Abstract
Speed and processivity of replicative DNA polymerases can be enhanced via coupling to a sliding clamp. Due to the closed ring shape of the clamp, a clamp loader protein, belonging to the AAA+ class of ATPases, needs to open the ring-shaped clamp before loading it to DNA. Here, we developed real-time fluorescence assays to study the clamp (PCNA) and the clamp loader (RFC) from the mesophilic archaeon Methanosarcina acetivorans. Unexpectedly, we discovered that RFC can assemble a PCNA ring from monomers in solution. A motion-based DNA polymerization assay showed that the PCNA assembled by RFC is functional. This PCNA assembly activity required the ATP-bound conformation of RFC. Our work demonstrates a reverse-chaperoning activity for an AAA+ protein that can act as a template for the assembly of another protein complex.
Introduction
Large chromosome replication requires fast and reliable polymerization of thousands of bases of DNA. To enhance the rate and the processivity with which a replicative polymerase can synthesize DNA, a ring-shaped sliding clamp follows the polymerase tethering it to the DNA (1–3). DNA sliding clamps are essential, structurally conserved proteins that encircle and slide along DNA (4). Because the clamp has the closed topology of a toroid and the DNA is essentially an infinitely long cylinder, the clamp must be opened before being loaded onto DNA. The proteins that load the clamp to the DNA, or the clamp loaders, are called the γ-complex in bacteria and replication factor C (RFC) in eukaryotes and archaea. The clamp loader belongs to the family of proteins called AAA+ (ATPases associated with a variety of cellular activities) (5) which function in diverse processes such as vesicle fusion (6), protein degradation (7), cargo transport, and nucleic acids translocation and unwinding (8,9).
The structure of the clamps is highly conserved across domains of life from Bacteria, to Archaea, to Eukarya and even bacteriophage (10–13). In bacteria, the clamp is called the β-clamp. For eukaryotes and archaea, the clamp is named proliferating cell nuclear antigen (PCNA). PCNA serves as a nexus for replication and other DNA metabolic transactions via interaction with other proteins, and is modified by ubiquitin and SUMO (14).
The clamp loader protein in all organisms is composed of five subunits that are members of the AAA+ protein family (15). The eukaryotic RFC is composed of five distinct subunits (one large and four similar small subunits), but most of the archaeal RFCs have only two. Here, we have studied a recently described mesophilic archaeal RFC from Methanosarcina acetivorans (Mac) using fluorescence-based biophysical assays. MacRFC is made of three gene products and a functional and stable RFC complex can form only when all three proteins, RFCL, RFCS1, and RFCS2, are coexpressed to form a pentameric complex (16,17). Remarkably, we found that MacRFC dramatically enhances the assembly of the PCNA trimer. This activity did not derive from stabilization of transiently formed PCNA trimers. Rather, ATP-bound MacRFC provided a template for the assembly of PCNA trimer from monomers in solution. This is an example of an AAA+ protein functioning as a reverse-chaperone where it provides a template for the assembly of another protein complex.
Methods
DNA constructs and proteins
All DNA were purchased from Integrated DNA Technologies (Coralville, IA). DNA sequences and annealing protocols are found in the Supporting Material. All proteins were expressed and purified as previously described in Chen et al. (16).
Primer extension and gel filtration chromatography
Primer extension was resolved on 1% agarose gel electrophoresis followed by visualization using autoradiography. An oligonucleotide that is complementary to position 6205–6234 of the M13mp18 (+) strand was 5′ end-labeled with [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs, Ipswich, MA). One pmol of the end-labeled primer was annealed to 0.5 μg of the template (M13mp18 (+) circular single-stranded DNA (ssDNA)). The primer extension reaction (20 μL) contained 20 mM Tris-HCl (pH 8.8), 100 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol, 1 mM ATP, 250 μM of each dNTP, 0.5 μg of MacPolBI, 0.4 μg of MacRFC complex, and different amounts (300, 200, 100, 50, 25, or 0 nM) of MacPCNA, nonlabeled or labeled, per reaction. Each reaction mixture was incubated at 37°C for 5 min, followed by termination through addition of 6 μL of stop solution (98% formamide, 1 mM EDTA, 0.1% xylene cyanol, and 0.1% bromophenol blue). After heating to 95°C for 5 min, the samples were resolved on 1% alkali agarose gel in 50 mM sodium hydroxide and visualized by autoradiography.
Gel filtration chromatography for estimation of the molecular sizes of the M. acetivorans PCNA at different concentrations in solution was performed with the SMART system (GE Healthcare, Kings Park, NY). The purified M. acetivorans PCNA was applied to a Superdex 200 3.2/30 column (GE Healthcare) and preequilibrated with buffer C (50 mM Tris-HCl at pH 8.0, 0.5 mM dithiothreitol (DTT), 0.1 mM EDTA, and 0.15 M NaCl). Elution of proteins was monitored by absorbance at 280 nm. Proteins in eluted fractions were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis. For visualization, the proteins at 30 μM and 5 μM were stained with Coomassie Brilliant Blue, whereas staining of fractions from the analysis at 0.5 μM was done through silver-staining. All protein concentrations are in PCNA trimers (except when otherwise noted).
