Abstract
Mesenchymal stem cells (MSCs) provide an appropriate model to study epigenetic changes during osteogenesis and bone regeneration due to their differentiation potential. Since there are no unique markers for MSCs, methods of identification are limited. The complex morphology of human embryonic palatal mesenchyme stem cell (HEPM) requires analysis of fractal dimensions to provide an objective quantification of self-similarity, a statistical transformation of cellular shape and border complexity. We propose the hypothesis of a study to compare and contrast sequential steps of osteogenic differentiation in HEPMs both phenotypically using immunocytochemistry, and morphometrically using fractal analysis from undifferentiated passage 1 (P1) to passage 7 (P7) cells. The proof-of-concept is provided by results we present here that identify and compare the modulation of expression of certain epigenetic biomarkers (alkaline phosphatase, ALP; stromal interaction molecule-1, STRO-1; runt-related transcription factor-2, RUNX2), which are established markers of osteogenesis in bone marrow studies, of osteoblastic/skeletal morphogenesis, and of osteoblast maturation. We show that Osteoinductive medium (OIM) modulates the rate of differentiation of HEPM into Run-2+ cells, the most differentiated subpopulation, followed by ALP+ and STRO-1+ cells. Taken together, our phenotypical and morphometric data demonstrate the feasibility of using HEPM to assess osteogenic differentiation from an early undifferentiated to a differentiated stage. This research model may lay the foundation for future studies aimed at characterizing the epigenetic characteristics of osteoimmunological disorders and dysfunctions (e.g., osteoarthritis, temporomandibular joint disorders), so that proteomic profiling can aid the diagnosis and monitor the prognosis of these and other osteoimmunopathologies.
Keywords: Human mesenchymal stem cells, Epigenetics, Fractal dimension, ALP, STRO 1, Runx 2, BGJb medium, Osteoinductive medium (OIM), human embryonic palatal mesenchyme stem cell (HEPM)
Background
Epigenetics is the study of changes in phenotype due to factors other than an alternation of the underlying DNA sequence. Environmental factors can influence gene expression by either turning specific genes on to express their phenotype or by turning them off. Mesenchymal stem cells (MSCs) provide an example of epigenetic changes, demonstrating the capability of differentiating into distinctive cell types. MSCs support the growth of hematopoietic cells and take part in the maintenance of the skeleton throughout life. Since there are no unique markers for MSCs, methods of identification are limited to some functional and morphological criteria such as growth on plastic, resistance to trypsin, presence of specific cell surface antigen, and the potential to differentiate into adipocytes, chondrocytes, and osteocytes. Over the past few decades, MSCs have been the subject of considerable research, and because of their differentiation potential, they became a valuable asset in many areas of cell therapy and regenerative and reconstructive medicine, specifically bone regeneration [1].
Osteoprogenitor cells, found at the external and internal surfaces of bone, are capable of giving rise to osteoblasts, which secrete the extracellular matrix of bone. Osteoblasts secrete the collagen fibers and ground substance, which together form the matrix of bone called osteoid, otherwise known as unmineralized bone. When an osteoblast is completely surrounded by matrix, it is called an osteocyte. An osteocyte can both secrete and resorb matrix, which is important in the maintenance of healthy blood calcium levels. Osteoblasts are responsible for the calcification of the matrix through the secretion of small membrane–limited vesicles that are secreted during the production of bone matrix. Matrix vesicles contain enzymes like alkaline phosphatase that cause the accumulation of calcium and phosphate inside the vesicles. Later, loaded vesicles rupture, increasing the local concentration of minerals and initiating mineralization. Thus, osteoprogenitor cells, osteoblasts, and osteocytes are all part of a series derived from MSCs and their differentiation capabilities [2–6].
Embryology and the Formation of Bone
In the embryo, bone tissue arises through two principal processes: Intramembranous ossification, and Endochondral ossification. In intramembranous ossification, bone is formed directly from mesenchymal tissue. In endochondral ossification, a cartilage model is formed first, and is later replaced by bone. The first step in intramembranous ossification is the aggregation of mesenchyme cells in the area where bone is to be formed. The tissue in this area becomes more vascularized, and the mesenchyme cells begin to differentiate into osteoblasts, secreting collagen and proteoglycans of the bone matrix. Over time the osteoid calcifies and the cell becomes enclosed in canaliculi. Some of the mesenchymal cells surrounding the developing bone spicules proliferate and differentiate into osteoprogenitor cells. Osteoprogenitor cells in contact with bone spicules become osteoblasts and secrete matrix, resulting in oppositional growth of the spicule [[3], [7–9]].
