Abstract
ExoU is a 74-kDa, water-soluble toxin injected directly into mammalian cells through the type III secretion system of the opportunistic pathogen, Pseudomonas aeruginosa. Previous studies have shown that ExoU is a Ca2+-independent phospholipase that requires a eukaryotic protein cofactor. One protein capable of activating ExoU and serving as a required cofactor was identified by biochemical and proteomic methods as superoxide dismutase (SOD1). In these studies, we carried out site-directed spin-labeling electron paramagnetic resonance spectroscopy to examine the effects of SOD1 and substrate liposomes on the structure and dynamics of ExoU. Local conformational changes within the catalytic site were observed in the presence of substrate liposomes, and were enhanced by the addition of SOD1 in a concentration-dependent manner. Conformational changes in the C-terminal domain of ExoU were observed upon addition of cofactor, even in the absence of liposomes. Double electron-electron resonance experiments indicated that ExoU samples multiple conformations in the resting state. In contrast, addition of SOD1 induced ExoU to adopt a single, well-defined conformation. These studies provide, to our knowledge, the first direct evidence for cofactor- and membrane-induced conformational changes in the mechanism of activation of ExoU.
Introduction
ExoU is a 74-kDa, water-soluble toxin expressed by the opportunistic pathogen Pseudomonas aeruginosa. P. aeruginosa is one of the leading causes of hospital-acquired infections (1–3), and persists chronically in individuals who suffer from cystic fibrosis, resulting in irreversible lung damage and mortality (4,5). The severity of these infections is related to the bacterium's ability to intoxicate epithelial cells with effector proteins that are directly translocated into the host cell cytosol using its type III secretion system (T3SS), a specialized injection system that bypasses receptor-mediated uptake and prevents exposure of the toxin to the extracellular milieu (6,7). ExoU is the most cytotoxic of these effector proteins, and clinical strains that secrete ExoU exhibit enhanced virulence and are directly associated with poor clinical outcomes in susceptible patients (3,8–12).
We and others have previously shown that ExoU is a Ca+2-independent phospholipase, with broad specificity against a wide range of phospholipid substrates (13–15). Although toxic during transfection/infection studies in both yeast and mammalian cells, the phospholipase activity of ExoU was initially not detectable in reactions using purified recombinant ExoU (rExoU) and liposome substrates. However, rExoU activity was detectable when the reaction was supplemented with yeast or mammalian cellular extracts (13). Further, protease treatment or heating of extracts reduced their capacity to activate rExoU (13,14), suggesting the presence of a protein cofactor. Other P. aeruginosa type III effectors also require a eukaryotic protein cofactor for activity (reviewed in Hauser (16)). Biochemical enrichments of yeast cytosolic fractions and in vitro activity studies led to the identification of Cu/Zn superoxide dismutase 1 (SOD1) as an activator of ExoU (17). Interestingly, neither bacterial SODs nor low-molecular-weight SOD-mimetics activate ExoU, and treatments that eliminate the superoxide dismutase activity of SOD1 (e.g., removal of metal ions) do not alter its ability to activate ExoU (17), indicating that the ability of SOD1 to act as a cofactor for ExoU is apparently unrelated to its enzymatic activity. Thus, the details of how SOD1 activates ExoU remain unknown.
Although little is currently known regarding the structural organization of ExoU, an N-terminal region encompassing residues 107–357 has been identified as the putative catalytic domain (13,18). Limited sequence homology with the plant patatins and human cytosolic phospholipases enabled the identification of serine 142 (S142) and aspartate 344 (D344) as catalytic residues, and alanine substitutions at either of these residues eliminated ExoU phospholipase activity, confirming their role in formation of the canonical phospholipase catalytic dyad (13,14). In contrast, the C-terminal half of ExoU (residues 351–687) extends beyond the region of homology with other phospholipases and has little similarity to other protein sequences within global databases. Nonetheless, the C-terminal domain of ExoU is essential for function. Truncation of more than nine amino acid residues from the C-terminus of ExoU dramatically abrogates activity (19), and linker-insertion mutagenesis studies and single amino-acid substitutions also indicate an essential role for the C-terminal domain of ExoU (20–23).
Additionally, suppressor mutation analysis in Saccharomyces cerevisiae identified residues 635–643 as a required motif for ExoU cytotoxicity (13). The above studies have led us to a model in which ExoU contains a relatively well-defined N-terminal catalytic domain and a C-terminal domain of unknown structure. The exact functional role of the C-terminal domain is also uncertain, but may involve mediating the interaction of ExoU with its obligatory cofactor and/or the target membrane. It is also unclear whether the cofactor acts to convert ExoU to an active conformation, recruits ExoU to its target membrane, or plays a role in both processes. To begin to address these questions, goals of this study were to determine whether interaction with cofactor and/or target liposomes produced conformational changes in either the C-terminal domain or active site region of ExoU.
To test the hypothesis that SOD1 alters the conformation of ExoU to an active state, we have carried out studies utilizing site-directed spin-labeling (SDSL) electron paramagnetic resonance (EPR) spectroscopy. SDSL is a powerful method for examining protein structure and dynamics that is not limited by a protein's size or the complexity of its environment (24–27). The SDSL technique involves modifying selectively introduced cysteine residues with a nitroxide spin label (Fig. 1), which gives rise to EPR spectra that are highly sensitive to local conformational dynamics. In addition, when two spin labels are introduced into a protein, double electron-electron resonance (DEER) methods can be used to determine the distance between the two nitroxides over a range of ∼20–60 Å (28,29), providing a complementary approach for investigating changes in protein structure (30–34). For our initial SDSL studies on ExoU, spin labels were introduced at sites near the catalytic residue S142 and within the functionally important E635–S643 motif. Both continuous-wave (cw) EPR and DEER studies demonstrate that interaction with SOD1 produces conformational changes near the active site as well as in the C-terminal domain of ExoU. These studies also indicate that ExoU is conformationally flexible in the absence of SOD1, adopting a well-defined structure only in the presence of the cofactor. To our knowledge, this is the first direct demonstration of a cofactor-induced conformational change in ExoU.