FRET measurements
All fluorescence measurements were performed on a Cary Eclipse fluorometer (Varian, Cary, NC) at 23(±1)°C except for those performed at 37(±1)°C as stated. Fluorescence data were collected in a reaction buffer of 25 mM Tris (pH 8.0), 10 mM MgCl2, 0.1 mM DTT, and 0.05 mg/mL bovine serum albumin. We confirmed that PCNA loading reaction also occurs efficiently when 50 mM NaCl is added to the reaction (data not shown). The donor, Cy3, was excited at 515 nm, and emission was collected at 565 nm (Cy3) and 665 nm (Cy5). FRET was calculated by Cy5 intensity divided by the sum of Cy3 and Cy5 intensities after subtracting acceptor emission due to direct excitation and background signal. Labeling heterogeneities of PCNA and DNA were not corrected for in this ensemble study. Therefore, the reported FRET values should be considered as a relative measure of proximity or intermolecular interaction rather than of absolute distance. In addition, different preparations of labeled PCNA had different labeling efficiencies. As a result, absolute FRET values could not be compared across different PCNA preparations. All FRET data shown in the same graph in this work were obtained from the same PCNA preparations so that comparison among the dataset within each graph is meaningful.
Labeling of PCNA
PCNA protein was labeled with Cy3 or Cy5 maleimide reactive fluorescent probes (Amersham Biosciences, GE Healthcare). The location of the cysteines was approximated by aligning the sequence of MacPCNA (GenBank ID: AAM03564) with human PCNA (GenBank ID: NP_872590) using the software ClustalX (http://www.clustal.org/). Using the crystal structure of the human PCNA (Protein DataBank ID: 1AXC (18)), the approximate location of the PCNA cysteines can be inferred to be near the surface of the protein and most likely available for labeling by maleimide reactive dyes.
The PCNA storage buffer was exchanged with a DTT-deficient storage buffer using a 10-kDa Centricon spin column (Millipore, Billerica, MA). Ten molar excess TCEP (Tris(2-carboxyethyl) phosphine) was added to the PCNA along with a 5–10-fold molar excess dye. The sample was deoxygenated and the labeling reaction was allowed to occur overnight at 4°C while rotating in the dark. Free dyes were removed using another spin column. The concentration of the protein was measured by the Bradford method protein assay (BioRad, Hercules, CA). The concentration of the dye was measured by absorption at 550 nm for Cy3 and 650 nm for Cy5 and using extinction coefficients of 150,000 cm−1 M−1 and 250,000 cm−1 M−1, respectively. The absorption spectrum of the labeled protein was fitted with a combination of calibration spectra of the fluorophores and the protein to determine the final fluorophore-protein ratio. The ratio ranged from 1 to 4 fluorophores per trimer and was controlled by adjusting the amount of dye added and the incubation time.
Results
Loading of PCNA onto DNA by RFC
To monitor the DNA loading of its cognate clamp by MacRFC, MacPCNA was labeled with a fluorophore, Cy3, at the native cysteine residues. Sequence alignments and comparison to the human PCNA structure (18) showed that six cysteines (two on each monomer) in MacPCNA would be located in nonconserved regions of the protein and approximately out on the tips of the six globular domains such that minimal perturbation is expected of interactions with the RFC or DNA. The labeling ratio varied between preparations and ranged from one to four Cy3 molecules per trimer. Consequently, the absolute FRET values differed for different preparations, and it was not meaningful to compare the absolute FRET values across different PCNA preparations. Therefore, we made sure that the FRET values and time courses were compared only among those experiments performed using the same preparation of labeled PCNA.
Fluorescent labeling did not affect MacPCNA's capacity to stimulate DNA synthesis by its cognate DNA polymerase in a RFC-dependent manner, as described below. To test the DNA-loading activity of labeled-PCNA, we used an ensemble fluorescence clamp loading assay based on fluorescence resonance energy transfer (FRET) (19). In this assay, FRET between Cy3-PCNA and Cy5-labeled DNA, Cy5-pdDNA, is used as a spectroscopic signal of PCNA loading onto the DNA (Fig. 1 a). The partial duplex (pd) DNA had an 18-basepair duplex and a 40-nucleotide 5′ ssDNA tail, and was labeled at the junction with a Cy5 (acceptor), and a biotin at the duplex end that is linked to a neutravidin to prevent PCNA loading/unloading through the duplex end. For all experiments below, we used low DNA concentrations (≤40 nM) such that passive DNA loading of PCNA is minimal. All PCNA concentrations reported here are trimer concentrations.
Figure 1.
