLR200. [DOI] [PubMed] [Google Scholar]
- 118.Lehman JJ, Boudina S, Banke NH, Sambandam N, Han X, Young DM, Leone TC, Gross RW, Lewandowski ED, Abel ED, Kelly DP. The transcriptional coactivator PGC-1alpha is essential for maximal and efficient cardiac mitochondrial fatty acid oxidation and lipid homeostasis. Am J Physiol Heart Circ Physiol. 2008;295:H185–H196. doi: 10.1152/ajpheart.00081.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Samra JS. Sir David Cuthbertson Medal Lecture. Regulation of lipid metabolism in adipose tissue. Proc Nutr Soc. 2000;59:441–446. doi: 10.1017/s0029665100000604. [DOI] [PubMed] [Google Scholar]
- 120.Wang L, Liu J, Saha P, Huang J, Chan L, Spiegelman B, Moore DD. The orphan nuclear receptor SHP regulates PGC-1alpha expression and energy production in brown adipocytes. Cell Metab. 2005;2:227–238. doi: 10.1016/j.cmet.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 121.Tabbi-Anneni I, Cooksey R, Gunda V, Liu S, Mueller A, Song G, McClain DA, Wang L. Overexpression of nuclear receptor SHP in adipose tissues affects diet-induced obesity and adaptive thermogenesis. Am J Physiol Endocrinol Metab. 2010;298:E961–E970. doi: 10.1152/ajpendo.00655.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jiao J, Friedman SL, Aloman C. Hepatic fibrosis. Curr Opin Gastroenterol. 2009;25:223–229. doi: 10.1097/mog.0b013e3283279668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mann DA, Smart DE. Transcriptional regulation of hepatic stellate cell activation. Gut. 2002;50:891–896. doi: 10.1136/gut.50.6.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Friedman SL. Evolving challenges in hepatic fibrosis. Nat Rev Gastroenterol Hepatol. 2010;7:425–436. doi: 10.1038/nrgastro.2010.97. [DOI] [PubMed] [Google Scholar]
- 125.Fiorucci S, Rizzo G, Antonelli E, Renga B, Mencarelli A, Riccardi L, Orlandi S, Pruzanski M, Morelli A, Pellicciari R. A farnesoid x receptor-small heterodimer partner regulatory cascade modulates tissue metalloproteinase inhibitor-1 and matrix metalloprotease expression in hepatic stellate cells and promotes resolution of liver fibrosis. J Pharmacol Exp Ther. 2005;314:584–595. doi: 10.1124/jpet.105.084905. [DOI] [PubMed] [Google Scholar]
- 126.Fickert P, Fuchsbichler A, Moustafa T, Wagner M, Zollner G, Halilbasic E, Stoger U, Arrese M, Pizarro M, Solis N, Carrasco G, Caligiuri A, Sombetzki M, Reisinger E, Tsybrovskyy O, Zatloukal K, Denk H, Jaeschke H, Pinzani M, Trauner M. Farnesoid X receptor critically determines the fibrotic response in mice but is expressed to a low extent in human hepatic stellate cells and periductal myofibroblasts. Am J Pathol. 2009;175:2392–2405. doi: 10.2353/ajpath.2009.090114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Park YJ, Qatanani M, Chua SS, LaRey JL, Johnson SA, Watanabe M, Moore DD, Lee YK. Loss of orphan receptor small heterodimer partner sensitizes mice to liver injury from obstructive cholestasis. Hepatology. 2008;47:1578–1586. doi: 10.1002/hep.22196. [DOI] [PubMed] [Google Scholar]
- 128.Chanda D, Lee CH, Kim YH, Noh JR, Kim DK, Park JH, Hwang JH, Lee MR, Jeong KH, Lee IK, Kweon GR, Shong M, Oh GT, Chiang JY, Choi HS. Fenofibrate differentially regulates plasminogen activator inhibitor-1 gene expression via adenosine monophosphate-activated protein kinase-dependent induction of orphan nuclear receptor small heterodimer partner. Hepatology. 2009;50:880–892. doi: 10.1002/hep.23049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zelent D, Golson ML, Koeberlein B, Quintens R, van Lommel L, Buettger C, Weik-Collins H, Taub R, Grimsby J, Schuit F, Kaestner KH, Matschinsky FM. A glucose sensor role for glucokinase in anterior pituitary cells. Diabetes. 2006;55:1923–1929. doi: 10.2337/db06-0151. [DOI] [PubMed] [Google Scholar]