Figure 1.
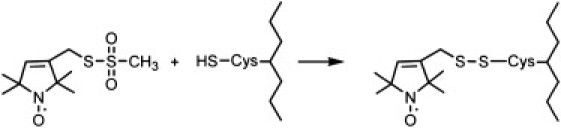
The MTSL spin label (left) reacts with the cysteine sulfhydryl group to yield the R1 side chain (right).
Materials and Methods
Reagents
Bovine Cu/Zn SOD1, Sephacryl S-200 HR resin (GE Healthcare, Piscataway, NJ), monosodium L-glutamic acid, and MES ((n-morpholino)ethanesulfonic acid) were purchased from Sigma Aldrich (St. Louis, MO). Talon cobalt affinity chromatography resin was obtained from Clontech (Mountain View, CA). PED6 (N-((6-(2,4-dinitrophenyl)amino)hexanoyl)-2-(4,4-difluoro-5,7-dimethyl-4-bora-2A,3A-diaza-s-indacene-3-pentanoyl)-1-hexadecanoyl-sn-glycero-3-phosphoethanolamine, triethylammonium salt) was from Molecular Probes (Carlsbad, CA). The methanethiosulfonate spin label MTSL (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methylmethanethiosulphonate) was purchased from Toronto Research Chemicals (North York, ON, Canada). Benzamidine Sepharose 6B resin was from GE Healthcare. Lipids POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) and POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine) were purchased from Avanti Polar Lipids (Alabaster, AL).
Bacterial strains, plasmids, and growth conditions
Escherichia coli BL21 pLysS pET15exoU and cysteine variants were cultivated in Luria-Bertani medium containing ampicillin (100 μg/mL) and chloramphenicol (30 μg/mL). Cysteine mutagenesis was performed using the Change-IT site-directed mutagenesis kit according to the manufacturer's protocol (USB, Cleveland, OH).
Protein expression and spin-labeling
The purification of rExoU and cysteine variants was performed as previously described (35), with modifications to incorporate the spin label. An overnight culture of BL21 pLysS pET15exoU was subcultured (1:60) in Luria-Bertani medium containing the appropriate antibiotics and grown at 37°C for 2 h. The expression of rExoU was induced with 0.25 mM Isopropyl β-D-1 thiogalactopyranoside for 2 h at 30°C. Soluble lysate was obtained by French press breakage in the presence of DNase, RNase, and protease inhibitors (35). After centrifugation, the cleared lysate containing histidine-tagged rExoU was incubated with Talon cobalt affinity resin that had been equilibrated with binding buffer (50 mM Na2HPO4, 5 mM imidazole, and 300 mM NaCl, pH 7) at 4°C, washed with 10 column volumes of binding buffer, and then eluted with a buffer containing 50 mM sodium phosphate, 300 mM NaCl, and 100 mM imidazole, pH 7.0. The eluted fraction was passed through Benzamidine Sepharose 6B resin (Amersham Pharmacia Biotech, Piscataway, NJ) to remove potential serine protease contaminants.
Spin labeling to convert cysteine residues to the nitroxide R1 side chain (Fig. 1) was carried out by addition of MTSL (10-fold molar excess relative to cysteine) and incubation overnight at 4°C in elution buffer containing a final concentration of 20% (v/v) glycerol. The N-terminal 6× histidine tag was removed by a 2 h digestion with 50 units of thrombin at 4°C, and the digested material was passed through Benzamidine Sepharose 6B resin (Amersham Pharmacia Biotech) to remove the thrombin. Spin-labeled rExoU was further purified and excess MTSL removed by passing the sample through a Sephacryl S-200 HR size exclusion column (1.6 × 40 cm; GE Healthcare) equilibrated with 10 mM Tris, 15 mM NaCl, and 20% glycerol, pH 7.0. Fractions with the highest protein content were pooled and concentrated at 4°C using centrifugal spin concentrators (30 kDa MWCO Amicon Ultra; Millipore, Billerica, MA) to a final concentration of ∼0.32 mM. Analysis by sodium dodecyl-sulfate polyacrylamide gel electrophoresis indicated that the protein was >95% pure as assessed by Coomassie-blue staining (see Fig. S1 in the Supporting Material).
Liposome preparation
Liposomes were prepared by drying appropriate amounts of chloroform stock solutions of POPC and POPS to give a 1:1 molar ratio under N2, followed by further removal of the solvent under vacuum. The dried lipid film was suspended at a concentration of 14 mM total lipid in 10 mM Tris, 15 mM NaCl, and 20% glycerol, pH 7.0 and sonicated (five times for 15 s each at 60% max power in a Misonix sonicator 3000; Qsonica, Newtown, CT) to produce a clear, opalescent solution of small unilamellar vesicles. Sonication debris was removed by centrifugation (5000 × g, 10 min, 4°C) in an Eppendorf microcentrifuge.
ExoU activity assay
ExoU phospholipase activity was measured using an optimized in vitro assay as recently described (35). PED6 is a nonfluorescent lipid analog containing a BIODIPY fluorophore on the sn-2 acyl chain and a dinitrophenyl quenching moiety in the headgroup. Hydrolysis of PED6 results in dequenching of the highly fluorescent BIODIPY group, producing an increase in fluorescence that is proportional to the amount of substrate cleaved. Relative phospholipase activity of a given ExoU mutant is compared to wild-type (wt) ExoU based on the maximum rate of PED6 hydrolysis.
