Abstract
Cells from many different tissues sense the stiffness and spatial patterning of their microenvironment to modulate their shape and cortical stiffness. It is currently unknown how substrate stiffness, cell shape, and cell stiffness modulate or interact with one another. Here, we use microcontact printing and microfabricated arrays of elastomeric posts to independently and simultaneously control cell shape and substrate stiffness. Our experiments show that cell cortical stiffness increases as a function of both substrate stiffness and spread area. For soft substrates, the influence of substrate stiffness on cell cortical stiffness is more prominent than that of cell shape, since increasing adherent area does not lead to cell stiffening. On the other hand, for cells constrained to a small area, cell shape effects are more dominant than substrate stiffness, since increasing substrate stiffness no longer affects cell stiffness. These results suggest that cell size and substrate stiffness can interact in a complex fashion to either enhance or antagonize each other's effect on cell morphology and mechanics.
A large number of cell types from many different tissues respond not only to biochemical signals but also to mechanical cues (1,2). In particular, cells sense the rigidity of their substrates and respond by regulating their cell shape, proliferation, internal cytoskeletal tension, and stiffness through a process known as mechanotransduction (3–6). Most cell types, with the exception of neurons, adhere and spread better on more-rigid substrates (7). As cells spread more, they also can increase cortical stiffness by a mechanism that is thought to depend on up-regulated cytoskeletal contractility (8). On the other hand, cells that are attached to rigid, uniform surfaces can also sense their shapes and respond by switching on apoptotic pathways when the cell adhesive area falls below certain critical values (9). Cell shape can specify human mesenchymal stem cell (hMSC) differentiation via a RhoA-ROCK pathway mediated through internal cytoskeletal tension (3). The fact that substrate rigidity also specifies hMSC differentiation raises questions about how the mechanical signals from substrate rigidity and cell shape are related (10).
A key to understanding mechanotransduction is the mechanism by which cells sense rigidity or geometric cues. One way to reveal this mechanism is to quantify the dependence of cell stiffness on cell shape and on substrate rigidity. Virtually all previous experimental studies that investigated the effects of mechanical cues on cell physiology focused on a single mechanical cue, such as cell shape, substrate stiffness, or surface topography (3,10). In this letter, we present evidence showing that cell cortical stiffness is controlled by both cell shape and substrate stiffness, and define the conditions under which one influence dominates the other.
We grew hMSCs on polyacrylamide gels of different rigidities coated with saturating concentrations of fibronectin. On soft gels with a rigidity of 1–2 kPa, the hMSCs were typically rounded and unspread, and exhibited few if any stress fibers. In contrast, on stiffer gels with a rigidity of ≥5 kPa, the hMSCs were well spread and amassed a large number of stress fibers. To quantify how hMSCs regulate their stiffness in response to the rigidity of their microenvironment, we performed atomic force microscopy by indenting the cells at a frequency of 1 Hz and a depth of 400 nm at regions far from both the nucleus and lamellopodia (see Fig. S1 in the Supporting Material). When grown on soft polyacrylamide gels with physiologically relevant stiffness ranging from 1 to 30 kPa, the cell cortical stiffness gradually increased from 1 kPa to 7 kPa (Fig. 1). However, when the substrate rigidity was increased beyond a critical level (20 kPa in the case of hMSC), cell stiffness was maintained at a saturating level of 7 kPa. This limit probably represents the maximum cortical stiffness that hMSCs can generate. As the substrate rigidity was increased from 1 to 30 kPa, the spread area of hMSCs also increased from ∼1500 to 6000 μm2. As in the case of cell stiffness, the spreading area of the hMSCs also saturated, and beyond a substrate rigidity of 20 kPa, the cell spread area remained roughly constant at 6000 μm2. Both cell area and cell stiffness increased as substrate rigidity was increased from 1 to 20 kPa, and both saturated at ∼20 kPa. This similarity raises a critical issue relevant to mechanobiology: it is unclear whether substrate rigidity modulates cell shape that in turn regulates cell stiffness, or whether substrate rigidity and cell shape independently regulate cell stiffness.
Figure 1.
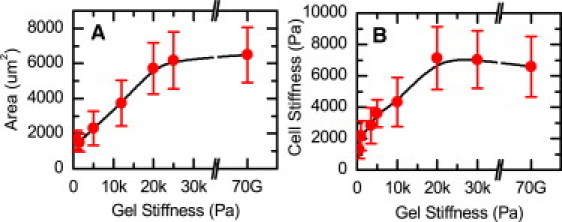
Spread area and stiffness of hMSC as a function of substrate stiffness. (A) hMSC area increased as a function of substrate stiffness and leveled off at a saturating level of ∼6000 μm2. (B) Similarly, hMSC stiffness increased as a function of substrate stiffness up to a level of ∼7 kPa. As controls, cell area and stiffness for cells grown on fibronectin-coated glass of stiffness 70 GPa are shown in A and B.