- 130.Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 131.Lee YS, Kim DK, Kim YD, Park KC, Shong M, Seong HA, Ha HJ, Choi HS. Orphan nuclear receptor SHP interacts with and represses hepatocyte nuclear factor-6 (HNF-6) transactivation. Biochem J. 2008;413:559–569. doi: 10.1042/BJ20071637. [DOI] [PubMed] [Google Scholar]
- 132.Ma K, Saha PK, Chan L, Moore DD. Farnesoid X receptor is essential for normal glucose homeostasis. J Clin Invest. 2006;116:1102–1109. doi: 10.1172/JCI25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yamagata K, Yoshimochi K, Daitoku H, Hirota K, Fukamizu A. Bile acid represses the peroxisome proliferator-activated receptor-gamma coactivator-1 promoter activity in a small heterodimer partner-dependent manner. Int J Mol Med. 2007;19:751–756. [PubMed] [Google Scholar]
- 134.Kobayashi M, Ikegami H, Fujisawa T, Nojima K, Kawabata Y, Noso S, Babaya N, Itoi-Babaya M, Yamaji K, Hiromine Y, Shibata M, Ogihara T. Prevention and treatment of obesity, insulin resistance, and diabetes by bile acid-binding resin. Diabetes. 2007;56:239–247. doi: 10.2337/db06-0353. [DOI] [PubMed] [Google Scholar]
- 135.Kim YD, Park KG, Lee YS, Park YY, Kim DK, Nedumaran B, Jang WG, Cho WJ, Ha J, Lee IK, Lee CH, Choi HS. Metformin inhibits hepatic gluconeogenesis through AMP-activated protein kinase-dependent regulation of the orphan nuclear receptor SHP. Diabetes. 2008;57:306–314. doi: 10.2337/db07-0381. [DOI] [PubMed] [Google Scholar]
- 136.Chanda D, Kim SJ, Lee IK, Shong M, Choi HS. Sodium arsenite induces orphan nuclear receptor SHP gene expression via AMP-activated protein kinase to inhibit gluconeogenic enzyme gene expression. Am J Physiol Endocrinol Metab. 2008;295:E368–E379. doi: 10.1152/ajpendo.00800.2007. [DOI] [PubMed] [Google Scholar]
- 137.Kim TH, Kim H, Park JM, Im SS, Bae JS, Kim MY, Yoon HG, Cha JY, Kim KS, Ahn YH. Interrelationship between liver X receptor alpha, sterol regulatory element-binding protein-1c, peroxisome proliferator-activated receptor gamma, and small heterodimer partner in the transcriptional regulation of glucokinase gene expression in liver. J Biol Chem. 2009;284:15071–15083. doi: 10.1074/jbc.M109.006742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gollamudi R, Gupta D, Goel S, Mani S. Novel orphan nuclear receptors-coregulator interactions controlling anti-cancer drug metabolism. Curr Drug Metab. 2008;9:611–613. doi: 10.2174/138920008785821701. [DOI] [PubMed] [Google Scholar]
- 139.Waxman DJ. P450 gene induction by structurally diverse xenochemicals: central role of nuclear receptors CAR, PXR, and PPAR. Arch Biochem Biophys. 1999;369:11–23. doi: 10.1006/abbi.1999.1351. [DOI] [PubMed] [Google Scholar]
- 140.Bae Y, Kemper JK, Kemper B. Repression of CAR-mediated transactivation of CYP2B genes by the orphan nuclear receptor, short heterodimer partner (SHP) DNA Cell Biol. 2004;23:81–91. doi: 10.1089/104454904322759894. [DOI] [PubMed] [Google Scholar]
- 141.Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest. 1998;102:1016–1023. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, Willson TM, Koller BH, Kliewer SA. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Staudinger J, Liu Y, Madan A, Habeebu S, Klaassen CD. Coordinate regulation of xenobiotic and bile acid homeostasis by pregnane X receptor. Drug Metab Dispos. 2001;29:1467–1472. [PubMed] [Google Scholar]