Briefly, between 100 and 400 ng ExoU was mixed in a 96-well microtiter plate with (final concentrations) 30 μM PED6, 750 mM monosodium glutamate, 50 mM MES pH 6.3, and 10 μg bovine liver SOD1 in a final volume of 50 μL. Blank wells contained all components except SOD1. The reaction was initiated by addition of ExoU and fluorescence was read at 15 min intervals for 6 h (excitation 488 nm, emission 511 nm) on a SpectraMax M2 plate reader (Molecular Dynamics, Mountain View, CA). Recombinant wt ExoU was included in each assay as a positive control. Aliquots of recombinant wt ExoU were stored at −80°C, and gave consistent rates of PED6 hydrolysis over several months.
Electron paramagnetic resonance spectroscopy
Continuous wave (cw) EPR spectroscopy was carried out at room temperature (23 ± 1°C) on an Elexsys E500 spectrometer (Bruker Biospin, Billerica, MA) equipped with a high-Q cavity operating at X-band. Samples (32 μL total volume contained in 50 μL glass capillaries) were prepared by mixing spin-labeled ExoU, POPC/POPS (1:1) liposomes, and SOD1 (from a 2 mM stock solution in 10 mM Tris, 15 mM NaCl, and 20% glycerol, pH 7.0) to give a final concentration of 80 μM ExoU and the desired final concentrations of liposomes and/or SOD1. Spectrometer conditions were: time constant 10.28 ms, conversion time 20.48 ms, scan time 20.97 s, a 100 kHz field modulation amplitude of 1.0 G, 10 mW microwave power, and a sweep width of 100 G.
Experimental spectra were fit to the MOMD model of Budil et al. (36) using a version of the program NLSL written in LABVIEW and generously provided by Christian Altenbach and Wayne L. Hubbell (Jules Stein Eye Institute, University of California, Los Angeles). Values of the g and A tensors were fixed at gxx = 2.0078, gyy = 2.0058, gzz = 2.0022, Axx = 6.2 G, Ayy = 5.9 G, and Azz = 37 G (37,38). Angles relating the diffusion tensor to the magnetic tensor of the nitroxide spin label (α, β, γ) were assigned values of (0°, 36°, 0°) as previously described for the R1 side chain attached to the soluble protein, T4 lysozyme (37).
Spectra were fit by varying the restoring potential coefficient c20 and rotational diffusion tensor parameters (Rx, Ry, Rz), initially assuming axial symmetry (Rx = Ry) and with particular emphasis on fitting the outer hyperfine extrema. Rotational correlation times (τc = 1/6 [10〈R〉], where 〈R〉 = −[(Rx)(Ry)(Rz)]1/3) and order parameters (S) were calculated as previously described (37).
Four-pulse double electron-electron resonance (DEER) spectroscopy (28) was carried out at ∼9.5 GHz on an E580 spectrometer (Bruker Biospin) equipped with a 3-mm split-ring resonator. Samples (24 μL total volume) containing double-labeled ExoU (∼200 μM) with or without SOD and/or liposomes and a final concentration of 25% (v/v) perdeuterated glycerol (Sigma Aldrich) as cryoprotectant were placed in 2 × 2.4 mm glass capillaries and flash-frozen by immersion into liquid N2. Sample temperature during the experiment was maintained at 80 K using an Oxford cryostat. Observer pulses were positioned at the low field maximum with π and π/2 pulse lengths of 32 ns and 16 ns, respectively. Pump pulses were positioned at the center field maximum. Data were corrected for background decay assuming a homogeneous three-dimensional protein distribution and analyzed by model free Tikhonov regularization using DeerAnalysis2010 software (http://www.epr.ethz.ch/) (39). DEER acquisition times ranged between 8 and 16 h.
Results
To initiate our SDSL studies of rExoU we created seven single-cysteine variants (wild-type ExoU contains no native cysteines), including four near the catalytic serine residue S142 (I135C, S137C, S139C, and S141C) and three (A641C, T642C, and S643C) within a functionally important motif (residues 635–643) previously identified in the C-terminal domain. Analysis of SOD1-stimulated phospholipase activity indicated that I135C was catalytically inactive. S137C, S139C, and S141C retained between 45 and 63% of wild-type activity in the free thiol state, consistent with the relatively conservative Ser to Cys mutation (Table 1).
Table 1.
Phospholipase activity of rExoU cysteine mutants before and after spin labeling
| Sample | free Cys | + MTSL |
|---|---|---|
| I135C | NA∗ | NA∗ |
| S137C | 63 ± 3 | 51 ± 2 |
| S139C | 56 ± 9 | 7 ± 1 |
| S141C | 45 ± 4 | NA∗ |
| A641C | 35 ± 7 | 19 ± 4 |
| T642C | 92 ± 6 | 95 ± 3 |
| S643C | 103 ± 8 | 98 ± 5 |
Phospholipase activities are % (mean ± SE) of recombinant wild-type ExoU.
NA = No activity.
Upon spin labeling, the phospholipase activities of S139R1 and S141R1 were substantially reduced; however, S137R1 retained ∼50% of wt activity (Table 1). Cysteine mutagenesis was reasonably well tolerated for A641C (35% wt activity, 19% after spin labeling), and cysteine substitution at T642 and S643 had essentially no effect on the phospholipase activity of rExoU. T642R1 and S643R1 also retained full activity after spin labeling. These results are consistent with a wide range of studies showing that site-specific cysteine mutagenesis is generally well tolerated (e.g., 27,33,40–44), and indicate that the majority of these sites are well suited for further study by SDSL.
Conformational changes in close proximity to the active site
EPR spectra of the spin-labeled rExoU single-cysteine variants located near the catalytic S142 residue are shown in Fig. 2. Each of these sites shows the presence of a population of spin labels that is relatively immobilized, with outer hyperfine splittings in the range of 65–69 G and rotational correlation times of ∼10–20 ns (see Fig. S3 and Table S1), indicating the presence of extensive tertiary contacts that limit the mobility of the R1 side chain. Sites S135R1, S139R1, and S141R1, which retain little or no catalytic activity after spin labeling, showed no change in their EPR spectra upon the addition of either SOD1 or liposomes alone, or with the combination of liposomes and SOD1 (data not shown). In contrast, obvious spectral changes occurred for S137R1 upon the addition of liposomes and SOD1 (Fig. 3 and Fig. S4).