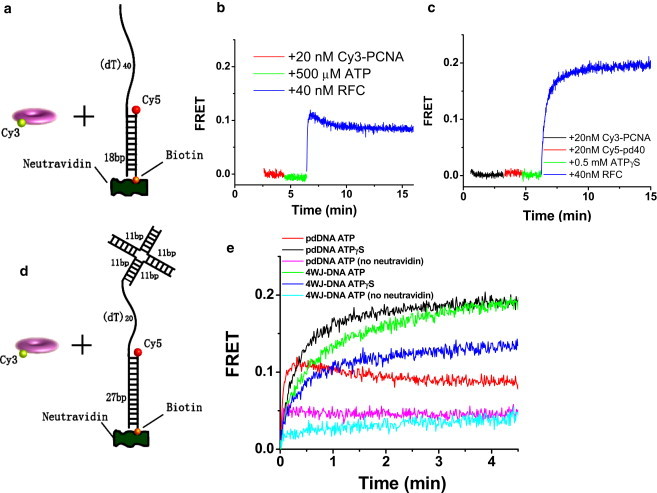
(a) Schematic of FRET assay of PCNA (labeled with Cy3 donor) loading to a 5′-tailed partial duplex DNA (labeled with Cy5 acceptor). (b) FRET measurements reveal loading of the PCNA onto DNA in an RFC-dependent manner. Rapid FRET increase upon RFC addition is due to PCNA loading onto DNA. Subsequent slow decrease in FRET is attributed to PCNA sliding off the single-stranded DNA end after it has been released upon ATP hydrolysis by RFC. (c) ATPγS instead of ATP is used to trap the RFC/DNA/PCNA complex resulting in more significant FRET increase without a slowly decreasing phase. (d) Schematic of the 4WJ-DNA used to trap the loaded PCNA. (e) FRET measurements comparing pdDNA and 4WJ-DNA show that 4WJ-DNA is able to trap the PCNA on the DNA. Experiments were done in a similar manner as in panels b and c and the time axes were shifted to denote t = 0 as the moment of RFC addition. The slower reaction with the 4WJ-DNA may be due to the steric hindrance imposed by the stacked-X structure of the four-way junction.
Fig. 1 b shows the time evolution of FRET in a typical loading experiment performed using a fluorometer. First, the Cy5-pdDNA (400 nM neutravidin is included unless otherwise mentioned) is added to a cuvette at 23°C. Next, 20 nM Cy3-PCNA is added followed by addition of 0.5 mM ATP. When 40 nM RFC is added to the solution, there is a rapid increase in FRET to ∼0.12 (half-lifetime ∼3 s) and a slow (∼2 min), small reduction to a final value of ∼0.1. The same experiment but with 500 μM ATPγS instead of ATP (Fig. 1 c) showed a similar jump in FRET when RFC is added, but this time FRET continued to increase and plateaus at ∼0.2. RFC is expected to eject from PCNA upon ATP hydrolysis (15) so that PCNA may slowly slide off the 5′ ssDNA tail of our DNA resulting in slow FRET decrease (Fig. 1 b). PCNA reloading by RFC presumably prevents FRET from reaching zero. With ATPγS, RFC remains bound to PCNA and DNA, because its conformational changes needed to eject from the PCNA cannot be achieved (20–23). We conclude that our assay using labeled PCNA and DNA recapitulates a biologically relevant function of RFC in loading PCNA onto DNA and ejecting from PCNA upon ATP hydrolysis.
To prevent the PCNA from sliding off the single-stranded end, we designed another DNA (4WJ-DNA) where the 5′ end of the tail is blocked by a DNA four-way junction (Fig. 1 d). In addition, the primer/template duplex is 27 basepairs and the single-stranded template region is 20 nucleotides. Fig. 1 e shows the Cy3-PCNA loading experiments with both types of Cy5-labeled DNA (pdDNA and 4WJ-DNA) and with ATP or ATPγS. 20 nM Cy3-PCNA, 40 nM DNA, and 0.5 mM ATP or ATPγS are prepared and then at t = 0, 40 nM RFC is added. The pdDNA shows the greatest increase in FRET with ATPγS and a smaller increase in FRET followed by slow FRET decrease with ATP, likely due to PCNA sliding off the single-stranded end as discussed above. The 4WJ-DNA, on the other hand, shows significant FRET increase without subsequent FRET decrease regardless of whether ATP or ATPγS is used. No FRET increase was observed if only the four-way junction part is used (Cy5 on the four-way junction), omitting the 3′ recessed end which is the putative site for PCNA loading by RFC (23,24) (see Fig. S1 in the Supporting Material).
If the neutravidin is not added to the DNA, much lower FRET is seen for both types of DNA, likely due to PCNA sliding off the duplex end (Fig. 1 e). Comparing the two DNA constructs with only one end blocked, pdDNA with neutravidin versus 4WJ-DNA without neutravidin, much reduced FRET amplitude is seen when the duplex end is unblocked. Therefore, PCNA must slide more easily on dsDNA than on ssDNA (4).
Labeled PCNA enhances DNA polymerization
To test the biological activity of the labeled PCNA after its loading on the DNA, we performed primer extension by a cognate DNA polymerase, MacPolBI (25), using the M13mp18 ssDNA as the template (Fig. S2). Labeling stoichiometry of 4:1 was used between Cy3 and PCNA trimer to make sure that a majority of PCNA monomers are fluorescently modified. Both unlabeled PCNA and labeled PCNA promoted the synthesis of up to ∼7-kilobase-pair-long DNA at all PCNA concentrations tested (25–300 nM) in a manner dependent on RFC, showing that labeling does not perturb MacPCNA's function to enhance the polymerase activity.