- 144.Paumgartner G. Pharmacotherapy of cholestatic liver diseases. J Dig Dis. 11:119–125. doi: 10.1111/j.1751-2980.2010.00427.x. [DOI] [PubMed] [Google Scholar]
- 145.Zein CO, Lindor KD. Latest and emerging therapies for primary biliary cirrhosis and primary sclerosing cholangitis. Curr Gastroenterol Rep. 12:13–22. doi: 10.1007/s11894-009-0079-2. [DOI] [PubMed] [Google Scholar]
- 146.Zhang Y, Xu P, Park K, Choi Y, Moore DD, Wang L. Orphan receptor small heterodimer partner suppresses tumorigenesis by modulating cyclin D1 expression and cellular proliferation. Hepatology. 2008;48:289–298. doi: 10.1002/hep.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.He N, Park K, Zhang Y, Huang J, Lu S, Wang L. Epigenetic inhibition of nuclear receptor small heterodimer partner is associated with and regulates hepatocellular carcinoma growth. Gastroenterology. 2008;134:793–802. doi: 10.1053/j.gastro.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 148.Song G, Zhang Y, Wang L. MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation. J Biol Chem. 2009;284:31921–31927. doi: 10.1074/jbc.M109.046862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Grasl-Kraupp B, Ruttkay-Nedecky B, Mullauer L, Taper H, Huber W, Bursch W, Schulte-Hermann R. Inherent increase of apoptosis in liver tumors: implications for carcinogenesis and tumor regression. Hepatology. 1997;25:906–912. doi: 10.1002/hep.510250420. [DOI] [PubMed] [Google Scholar]
- 150.Kim K, Kim KH, Cho HK, Kim HY, Kim HH, Cheong J. SHP (small heterodimer partner) suppresses the transcriptional activity and nuclear localization of Hedgehog signalling protein Gli1. Biochem J. 2010;27:413–422. doi: 10.1042/BJ20091445. [DOI] [PubMed] [Google Scholar]
- 151.Seol W, Hanstein B, Brown M, Moore DD. Inhibition of estrogen receptor action by the orphan receptor SHP (short heterodimer partner) Mol Endocrinol. 1998;12:1551–1557. doi: 10.1210/mend.12.10.0184. [DOI] [PubMed] [Google Scholar]
- 152.Simpson E, Rubin G, Clyne C, Robertson K, O'Donnell L, Jones M, Davis S. The role of local estrogen biosynthesis in males and females. Trends Endocrinol Metab. 2000;11:184–188. doi: 10.1016/s1043-2760(00)00254-x. [DOI] [PubMed] [Google Scholar]
- 153.Mouridsen H, Gershanovich M, Sun Y, Perez-Carrion R, Boni C, Monnier A, Apffelstaedt J, Smith R, Sleeboom HP, Janicke F, Pluzanska A, Dank M, Becquart D, Bapsy PP, Salminen E, Snyder R, Lassus M, Verbeek JA, Staffler B, Chaudri-Ross HA, Dugan M. Superior efficacy of letrozole versus tamoxifen as first-line therapy for postmenopausal women with advanced breast cancer: results of a phase III study of the International Letrozole Breast Cancer Group. J Clin Oncol. 2001;19:2596–2606. doi: 10.1200/JCO.2001.19.10.2596. [DOI] [PubMed] [Google Scholar]
- 154.Rubin GL, Duong JH, Clyne CD, Speed CJ, Murata Y, Gong C, Simpson ER. Ligands for the peroxisomal proliferator-activated receptor gamma and the retinoid X receptor inhibit aromatase cytochrome P450 (CYP19) expression mediated by promoter II in human breast adipose. Endocrinology. 2002;143:2863–2871. doi: 10.1210/endo.143.8.8932. [DOI] [PubMed] [Google Scholar]
- 155.Kovacic A, Speed CJ, Simpson ER, Clyne CD. Inhibition of aromatase transcription via promoter II by short heterodimer partner in human preadipocytes. Mol Endocrinol. 2004;18:252–259. doi: 10.1210/me.2003-0211. [DOI] [PubMed] [Google Scholar]
- 156.Swales KE, Korbonits M, Carpenter R, Walsh DT, Warner TD, Bishop-Bailey D. The farnesoid X receptor is expressed in breast cancer and regulates apoptosis and aromatase expression. Cancer Res. 2006;66:10120–10126. doi: 10.1158/0008-5472.CAN-06-2399. [DOI] [PubMed] [Google Scholar]