Figure 2.
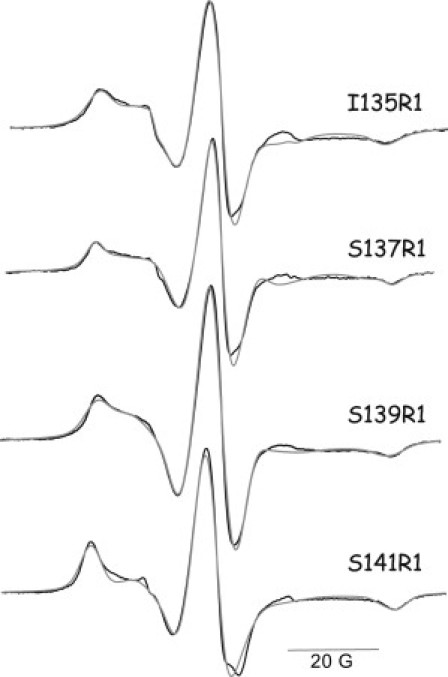
EPR spectra of MTSL-labeled cysteine variants near catalytic residue S142 of ExoU. (Solid lines) Experimental spectra. (Shaded lines) Two-component simulations. Spectra are scaled to approximately equal amplitude of the center line.
Figure 3.
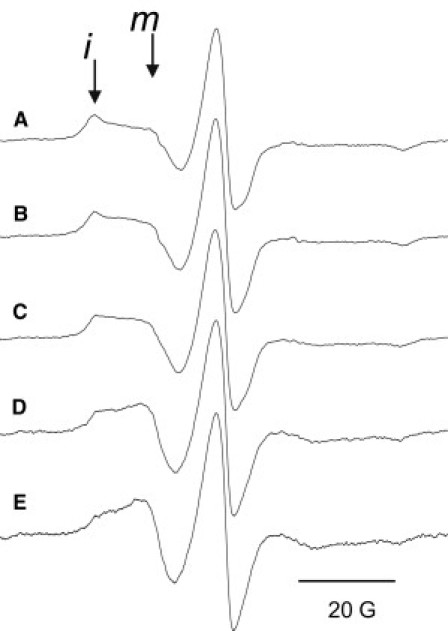
Liposome and SOD1-induced conformational changes in S137R1. Samples contained ExoU-S137R1 in 10 mM Tris pH 7.0, 15 mM NaCl, and 20% glycerol. (A) Buffer control; (B) after addition of POPC/POPS (1:1) liposomes; and (C–E) after addition of liposomes and SOD1. The SOD1:ExoU molar ratios were (C) 6.25:1, (D) 12.5:1, and (E) 25:1. The lipid/ExoU molar ratio was held constant at 44:1. Spectra are scaled to equal amplitude of the center line. Positions of the more immobile (i) and mobile (m) motional states of the nitroxide side chain are indicated. Overlays of spectra A and B and of spectra A and E are provided in Fig. S4.
As noted above, the EPR spectrum of S137R1 reflects the presence of a least two motional components, corresponding to relatively mobile (m) and immobile (i) states of the nitroxide side chain (Fig. 3, see also Fig. S3). Addition of POPC/POPS (1:1) liposomes alone produced a very slight shift toward increased mobility (Fig. 3 B and Fig. S4); no changes were observed upon addition of SOD1 alone (data not shown). However, the addition of substrate liposomes in combination with SOD1 produced substantial changes in the rotational mobility of S137R1 (Fig. 3, C–E, see also Fig. S4), changing both the motional parameters and relative populations of the mobile and immobile spin states. In the absence of either liposomes or SOD1 (i.e., Fig. 3 A), the more mobile population accounted for ∼56% of the total spin concentration, increasing to ∼80% in the presence of a 25-fold excess of SOD1 (Fig. S5). These results clearly demonstrate the occurrence of a conformational change in close proximity to the catalytic site of ExoU, indicating that the S137R1 side chain experiences increased motional freedom in the presence of SOD1 and substrate liposomes.
Conformational changes in the C-terminal domain
EPR spectra of A641R1, T642R1 and S643R1, located in the C-terminal domain of rExoU, are shown in Fig. 4. T642C and S643C retain essentially full catalytic activity as compared to wt rExoU, both before and after spin labeling, while A641C retains ∼35% of wt activity (19% after labeling) (Table 1). As shown in Fig. 4, these sites are also characterized by complex EPR spectra with relatively mobile and immobile states of the spin-label side chain. The EPR spectra of A641R1 and T642R1 were unchanged upon the addition of liposomes (not shown), and showed only small increases in the more mobile spin population in the presence of SOD1 (see Fig. S6). S643R1 also showed no change upon the addition of liposomes alone (Fig. 5 B). However, addition of SOD1 to S643R1 produced a dramatic change in which the more immobile component disappeared completely (Fig. 5 C, see also Fig. S6). The EPR spectrum of S643R1 in the presence of both liposomes and SOD1 (Fig. 5 D) was identical to that in the presence of SOD1 alone. Thus, even in the absence of substrate liposomes, interaction with SOD1 produces a conformational change that alters the structure and/or dynamics in the C-terminal domain of ExoU.
Figure 4.