Cell Markers
For the purpose of this proof-of-concept study, the cells used in this experiment were analyzed by indirect immunocytochemistry. AML3/Runx2 was one of the primary antibodies used and is a transcriptional factor involved in osteoblastic and skeletal morphogenesis. It is essential for the maturation of osteoblasts and for intramembranous ossification. Runx2 induces osteoblastic function and differentiation in non-osteoblastic cells by expression of osteoblast specific genes. ALP, another primary antibody that was used, is a hydrolase enzyme that removes phosphate groups from the 5’ end of DNA and RNA, and from proteins at high pH. High expression of ALP is observed in pre-osteoblasts and osteoblasts. Lastly, STRO-1 is a cell membrane single pass type I protein that translocates from the endoplasmic reticulum to the cell membrane in response to a depletion of intracellular calcium. This monoclonal antibody recognizes the progenitor cell of the fibroblast colony-forming units. High expression of STRO-1 could be found in osteoprogenitor cells and pre-osteoblasts; by utilizing these antibodies, specific cells can be selected. The secondary antibody was labeled with horseradish peroxidase (HRP) [5], [10–17].
Fractal Dimensions
The complex morphology of HEPM is not easily quantified using classical measures such as length, diameter or surface. However, the complex morphology of natural objects can be quantified using the statistical selfsimilarity concept of fractal geometry introduced by Mandelbrot. We have used this method to obtain objective quantification of shape border complexity of immunostained lymphocytes by measuring its fractal dimension (FD) [17]. Fractal dimension is a measure of how complicated a self-similar figure is. Since cells are self-similar objects, fractal analysis can be used to compute cell complexity for the means of comparison [9, 17, 18].
Methodology
Commercially obtained frozen HEPM stem cells were plated in T75 culture flask using control culture medium (BGJb). Once cells were attached the medium was replaced in order to remove preservatives and possible non-adherent cells. Medium was replaced every 3-4 days until cell density reached 70% -80% confluence. Half of the cells in the first passage (P1) were shifted from BGJb medium to Osteoinductive medium (OIM). To have a back up, cells were frozen at each passage to -80° C. Osteogenically differentiated and control cells were seeded at ˜1.2X104 cells per well in 24 well plates using either BGJb medium or OIM. Cells were collected at different passages and were subjected to immunocytochemical analysis once they were at 50% confluence. Immunocytochemical and morphometric analysis were performed using: STRO-1, ALP, and Runx 2 markers.
Immunocytochemistry on Cultured Cells
When cells were attached and reached a 50% confluence level, transfer pipettes were used to discard the spent BGJb medium and OIM. Then, cells were gently washed with 1X phosphate-buffered saline (PBS) three times with a 5 minute intervals. 200µL of fixative solution (0.1% Formaldehyde and 0.01% Triton-X) was added to each well of the 24-well plates, which covered the cell surface for ˜15 minutes. This procedure was followed by repeated washing three times with 1X PBS and 5 minute intervals [17].
Mouse monoclonal antibody (for Runx-2 and STRO-1) and rabbit polyclonal antibody (for ALP) were the primary antibodies used. Antibodies were diluted (1/500 dilution) for overnight incubation and washed three times with 1X PBS and 5 minute intervals. The secondary antibodies used were anti-rabbit IgG-peroxidase produced in goat, and anti-mouse IgG-peroxidase produced in rabbit. Appropriate dilutions of the secondary antibodies were prepared using PBS supplemented with 10% fetal bovine serum (FBS). 200µL of the diluted antibodies were added to each corresponding well and incubated overnight (1/1000 dilution). Cells were then washed three times using copious amounts of 1X PBS with 5 minute intervals as above. In order to remove the remaining PBS from the cultures, cells were further washed with a 0.05 molar Tris (hydromethylaminomethane) buffer (pH 7.4) [17].
Color Development
Color development was done using the EnVision Dako Kit. One drop of diamino benzidine (DAB)/Chromogen solution (5% 3, 3'- diaminobenzidine and tetrahydrochloride chromogen solution) was added to each mL of the provided substrate buffer solution. 200µL of the mixture was added to the wells, and the color was developed for 45 minutes. Cultures were then washed three times with cold 1X PBS to stop the reaction, and stored in 1X PBS until microphotography (Nikon microscope, 20X & 40X magnification, with 10X eyepiece) [17].