RFC-induced assembly of PCNA
Next, we prepared Cy5-labeled PCNA (Cy5-PCNA) so that PCNA oligomerization can be monitored via FRET by mixing with Cy3-PCNA. We first examined the self-association of PCNA (Fig. 2 a). Fig. 2 b shows time evolution of FRET after Cy3-PCNA and Cy5-PCNA are mixed at 1:1 ratio for various total concentrations of PCNA. FRET starts at zero and increases with time. FRET increase was negligible at 10 nM PCNA but becomes progressively larger as the PCNA concentration increases up to 200 nM, likely as a result of PCNA oligomerization. The half-lifetime of oligomer assembly, defined as the time required to reach half the maximum, decreased from 6.8 min at 40 nM PCNA to 4.0 min at 200 nM PCNA (see Table S1 in the Supporting Material).
Figure 2.
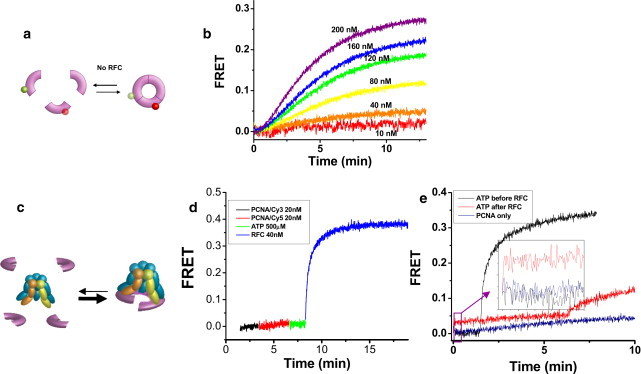
Spontaneous and RFC-induced assembly of PCNA measured via FRET. (a) PCNA trimers' spontaneous assembly and disassembly monitored by FRET between the donor and acceptor fluorophores attached to PCNA monomers. (b) Cy3-PCNA and Cy5-PCNA can spontaneously assemble (oligomerize) upon mixing, giving rise to FRET increase. Cy3-PCNA is first added to the sample in half the indicated concentration and at time equal to zero, Cy5-PCNA is added to make the total indicated concentration of PCNA. Measurements were done at 37°C. (c) RFC-templated assembly of PCNA trimers monitored by FRET. PCNA assembly is accelerated by at least 20-fold by ATP-bound RFC (as denoted by the thickness of the arrows). The cartoon also represents the model that the PCNA is held by the RFC instead of being released from RFC as discussed in the text. (d) Rapid FRET increase between Cy3-PCNA and Cy5-PCNA upon addition of 0.5 mM ATP and RFC. (e) 20 nM Cy3-PCNA and 20 nM Cy5-PCNA are in solution before time t = 0. (Black) 0.5 mM ATP is added at t = 0 followed by RFC at t = 1.5 min. (Red) RFC is added at t = 0 and 0.5 mM ATP at 6.5 min. (Blue) The self-assembly of PCNA. (Inset) Same curves, but zoomed in to ∼t = 0, to show the rapid jump in FRET for the red curve.
To examine PCNA assembly by RFC (Fig. 2 c), we prepared a solution with 20 nM Cy3-PCNA, 20 nM Cy5-PCNA, and 0.5 mM ATP. When 40 nM RFC is added, FRET starts to increase immediately with a half-lifetime of 0.3 min (Fig. 2 d), indicating that RFC enhances PCNA assembly by at least 20-fold. Control experiments performed without ATP did not show any rapid FRET increase. As we shall argue below, the assembled PCNA is very likely to be a functional trimer.
The average lifetimes of the PCNA trimer were determined by adding 1 μM unlabeled PCNA to the preassembled labeled PCNA and fitting the resulting FRET decay curve using an exponential function (Fig. S3 a). Labeled PCNA monomers, once disassembled, will be captured by the unlabeled PCNA in high excess, leading to FRET decrease. We obtained very similar (∼7 min) lifetimes for PCNA assembled spontaneously and for PCNA assembled by RFC (Table S2). ATP hydrolysis was not required for PCNA disassembly because nearly identical results were obtained with ATPγS (Fig. S3 a). To make sure that the added PCNA molecules are not inducing the disassembly of the PCNA trimer, we performed a dilution experiment where the PCNA trimer assembled by RFC was diluted by 15 folds into a buffer while FRET was being measured (Fig. S3 b). FRET decayed with a timescale very similar to that obtained by adding unlabeled PCNA. We can therefore rule out the possibility that RFC simply increases the lifetime of a spontaneously assembled PCNA trimer. Rather, RFC must be accelerating the assembly of PCNA trimers by binding individual monomers in solution, an activity that has not been described previously (to our knowledge).
ATP-bound RFC conformation is required for PCNA assembly
When RFC was added to the PCNA mixture in the absence of ATP, we observed only a small jump in FRET, <10% of that observed when ATP was present before RFC addition (Fig. 2 e, inset). Furthermore, even after a subsequent addition of ATP, a rapid FRET increase was not observed (Fig. 2 e). Without ATP, RFC appears to bind to PCNA but in a nonproductive manner and this nonproductive RFC/PCNA complex cannot be quickly rescued by adding ATP. Therefore, ATP binding is required to induce an RFC conformation that can template the efficient assembly of PCNA.