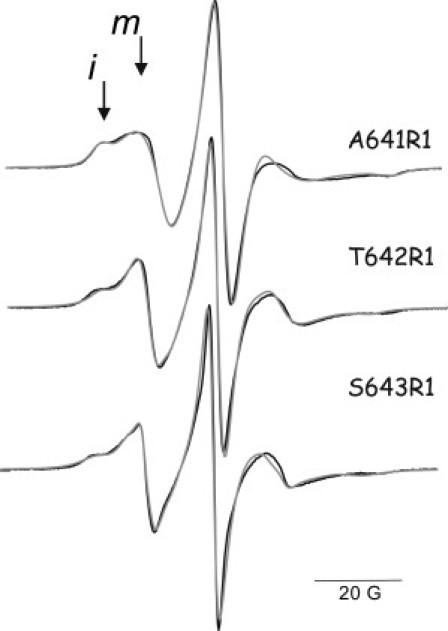
EPR spectra of MTSL-labeled cysteine mutants in the C-terminal domain of ExoU. (Solid lines) Experimental spectra. (Shaded lines) Multicomponent simulations. (Arrows) Positions of relatively mobile (m) and immobile (i) motional states of the R1 side chain.
Figure 5.
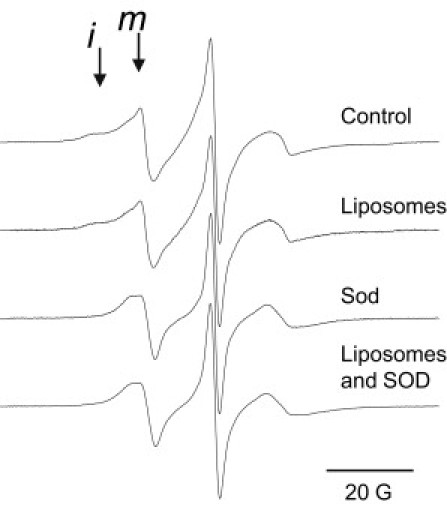
Liposome and SOD1-induced conformational changes in S643R1. Samples contained ExoU-S643R1 in buffer, after addition of liposomes, after addition of SOD1, and after addition of both liposomes and SOD1. The lipid/ExoU and SOD1/ExoU molar ratios were 44:1 and 15:1, respectively. Positions of the more immobile (i) and mobile (m) motional states of the nitroxide side chain are indicated.
Two-component EPR spectra such as those observed for A641R1, T642R1, and S643R1 can arise from either different conformational states of the protein, or from different rotameric states of the nitroxide side chain relative to the protein backbone. To distinguish between these alternatives for S643R1, we examined the effects of sucrose on the relative populations of the more mobile and immobile motional states of the spin label. It has been previously shown that protein conformational states are sensitive to the presence of osmolytes in the surrounding medium, with higher osmolyte concentrations favoring more-compact structures that typically correspond to the more-immobilized state of the spin label (45–47). In contrast, studies in several well-defined model systems have indicated that equilibria between different rotameric states of the R1 side chain are generally not sensitive to osmolytes (47). The effect of increasing sucrose concentrations on the EPR spectrum of S643R1 is shown in Fig. 6. As can be seen, the addition of sucrose caused a pronounced increase in the relative population of the more immobilized state of the R1 side chain, from 42% in the control sample to 54% in the presence of 45% (w/v) sucrose. This result suggests that the complexity observed in the EPR spectra of S643R1 arises due to the presence of distinct conformational states in the C-terminal domain.
Figure 6.
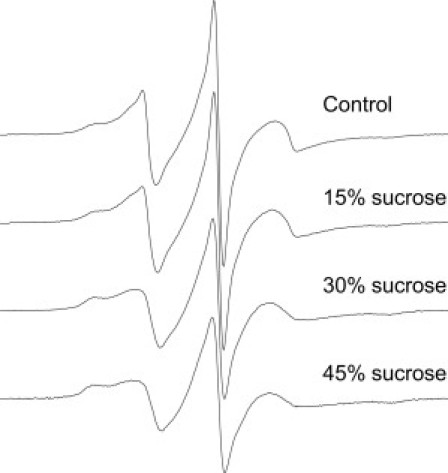
Effect of sucrose on S643R1. Comparison of the EPR spectra of S643R1 in buffer alone and in the presence of various concentrations (w/v) of sucrose. Spectra were normalized to the same integrated intensity.
DEER analysis of SOD1-induced conformational changes
To further characterize SOD1-induced conformational changes in ExoU, we used DEER spectroscopy to measure the distance between S137R1 and S643R1. In both the presence and absence of SOD1, dipole-dipole interaction between the two spin labels gave rise to modulation of the electron spin echo amplitude (Fig. 7). Model-free analysis of the dipolar evolution by Tikhonov regularization indicated that in the absence of SOD1 there exists a complex distance distribution between the two sites with a primary distance of 22 Å and lesser populations at 26.5 Å, 30.4 Å, and 38 Å. Given that the DEER echo curve for the control extended only to 1.2 μs, the peak observed at 38 Å is somewhat questionable. However, the dipolar evolution curve was poorly-fit if this distance was suppressed during the analysis (see Fig. S9). Thus, the peak at 38 Å appears to represent a population of spins with a separation distance of 38 Å or greater.
Figure 7.
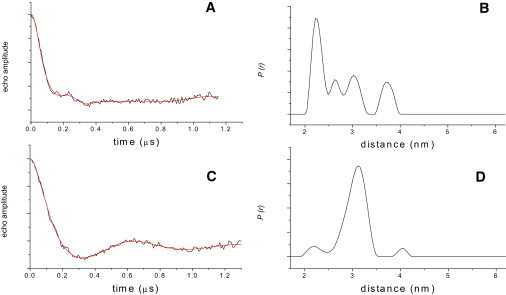
DEER evidence for a SOD1-induced conformational change in ExoU. (A and C) Dipolar evolution curves (black) and fits (red) obtained using model-free Tikhonov regularization for the S137R1-S643R1 rExoU double cysteine variant in the (A) absence and (C) presence of SOD1. (B and D) Corresponding distance distributions in the (B) absence and (D) presence of SOD1. The regularization parameter (α) was 10 for both fits. L-curves are given in Fig. S8.