To address this issue, it is essential to control both substrate rigidity and cell shape simultaneously. Thus, we employed an array of elastomeric polydimethylsiloxane postarray detectors to change the substrate mechanics, and microcontact printing to change the cell shape (11–13). The microposts act as springs to which the cells attach. By measuring the deflection of these microposts, we were able to calculate and spatially map the local cellular traction forces. To control the substrate mechanics, we used microposts of three different heights (tall, medium, and short) corresponding to spring constants of 3.8, 18, and 1500 nN/μm, respectively. The effective shear moduli of continuous substrates equivalent to these discrete substrates were calculated to be 3, 14, and 3500 kPa, respectively (13). To modulate cell shape, we used microcontact printing to pattern the micropost arrays with adhesive islands of different areas with saturating concentrations of fibronectin. hMSCs were constrained onto these islands, as can be seen in the phase contrast images in Fig. 2. On large adhesive islands (100 × 100 μm), vinculin staining (red) was more pronounced on stiffer posts than on softer posts. Fibrillar adhesions were clearly developed on stiffer posts. In contrast, vinculin-containing adhesions were much smaller on smaller adhesive islands (30 × 30 μm) regardless of the rigidity of the post. Actin stress fibers were clearly visible on the large islands, whereas only diffuse cortical actin was formed on small islands. This difference was observed on microposts of different rigidities, suggesting that actin stress fiber formation is largely controlled by cell shape and is relatively unaffected by substrate stiffness.
Figure 2.
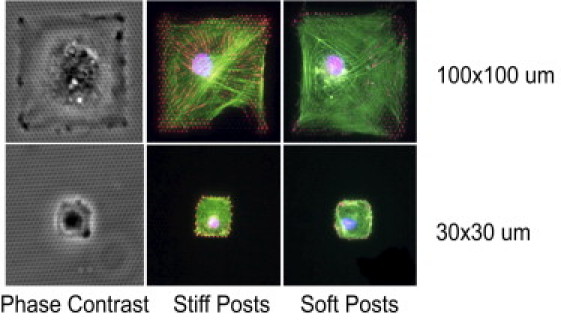
hMSC constrained on square adhesive islands formed by stamping fibronectin on polydimethylsiloxane microposts. Cells are stained with phalloidin (green) and vinculin (red). When spreading areas are large, hMSCs on posts of low (3.8 nN/μm) and high (18 nN/μm) spring constants develop stress fibers, whereas when spreading areas are small, no stress fibers develop. Cells grown on stiffer posts show more intense vinculin staining than those grown on soft posts. However, regardless of post stiffness, larger spread areas result in larger and more mature vinculin patches.
Previous studies showed that cells can adjust their cortical stiffness to match that of their underlying substrates (6). In hMSCs that were grown on soft posts but constrained to different cell shapes, cell cortical stiffness remained relatively constant at ∼2000 Pa (Fig. 3 A). However, on posts of medium and high stiffness, the cortical stiffness increased as the cell spread area was allowed to increase. This result suggests that cell shape may play an important role in controlling cell cortical stiffness, and this effect of cell shape on cell cortical stiffness may be regulated by substrate stiffness. When the cell spread area was kept small at ∼900 μm2, cell cortical stiffness remained low despite a large change in micropost stiffness. In contrast, when the spread area was kept at ∼5600 μm2, cortical stiffness increased as a function of micropost stiffness. These results suggest that both cell shape and substrate stiffness are important modulators of cell cortical stiffness. Concurrently, we measured the total contractile forces of cells as a function of cell spreading area and substrate stiffness. As shown in Fig. 3 B, the traction forces exerted by the cells increased in manner similar to that observed for cell cortical stiffness. Traction forces are related to cortical stiffness by the finding that treatment of cells with blebbistatin to inactivate myosin II decreased the stiffness of cells to the minimum level of ∼2 kPa, regardless of initial cell stiffness, substrate stiffness, or adhesive area (Fig. 4).
Figure 3.