Trimer/monomer equilibrium of PCNA
To further investigate trimer formation of PCNA, we subjected various concentrations of PCNA to gel filtration chromatography (Fig. 3 a). At 10 μM and 1.7 μM concentrations of PCNA trimer (30 μM and 5 μM in monomer concentrations, respectively), PCNA existed mostly as trimers, whereas at 170 nM trimer concentration (500 nM monomer) the proteins existed mostly as monomers under the solution condition used for gel filtration. Gel filtration chromatography required high salt concentration whereas our standard reaction buffer has no added monovalent ion. Therefore, we tested whether RFC can assemble PCNA trimers under the high salt concentration (150 mM NaCl) used for the gel filtration chromatography (Fig. 3 b). Even at 40 nM trimer PCNA concentration for which the PCNA must exist primarily as monomers according to the gel filtration chromatography, we could observe evidence of rapid PCNA assembly by RFC although the reaction rate and amplitude are reduced compared to the assembly reaction performed using our standard salt condition.
Figure 3.
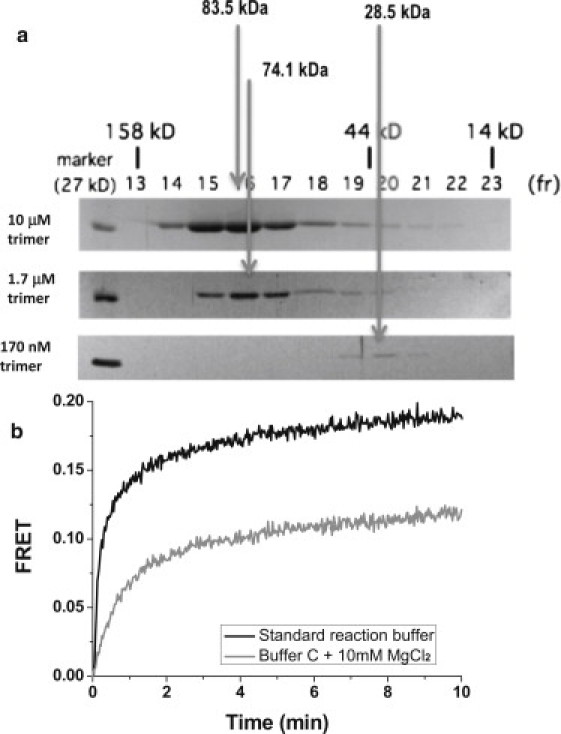
Oligomerization state of PCNA at various monomer concentrations and buffer conditions. (a) Gel filtration chromatography for estimation of the molecular sizes of the M. acetivorans PCNA at different concentrations by SMART system (GE Healthcare). The elution volume of the major peak at a concentration of 10 μM trimer was estimated as 83.5 kDa, and as the estimated molecular mass of a monomer of MacPCNA, based on the polypeptide sequence, is 26.6 kDa, the elution volume suggested a protein existing as trimers (83.5:26.6 = 3.1) in solution. The elution volume of the peak at a trimer concentration of 1.7 μM was estimated as representing a protein of molecular mass 74.1 kDa, hence this represented 2.7 monomers of PCNA, which one will assume represents a trimer, and finally the elution volume of the protein injected at 170 nM trimer concentration suggested a molecular mass of 28.5 kDa, which we assumed is a monomer. (b) RFC-induced assembly in our standard reaction buffer (black) and in the gel filtration high salt buffer C plus 10 mM MgCl2 (gray). Total PCNA concentration is 40 nM. RFC concentration is 40 nM. ATP concentration is 0.5 mM.
To test the possibility that PCNA may remain a trimer even at low concentrations and FRET changes are due to collision-induced monomer exchange between trimers (26), we carried out the spontaneous assembly experiment of labeled PCNA at 400 nM concentration and then diluted them into a cuvette down to 20, 40, or 80 nM final concentrations (Fig. S4). FRET decreased over a 3-min timescale, and more so for a larger dilution factor. If collision with trimers of unlabeled PCNA is required for FRET change as is expected according to the collision-induced monomer exchange model, a simple dilution without adding unlabeled PCNA should not give any FRET decrease. Furthermore, the monomer exchange model cannot account for the 20-fold acceleration of PCNA assembly induced by addition of RFC. Therefore, we conclude that PCNA is a monomer at low concentrations and that the FRET changes observed are due to monomer-trimer transitions.
Stoichiometric and cooperative assembly of PCNA by RFC
When RFC assembles a PCNA trimer, does it bind the first, second, and third monomers with equal affinity? Or, does the binding of the first monomer increase the subsequent binding of additional monomers (that is, is the binding cooperative)?
We addressed these questions by carrying out the FRET-based PCNA assembly experiment at a fixed PCNA concentration (20 nM Cy3-PCNA and 20 nM Cy5-PCNA trimers) for RFC concentrations ranging from 0 to 200 nM (Fig. 4 a). RFC-dependent acceleration of PCNA assembly is evident for all RFC concentrations ≥10 nM. In addition, the final amplitude is RFC-concentration-dependent; it increases linearly for up to 40 nM RFC and then plateaus at higher concentrations (Fig. 4 b). Because the total PCNA concentration is 40 nM, RFC likely assembles the stoichiometric amount of PCNA trimer rapidly and holds on to the assembled PCNA (Fig. 2 c). If RFC were to release the assembled trimer into solution shortly after the assembly, because of the slow disassembly of PCNA trimer and 20-fold faster assembly by RFC, we would have observed super-stoichiometric (or catalytic) assembly of PCNA by RFC.