In marked contrast, upon addition of SOD1 the distance distribution was dominated by a single distance at 31 Å (Fig. 7). In this case, suppression of the two minor peaks caused only a slight change in the quality of the fits (see Fig. S10), so that the presence of conformations corresponding to these distances must be considered questionable. These peaks may arise from small populations of ExoU that retain native conformations (i.e., with nitroxide-nitroxide distances of 22 Å and >38 Å, as observed in the absence of SOD1), or may arise due to artifacts of the Tikhonov regularization analysis. Nonetheless, the substantial alterations in the dipolar evolution curves and their associated distance distributions seen in Fig. 7 clearly indicate the occurrence of a SOD1-induced conformational change in ExoU. Further, these results suggest that ExoU samples multiple conformational states in the resting state, and adopts a single predominant conformation only upon interaction with its cofactor.
Discussion
ExoU, a potent phospholipase toxin produced by P. aeruginosa and injected into target cells through the type III secretion system, requires interaction with a eukaryotic cofactor for activity (13,14). The requirement of a eukaryotic activator provides an important safeguard for the bacterium, as ExoU is fully capable of hydrolyzing bacterial membrane lipids (15). We and others have previously shown that Cu/Zn SOD1 isolated from eukaryotic sources activates ExoU in vitro (17,22,23,35), by an unknown mechanism that is independent of SOD1 enzymatic activity (17). We have suggested that the ability of SOD1 to activate ExoU involves protein-protein interactions that induce a conformational change in ExoU. To our knowledge, these studies provide the first direct demonstration of conformational changes in ExoU upon the addition of SOD1 and/or substrate liposomes.
Conformational changes were observed both near the catalytic active site and at sites located in the C-terminal domain of ExoU. For S137R1, located in close proximity to catalytic residue S142, a slight increase in rotational mobility was observed in the presence of liposomes alone, suggesting that the structure of the catalytic site is sensitive to the presence of a target membrane. This increase in mobility became much more pronounced upon addition of SOD1 and liposomes in combination, and exhibited a concentration dependence on the amount of added SOD1, demonstrating the occurrence of SOD1/liposome-induced structural changes within the ExoU catalytic site.
We also observed SOD1-induced conformational changes in the C-terminal domain of ExoU, at sites A641, T642, and S643. These sites are located in a region of the protein previously identified as being important for ExoU cytotoxicity (13), despite being distal from the active site. In contrast to the effects observed with S137R1, addition of SOD1 alone was able to induce changes in the rotational mobility of a spin-label side chain introduced at these sites, without the requirement for substrate liposomes. For each of these sites, the cw EPR spectrum indicates the presence of at least two conformational states and interaction with SOD1 altered the relative populations of these states. In the case of S643R1, interaction with SOD1 completely eliminated the more restricted (immobile) conformation.
DEER results also indicate that ExoU samples multiple conformations in the absence of cofactor. For the S137R1-S643R1 pair, analysis of the dipolar evolution by Tikhonov regularization indicated multiple distinct distances, ranging from 22 Å to 38 Å with the most prominent peak at an interspin distance of 22.4 (±1.2) Å. In sharp contrast, after addition of SOD1 to the 137–643 pair we observed a single predominant peak in the DEER distance distribution, with an interspin distance of 30.6 (±2.0) Å. These results provide strong evidence that interaction of ExoU with its protein cofactor alone is sufficient to generate a substantial conformational change in ExoU.
A schematic, working model representing our current understanding of cofactor-induced conformational changes and membrane interactions for ExoU is shown in Fig. 8. In the unliganded state, ExoU exhibits conformational flexibility resulting in the observation of multiple distances between S137 (near or within the active site) and S643 in the C-terminal domain (Fig. 8 A). Similarly, several of the spin-labeling sites examined in this study, including all three in the C-terminal domain, exhibited cw EPR spectra in the resting state indicative of conformational heterogeneity. This is indicated in Fig. 8, A and B, as multiple orientations of the C-terminal domain, although the exact nature of conformational heterogeneity in the absence of cofactor (i.e., structural flexibility within either or both domains, heterogeneity in the spatial relationship between domains) is currently unknown. ExoU is apparently capable of some degree of interaction with the target membrane in the absence of cofactor, as indicated by our observation of changes in the local environment surrounding S137 upon addition of liposomes (Fig. 8 B). However, in the absence of cofactor this interaction does not result in productive phospholipase activity, either because the active site conformation is not fully formed, is not accessible to substrate, or because the phospholipid substrate cannot be properly engaged. Such a catalytically inactive state would preclude degradation of the bacterial cell membrane before export, serving as an important mechanism of protection for the bacterium from its own toxin.
Figure 8.
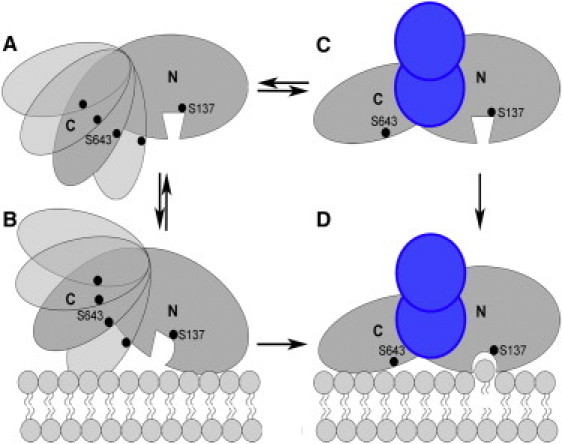
Model for the interaction of ExoU (gray) with its activating cofactor (blue) and the membrane bilayer. The observation of multiple distances between S643R1 and S137R1 suggests that the C-terminal domain may be conformationally flexible in the absence of the cofactor. SOD1 is envisioned as causing a realignment of the N- and C-terminal domains that facilitates membrane binding and exposure of the catalytic site to its lipid substrate. The conformational change in S137R1 observed in the presence of liposomes and SOD1 is represented as a change in the shape of the active site.