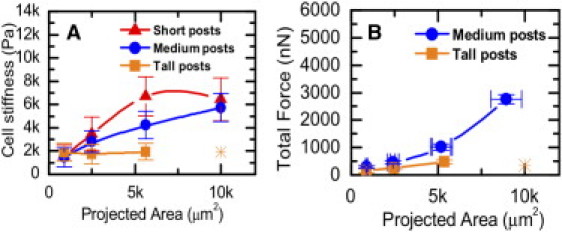
Stiffness and contractile forces of hMSCs as functions of their projected area. hMSCs are plated on microposts of different spring constants with various sizes of square islands of fibronectin. (A) Cell stiffness as measured by atomic force microscopy. For tall posts (3.8 nN/μm), cell stiffness remains constantly soft (square symbols), whereas for medium (18 nN/μm) and short (1500 nN/μm) posts, cell stiffness increases with stamped projected area (circles and triangles). (B) The total contractile forces of cells on tall posts remain small (<500 nN) but increase as cell area is increased on medium posts. Asterisks indicate that no cells of 10,000 μm2 were found on tall posts.
Figure 4.
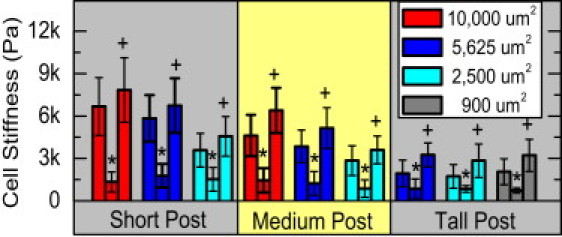
hMSC stiffness is mediated through prestress generated by myosin motors. Treatment with the myosin II antagonist blebbistatin (25 μm) softens cortical stiffness for all cells (∗). This holds true for all substrate rigidities and all constrained areas. Treatment with the tension agonist nocodazole increases cortical stiffness (+).
These results suggest that cell stiffness is modulated by substrate rigidity and cell shape in a more complex manner than previously documented. In particular, in the limit where hMSCs are constrained on small 900 μm2 islands, their cell stiffness is always low (∼2000 Pa) despite the change in substrate stiffness by >2 orders of magnitude (Fig. 3 A). Conversely, in the limit where cells are grown on very soft substrates, even when the cell shape is changed from unspread to well spread, the cell stiffness does not increase (Fig. 3 A, square symbols). However, when cell shape and substrate stiffness are allowed to assume larger magnitudes, these two physical cues interact and both modulate cell stiffness.
The picture presented here suggests that cells actively monitor cell shape and substrate rigidity cues, and integrate this information to modulate focal adhesion formation, cytoskeletal structure, and contractile force to dynamically alter their own stiffness. Thus, cell shape and substrate rigidity act as inputs to gate cell stiffness output.
Acknowledgments
S-Y.T. and P.A.J. were supported by National Institutes of Health grant GM083272-02S1 (funded by the American Recovery and Reinvestment Act). J.F. and C.S.C. were supported in part by the RESBIO Technology Resource for Polymeric Biomaterials, National Institutes of Health (EB00262 and HL90747). J.F. was also partially supported by a postdoctoral fellowship from the American Heart Association.
Footnotes
Jianping Fu's present address is Department of Mechanical Engineering, University of Michigan, Ann Arbor, Michigan.
Supporting Material
References and Footnotes
- 1.Pelham R.J., Jr., Wang Y.L. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Discher D.E., Janmey P., Wang Y.L. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 3.McBeath R., Pirone D.M., Chen C.S. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 4.Solon J., Levental I., Janmey P.A. Fibroblast adaptation and stiffness matching to soft elastic substrates. Biophys. J. 2007;93:4453–4461. doi: 10.1529/biophysj.106.101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajagopalan P., Marganski W.A., Wong J.Y. Direct comparison of the spread area, contractility, and migration of balb/c 3T3 fibroblasts adhered to fibronectin- and RGD-modified substrata. Biophys. J. 2004;87:2818–2827. doi: 10.1529/biophysj.103.037218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tee S., Bausch A., Janmey P. The mechanical cell. Curr. Biol. 2009;19:745–748. doi: 10.1016/j.cub.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Georges P.C., Miller W.J., Janmey P.A. Matrices with compliance comparable to that of brain tissue select neuronal over glial growth in mixed cortical cultures. Biophys. J. 2006;90:3012–3018. doi: 10.1529/biophysj.105.073114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasza K.E., Nakamura F., Weitz D.A. Filamin A is essential for active cell stiffening but not passive stiffening under external force. Biophys. J. 2009;96:4326–4335. doi: 10.1016/j.bpj.2009.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C.S., Mrksich M., Ingber D.E. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 10.Engler A.J., Sen S., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 11.Tan J.L., Tien J., Chen C.S. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc. Natl. Acad. Sci. USA. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu J., Wang Y.-K., Chen C.S. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ladoux Traction forces and rigidity sensing regulate cell functions. Soft Matter. 2008;4:1836–1843. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