Figure 4.
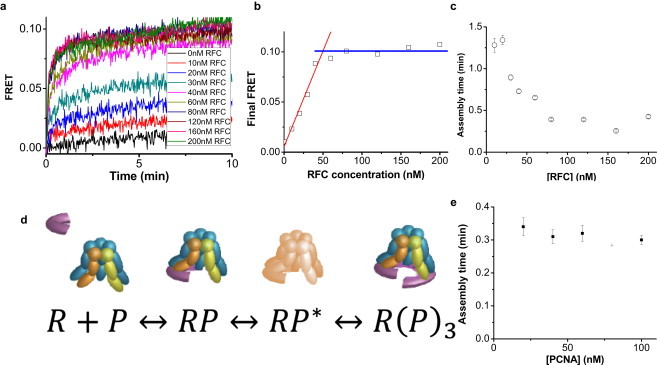
Cooperative and stoichiometric assembly of PCNA by RFC. (a) FRET versus time after adding RFC of indicated concentration to a solution containing 20 nM Cy3-PCNA, 20 nM Cy5-PCNA, and 0.5 mM ATP. (b) Amplitude of FRET change (obtained by single exponential fits to the FRET versus time curve) versus RFC concentration. (c) Average lifetime of RFC-induced PCNA assembly as measured in panel b as a function of RFC concentration. (d) Proposed kinetic mechanism of PCNA trimer assembly templated by RFC. A free RFC (R) and an RFC bound by a PCNA monomer (RP) are in rapid equilibrium. RP undergoes a conformational change to form the activated complex (RP∗), and two additional monomers bind to form R(P)3. (e) Average lifetime of the PCNA assembly reaction versus PCNA concentration at 200 nM RFC.
Because Cy3-PCNA and Cy5-PCNA are present in equimolar concentrations in 40 nM trimer concentration, the reaction mixture contained 120 nM PCNA monomers. If PCNA monomer binding to RFC is independent of how many monomers are already bound to RFC, there would be, on average, 3, 2, 1.5, 1, 0.75, and 0.6 PCNA monomers per RFC at 40, 60, 80, 120, 160, and 200 nM RFC concentrations, respectively. Therefore, one would expect a decrease in final FRET values as RFC concentration is increased above 40 nM. In contrast, we observed that the amplitude of FRET change plateaus above 40 nM (Fig. 4 b), showing that the binding of PCNA monomers to RFC is cooperative.
The average time for FRET increase saturated at RFC concentrations is >60 nM (Fig. 4 c), indicating that initial binding between RFC and PCNA monomer is not rate-limiting for the assembly of PCNA trimer under such conditions. The current model we favor is as follows (Fig. 4 d):
There is rapid equilibrium between a free RFC (R) and an RFC bound by a PCNA monomer (RP). Before binding of additional monomers to complete the assembly process, RP must undergo a conformational change to form the activated complex (RP∗) with a rate constant of ∼3 min−1, which is rate-limiting at high protein concentrations. Finally, two additional monomers bind to RP∗ to form R(P)3. The possibility that such a preactivation complex has two PCNA monomers, instead of one, bound to an RFC can be disregarded because R(P)2 would have displayed a substantial FRET. Consistent with this model where a conformational change, not binding, is rate-limiting, the time required for PCNA assembly by 200 nM RFC stayed constant within 20% when the PCNA concentration was varied by a factor of five, from 20 nM to 100 nM (Fig. 4 e).
Kinetic modeling of PCNA assembly dynamics
As a consistency check for our model of PCNA assembly by RFC, we modified the model of the previous section to include the parallel self-assembly and to include the possibility that the assembly may occur sequentially, one monomer at a time:
As detailed in Supporting Methods in the Supporting Material, we followed the statistical mechanical approach of Brown and Sethna (27) to fit the ensemble FRET data to the proposed model using the experimentally estimated constraints (kd′ = 0.15 min−1, kact = 3 min−1). A set of parameters could be determined that reproduced all experimental results reasonably well (see Fig. S6 and Table S3). Although the success in fitting the data to a model with many free parameters does not guarantee that the obtained parameters are correct or even the overall model is correct, this exercise still provides a minimal check that the model is not mathematically unfeasible. In addition, using the obtained parameters, we could deduce that, for PCNA self-assembly, the PCNA trimer concentration required for half the PCNA monomers to be in the trimeric form is ∼600 nM. This is consistent with our finding based on gel-filtration analysis that showed that PCNA is primarily a monomer at 170 nM but is primarily a trimer at 1.7 μM.