Our data suggests that, upon interaction with a eukaryotic cofactor, a catalytically active conformation of ExoU is selected or stabilized, with a single, well-defined distance between S137 and S643, even in the absence of liposomes (Fig. 8 C). Finally, membrane association of ExoU in the presence of the activating cofactor results in full catalytic activity, with evident conformational changes in the active site that are dependent on the presence of the cofactor (Fig. 8 D). In Fig. 8 D, we also indicate an interaction between the C-terminal domain and the target membrane, reflecting several studies that have implicated the C-terminal domain in membrane localization (e.g., (20–22)). Thus, we propose that the co-factor-induced conformational change in ExoU serves two functions in the activation mechanism: facilitating membrane association and establishing an open, catalytically competent conformation at the active site.
In our estimation, this study provides the first direct structural evidence showing that interaction of ExoU with an activating cofactor results in conformational changes in both its C-terminal domain and in close proximity to its active site. By understanding the detailed mechanism of the cofactor-mediated activation of ExoU, specific therapies that target this mechanism may be developed to reduce or prevent tissue damage due to this potent virulence factor. Additional studies to further define the structural arrangement of the C-terminal domain with respect to the active site are in progress.
Acknowledgments
We thank Dr. Candice Klug for critical reading of the manuscript.
This work was supported in part by National Institutes of Health grant No. AI49577 (to D.W.F.). The National Biomedical EPR Center is supported by National Institutes of Health grant No. EB001980.
Footnotes
Marc A. Benson's present address is Department of Molecular Microbiology and Immunology, University of Missouri School of Medicine, One Hospital Dr., Columbia, MO 65212.
Supporting Material
References
- 1.Armour A.D., Shankowsky H.A., Tredget E.E. The impact of nosocomially-acquired resistant Pseudomonas aeruginosa infection in a burn unit. J. Trauma. 2007;63:164–171. doi: 10.1097/01.ta.0000240175.18189.af. [DOI] [PubMed] [Google Scholar]
- 2.Wisplinghoff H., Bischoff T., Edmond M.B. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 3.Hauser A.R., Cobb E., Rello J. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit. Care Med. 2002;30:521–528. doi: 10.1097/00003246-200203000-00005. [DOI] [PubMed] [Google Scholar]
- 4.Lyczak J.B., Cannon C.L., Pier G.B. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pier G.B. CFTR mutations and host susceptibility to Pseudomonas aeruginosa lung infection. Curr. Opin. Microbiol. 2002;5:81–86. doi: 10.1016/s1369-5274(02)00290-4. [DOI] [PubMed] [Google Scholar]
- 6.Galan J.E., Collmer A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 7.Galan J.E., Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 8.Finck-Barbancon V., Goranson J., Frank D.W. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 9.Roy-Burman A., Savel R.H., Wiener-Kronish J.P. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 2001;183:1767–1774. doi: 10.1086/320737. [DOI] [PubMed] [Google Scholar]
- 10.Holder I.A., Neely A.N., Frank D.W. Type III secretion/intoxication system important in virulence of Pseudomonas aeruginosa infections in burns. Burns. 2001;27:129–130. doi: 10.1016/s0305-4179(00)00142-x. [DOI] [PubMed] [Google Scholar]
- 11.Holder I.A., Neely A.N., Frank D.W. PcrV immunization enhances survival of burned Pseudomonas aeruginosa-infected mice. Infect. Immun. 2001;69:5908–5910. doi: 10.1128/IAI.69.9.5908-5910.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheetz M.H., Hoffman M., Hauser A.R. Morbidity associated with Pseudomonas aeruginosa bloodstream infections. Diagn. Microbiol. Infect. Dis. 2009;64:311–319. doi: 10.1016/j.diagmicrobio.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato H., Frank D.W., Sawa T. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 2003;22:2959–2969. doi: 10.1093/emboj/cdg290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phillips R.M., Six D.A., Ghosh P. In vivo phospholipase activity of the Pseudomonas aeruginosa cytotoxin ExoU and protection of mammalian cells with phospholipase A2 inhibitors. J. Biol. Chem. 2003;278:41326–41332. doi: 10.1074/jbc.M302472200. [DOI] [PubMed] [Google Scholar]
- 15.Sato H., Feix J.B., Frank D.W. Characterization of phospholipase activity of the Pseudomonas aeruginosa type III cytotoxin, ExoU. J. Bacteriol. 2005;187:1192–1195. doi: 10.1128/JB.187.3.1192-1195.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hauser A.R. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 2009;7:654–665. doi: 10.1038/nrmicro2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato H., Feix J.B., Frank D.W. Identification of superoxide dismutase as a cofactor for the Pseudomonas type III toxin, ExoU. Biochemistry. 2006;45:10368–10375. doi: 10.1021/bi060788j. [DOI] [PubMed] [Google Scholar]
- 18.Finck-Barbancon V., Frank D.W. Multiple domains are required for the toxic activity of Pseudomonas aeruginosa ExoU. J. Bacteriol. 2001;183:4330–4344. doi: 10.1128/JB.183.14.4330-4344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stirling F.R., Cuzick A., Evans T.J. Eukaryotic localization, activation and ubiquitinylation of a bacterial type III secreted toxin. Cell Microbiol. 2006;8:1294–1309. doi: 10.1111/j.1462-5822.2006.00710.x. [DOI] [PubMed] [Google Scholar]