FRET-based DNA polymerization assay
As a further functional test of the PCNA trimer assembled by RFC, we asked whether the DNA polymerization reaction can be detected as an associated, physical movement of the PCNA (Fig. 5 a). After RFC loads Cy3-PCNA onto the 4WJ-DNA labeled with Cy5 at the 4WJ, an M. acetivorans translesion DNA polymerase, MacDinB, is added. Subsequently, dATP is added to start polymerization against the (dT)20 template, causing a sharp increase in FRET attributed to the movement of PCNA/DinB complex downstream toward the 4WJ (Fig. 5 b). If a mixture of dCTP, dGTP, and dTTP is added instead of dATP, no such FRET increase is observed, further supporting our interpretation (Fig. 5 c). Finally, if ATPγS is used instead of ATP such that RFC cannot release the PCNA onto the DNA and dissociate, no FRET increase is observed upon addition of dATP (Fig. S5). These FRET data show that the fluorescently labeled PCNA assembled and loaded by RFC is able to interact with the DNA polymerase during polymerization.
Figure 5.
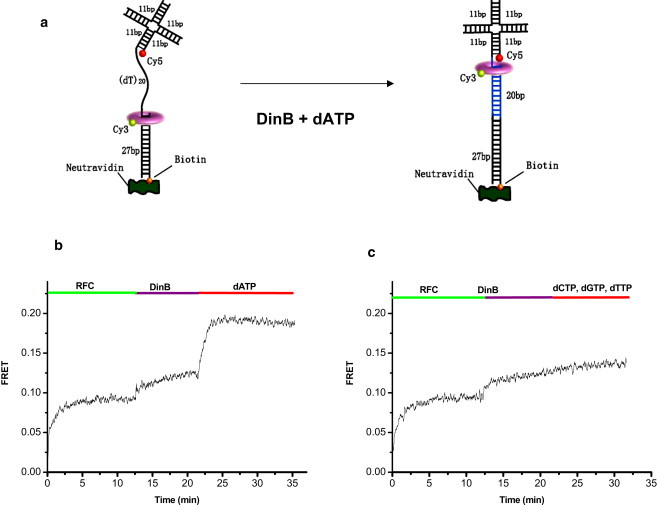
(a) A schematic of FRET-based assay for DNA polymerization by M. acetivorans DinB. If DinB polymerizes DNA and if it interacts with PCNA labeled with Cy3 during polymerization, FRET increase is expected between Cy3 on PCNA and Cy5 attached at the downstream location on the DNA. (b) Addition of RFC (80 nM) to the reaction mixture, which contained 1 mM ATP, leads to significant FRET increase due to Cy3-PCNA (20 nM) loading onto the 4WJ-DNA (50 nM). DinB (2 μM) addition causes a slight FRET increase possibly due to PCNA localization to the recessed 3′ end. Then with dATP (10 μM), a significant jump in FRET is observed. (c) In contrast, if a mixture of dCTP, dGTP, and dTTP (10 μM each) is added (that is, without dATP needed for polymerization on the poly-dT template), almost no FRET change is observed.
The PCNA concentration used in the above polymerization experiment was 20 nM at which PCNA concentration by itself exists primarily as monomers. Under this condition, PCNA loaded by RFC functionally associates with MacDinB during polymerization and therefore must be a trimer. We can therefore infer that the PCNA assembled by MacRFC is a functional trimer.
Discussion
Using FRET between labeled PCNA molecules, we could detect the spontaneous and RFC-induced assembly of PCNA and its disassembly. Low FRET observed at low PCNA concentrations in the absence of RFC is reminiscent of the observation of Yao et al. (28) that the yeast PCNA free in solution as well as those PCNA molecules that had been loaded onto circular DNA are able to dissociate into its constituent monomers at PCNA concentrations below 20 nM. Remarkably, we found that the PCNA assembly was accelerated by 20-fold by the MacRFC action that required ATP binding, but not hydrolysis.
This is the first demonstration, to our knowledge, that a clamp loader can serve as a template for assembling a replication clamp. If this observation holds true in vivo, our results suggest that MacRFC and perhaps other RFC homologs in the archaeal/eukaryotic sister lineages have evolved not only to open and load the clamp onto DNA, but also to aid in efficient assembly of the clamp. Intracellular concentration of PCNA is likely to be in the micromolar range. If most of the PCNA in vivo are in solution, that is, not bound to DNA, PCNA would primarily exist as a trimer, obviating the need for templated assembly. However, it is not known what fraction of PCNA in the cell is freely diffusing in solution instead of being bound to the DNA. If the number of freely available PCNA molecules is small in vivo then they may primarily exist as monomers. If so, our discovery of RFC-templated assembly of PCNA may have physiological relevance. Regardless, our study demonstrates what we believe to be a new intrinsic activity of an AAA+ protein that can act as a template for the assembly of another protein complex. This may be called the reverse-chaperone activity which contrasts well with the chaperoning activities of some AAA+ proteins that can disassemble protein complexes (5). Whether the reverse-chaperone activity is utilized in vivo remains to be tested.
Acknowledgments
This work was supported by the National Science Foundation (grant Nos. MCB-0238451, PHY-0822613, and PHY-0646550), the National Institutes of Health (grant No. GM065367), and the Human Frontiers Science Program. M.C.M. was supported by Agricultural Genome Sciences and Public Policy training grant No. 2001-52100-11527.
Supporting Material
References
- 1.Kelman Z., O'Donnell M. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu. Rev. Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 2.Waga S., Stillman B. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 3.Indiani C., O'Donnell M. The replication clamp-loading machine at work in the three domains of life. Nat. Rev. Mol. Cell Biol. 2006;7:751–761. doi: 10.1038/nrm2022. [DOI] [PubMed] [Google Scholar]
- 4.Stukenberg P.T., Studwell-Vaughan P.S., O'Donnell M. Mechanism of the sliding β-clamp of DNA polymerase III holoenzyme. J. Biol. Chem. 1991;266:11328–11334. [PubMed] [Google Scholar]
- 5.Ogura T., Wilkinson A.J. AAA+ superfamily ATPases: common structure—diverse function. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 6.Brunger A.T., DeLaBarre B. NSF and p97/VCP: similar at first, different at last. FEBS Lett. 2003;555:126–133. doi: 10.1016/s0014-5793(03)01107-4. [DOI] [PubMed] [Google Scholar]
- 7.Martin A., Baker T.A., Sauer R.T. Rebuilt AAA+ motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- 8.Li D., Zhao R., Chen X.S. Structure of the replicative helicase of the oncoprotein SV40 large tumor antigen. Nature. 2003;423:512–518. doi: 10.1038/nature01691. [DOI] [PubMed] [Google Scholar]
- 9.Enemark E.J., Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 10.Kong X.P., Onrust R., Kuriyan J. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 11.Krishna T.S., Kong X.P., Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 12.Matsumiya S., Ishino Y., Morikawa K. Crystal structure of an archaeal DNA sliding clamp: proliferating cell nuclear antigen from Pyrococcus furiosus. Protein Sci. 2001;10:17–23. doi: 10.1110/ps.36401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moarefi I., Jeruzalmi D., Kuriyan J. Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J. Mol. Biol. 2000;296:1215–1223. doi: 10.1006/jmbi.1999.3511. [DOI] [PubMed] [Google Scholar]
- 14.Moldovan G.L., Pfander B., Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Johnson A., O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y.H., Kocherginskaya S.A., Cann I.K. Biochemical and mutational analyses of a unique clamp loader complex in the archaeon Methanosarcina acetivorans. J. Biol. Chem. 2005;280:41852–41863. doi: 10.1074/jbc.M508684200. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y.H., Lin Y., Cann I.K. Molecular analyses of a three-subunit euryarchaeal clamp loader complex from Methanosarcina acetivorans. J. Bacteriol. 2009;191:6539–6549. doi: 10.1128/JB.00414-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gulbis J.M., Kelman Z., Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 19.Kumar R., Nashine V.C., Lee T.H. Stepwise loading of yeast clamp revealed by ensemble and single-molecule studies. Proc. Natl. Acad. Sci. USA. 2010;107:19736–19741. doi: 10.1073/pnas.1014139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgers P.M. Saccharomyces cerevisiae replication factor C. II. Formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerases δ and ɛ. J. Biol. Chem. 1991;266:22698–22706. [PubMed] [Google Scholar]
- 21.Lee S.H., Hurwitz J. Mechanism of elongation of primed DNA by DNA polymerase δ, proliferating cell nuclear antigen, and activator 1. Proc. Natl. Acad. Sci. USA. 1990;87:5672–5676. doi: 10.1073/pnas.87.15.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao N.Y., Johnson A., O'Donnell M. Mechanism of proliferating cell nuclear antigen clamp opening by replication factor C. J. Biol. Chem. 2006;281:17528–17539. doi: 10.1074/jbc.M601273200. [DOI] [PubMed] [Google Scholar]
- 23.Gomes X.V., Burgers P.M. ATP utilization by yeast replication factor C. I. ATP-mediated interaction with DNA and with proliferating cell nuclear antigen. J. Biol. Chem. 2001;276:34768–34775. doi: 10.1074/jbc.M011631200. [DOI] [PubMed] [Google Scholar]
- 24.Simonetta K.R., Kazmirski S.L., Kuriyan J. The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell. 2009;137:659–671. doi: 10.1016/j.cell.2009.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robbins J.B., Murphy M.C., Cann I.K. Functional analysis of multiple single-stranded DNA-binding proteins from Methanosarcina acetivorans and their effects on DNA synthesis by DNA polymerase BI. J. Biol. Chem. 2004;279:6315–6326. doi: 10.1074/jbc.M304491200. [DOI] [PubMed] [Google Scholar]
- 26.Stewart J., Hingorani M.M., O'Donnell M. Mechanism of β-clamp opening by the δ-subunit of Escherichia coli DNA polymerase III holoenzyme. J. Biol. Chem. 2001;276:19182–19189. doi: 10.1074/jbc.M100592200. [DOI] [PubMed] [Google Scholar]
- 27.Brown K.S., Sethna J.P. Statistical mechanical approaches to models with many poorly known parameters. Phys. Rev. E. 2003;68:021904. doi: 10.1103/PhysRevE.68.021904. [DOI] [PubMed] [Google Scholar]
- 28.Yao N., Turner J., O'Donnell M. Clamp loading, unloading and intrinsic stability of the PCNA, β and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells. 1996;1:101–113. doi: 10.1046/j.1365-2443.1996.07007.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