- 20.Rabin S.D., Hauser A.R. Functional regions of the Pseudomonas aeruginosa cytotoxin ExoU. Infect. Immun. 2005;73:573–582. doi: 10.1128/IAI.73.1.573-582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabin S.D., Veesenmeyer J.L., Hauser A.R. A C-terminal domain targets the Pseudomonas aeruginosa cytotoxin ExoU to the plasma membrane of host cells. Infect. Immun. 2006;74:2552–2561. doi: 10.1128/IAI.74.5.2552-2561.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veesenmeyer J.L., Howel H., Hauser A.R. Role of the membrane localization domain of the Pseudomonas aeruginosa effector protein ExoU in cytotoxicity. Infect. Immun. 2010;78:3346–3357. doi: 10.1128/IAI.00223-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmalzer K.M., Benson M.A., Frank D.W. Activation of ExoU phospholipase activity requires specific C-terminal regions. J. Bacteriol. 2010;192:1801–1812. doi: 10.1128/JB.00904-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hubbell W.L., Cafiso D.S., Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 25.Columbus L., Hubbell W.L. A new spin on protein dynamics. Trends Biochem. Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 26.Fanucci G.E., Cafiso D.S. Recent advances and applications of site-directed spin labeling. Curr. Opin. Struct. Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Klug C.S., Feix J.B. Methods and applications of site-directed spin labeling EPR spectroscopy. Methods Cell Biol. 2008;84:617–658. doi: 10.1016/S0091-679X(07)84020-9. [DOI] [PubMed] [Google Scholar]
- 28.Pannier M., Veit S., Spiess H.W. Dead-time free measurement of dipole-dipole interactions between electron spins. J. Magn. Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 29.Borbat P.P., Freed J.H. Measuring distances by pulsed dipolar ESR spectroscopy: spin-labeled histidine kinases. Methods Enzymol. 2007;423:52–116. doi: 10.1016/S0076-6879(07)23003-4. [DOI] [PubMed] [Google Scholar]
- 30.Xu Q., Ellena J.F., Cafiso D.S. Substrate-dependent unfolding of the energy coupling motif of a membrane transport protein determined by double electron-electron resonance. Biochemistry. 2006;45:10847–10854. doi: 10.1021/bi061051x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Z., DeSensi S.C., Beth A.H. Structure of the cytoplasmic domain of erythrocyte band 3 hereditary spherocytosis variant P327R: band 3 Tuscaloosa. Biochemistry. 2007;46:10248–10257. doi: 10.1021/bi700948p. [DOI] [PubMed] [Google Scholar]
- 32.Altenbach C., Kusnetzow A.K., Hubbell W.L. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc. Natl. Acad. Sci. USA. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou P., Bortolus M., Mchaourab H.S. Conformational cycle of the ABC transporter MsbA in liposomes: detailed analysis using double electron-electron resonance spectroscopy. J. Mol. Biol. 2009;393:586–597. doi: 10.1016/j.jmb.2009.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blackburn M.E., Veloro A.M., Fanucci G.E. Monitoring inhibitor-induced conformational population shifts in HIV-1 protease by pulsed EPR spectroscopy. Biochemistry. 2009;48:8765–8767. doi: 10.1021/bi901201q. [DOI] [PubMed] [Google Scholar]
- 35.Benson M.A., Schmalzer K.M., Frank D.W. A sensitive fluorescence-based assay for the detection of ExoU-mediated PLA2 activity. Clin. Chim. Acta. 2010;411:190–197. doi: 10.1016/j.cca.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Budil D.E., Lee S., Freed J.H. Nonlinear-least-squares analysis of slow motion EPR spectra in one and two dimensions using a modified Levenberg-Marquardt algorithm. J. Magn. Res. A. 1996;120:155–189. [Google Scholar]
- 37.Columbus L., Kálai T., Hubbell W.L. Molecular motion of spin labeled side chains in α-helices: analysis by variation of side chain structure. Biochemistry. 2001;40:3828–3846. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- 38.Kusnetzow A.K., Altenbach C., Hubbell W.L. Conformational states and dynamics of rhodopsin in micelles and bilayers. Biochemistry. 2006;45:5538–5550. doi: 10.1021/bi060101v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeschke G., Chechik V., Jung H. DEER Analysis 2006—a comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Res. 2006;30:473–498. [Google Scholar]
- 40.Altenbach C., Flitsch S.L., Hubbell W.L. Structural studies on transmembrane proteins. 2. Spin labeling of bacteriorhodopsin mutants at unique cysteines. Biochemistry. 1989;28:7806–7812. doi: 10.1021/bi00445a042. [DOI] [PubMed] [Google Scholar]
- 41.Isas J.M., Langen R., Hubbell W.L. Structure and dynamics of a helical hairpin and loop region in annexin 12: a site-directed spin labeling study. Biochemistry. 2002;41:1464–1473. doi: 10.1021/bi011856z. [DOI] [PubMed] [Google Scholar]
- 42.Cuello L.G., Cortes D.M., Perozo E. Molecular architecture of the KvAP voltage-dependent K+ channel in a lipid bilayer. Science. 2004;306:491–495. doi: 10.1126/science.1101373. [DOI] [PubMed] [Google Scholar]
- 43.Jao C.C., Hegde B.G., Langen R. Structure of membrane-bound α-synuclein from site-directed spin labeling and computational refinement. Proc. Natl. Acad. Sci. USA. 2008;105:19666–19671. doi: 10.1073/pnas.0807826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Westfahl K.M., Merten J.A., Klug C.S. Functionally important ATP binding and hydrolysis sites in Escherichia coli MsbA. Biochemistry. 2008;47:13878–13886. doi: 10.1021/bi801745u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fanucci G.E., Lee J.Y., Cafiso D.S. Spectroscopic evidence that osmolytes used in crystallization buffers inhibit a conformation change in a membrane protein. Biochemistry. 2003;42:13106–13112. doi: 10.1021/bi035439t. [DOI] [PubMed] [Google Scholar]
- 46.Kim M., Xu Q., Cafiso D.S. Solutes modify a conformational transition in a membrane transport protein. Biophys. J. 2006;90:2922–2929. doi: 10.1529/biophysj.105.078246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez C.J., Fleissner M.R., Hubbell W.L. Osmolyte perturbation reveals conformational equilibria in spin-labeled proteins. Protein Sci. 2009;18:1637–1652. doi: 10.1002/pro.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