Fractal Analysis
Using Benoit 1.3 Fractal Analysis Software, 40X black/white pictures of the cells were binarized. The fractal dimensions were calculated by evaluating the number of boxes needed to cover the fractal completely, as previously described [17]. Repeating this measurement with different sized boxes resulted in logarithmical function of box size (x-axis) versus the number of boxes needed to cover the fractal (y-axis). The slope of the function is referred to as box dimension. In other words, the fractal dimension was calculated from the slope of the linear regression between the log of the number of occupied boxes and the log of the corresponding size of the boxes. Box dimension was taken as an appropriate approximation of fractal dimension [17].
Discussion
This proof-of-concept study was designed to obtain preliminary support for the hypothesis of using HEPM to assess osteogenic differentiation from an early undifferentiated to a differentiated stage. Specifically, we presented phenotypical and morphometric comparison of sequential steps towards osteogenic differentiation of mesenchymal stem cells from an early undifferentiated passage (P1) to a late more differentiated passage (P7). The goal of this research project was to identify and compare epigenetic biomarkers that characterize human embryonic palatal mesenchyme stem cells (HEPM) undergoing progression through osteogenic differentiation. The purpose of this study was to further understand and increase our knowledge of the biological variabilities that control and determine osteogenic progression and differentiation. This was obtained by investigating time dependent regulation of ontogenesis in commercially available cultured HEPM stem cells through monitoring the phenotypic expression of certain epigenetic biomarkers (i.e., ALP, STRO-1, Runx-2), established key signatiures of osteogenesis in bone marrow studies. The patterns of marker expression during the progression of osteogenic differentiation from an early undifferentiated passage (P1) to a late or more differentiated passage (P7) were investigated using recognized inducers of osteogenesis, such as dexamethasone, and beta-glycerolphosphate, in osteogenesis-inducing growth medium. In addition to comparing the expression of these biomarkers between the control BGJb and the experimental OIM at P1 versus P7, we quantified and compared cell shape complexity by means of fractal dimension analysis [17].
Using immunocytochemical staining techniques HEPM stem cells were marked with the aid of commercially available mono- and poly-clonal antibodies. We determined that immunostaining of cells cultured in BGJb medium using different biomarkers resulted in a pattern of relative infrequent and less prominent marker expression in cells from P1 to P7. The intensity of marker expression from P1 to P7 in BGJb medium was more visible for P7 Runx-2 positive cells. According to the literature, high expression of STRO-1 marker could be found in osteoprogenitor cells and pre-osteoblasts while ALP markers are observed in pre-osteoblasts and osteoblasts [[12, 15, 16, 19]]. Runx-2 is a marker of osteogenic differentiation and is strongly expressed in osteoblasts of bone marrow and subpopulation of mature endosteal osteoblasts in extraction sockets [[10, 18, 20, 21]]. The P1 Runx2-positive (Runx-2+) cells cultured in BGJb medium had lower initial marker expression compared to P1 cells expressing other markers; they were more intensely expressed throughout all seven passages. It is possible that the difference in staining of the P7 Runx2+ cells may be attributable to the fact that they are nucleus markers in comparison to the ALP and STRO-1 markers that are cell surface markers. We interpret these observations as putatively suggesting that the staining may be concentrated in the cell nuclear components.
As summarized in Figure 1, the difference in the staining pattern of biomarker expressing cells from P1 to P7 generally increased for cells cultured in OIM, compared to BGJb medium. This difference in staining was more noticeable in Runx2 expressing cells. Most of the P1 cells cultured in OIM were stained more frequently and intensely than the P1 cells cultured in BGJb medium. This may have derived from the fact that cells cultured in OIM differentiated in a much faster rate than the cells cultured in BGJb medium; hence, expressing Runx-2 osteoblastic markers.
Figure 1.
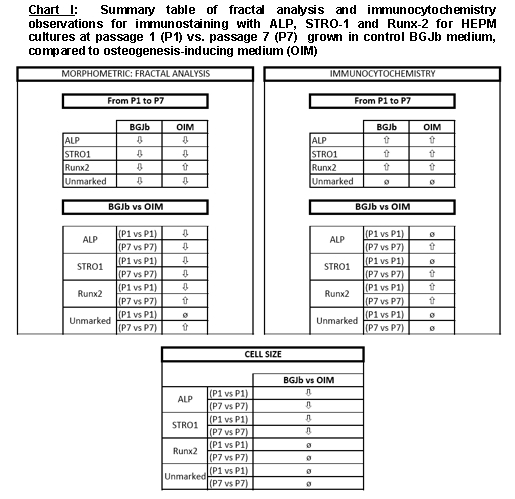
Summary table of fractal analysis and immunocytochemistry observations for immunostaining with ALP, STRO-1 and Runx-2 for HEPM cultures at passage 1 (P1) vs. passage 7 (P7) grown in control BGJb medium, compared to Osteoinductive medium (OIM).
In order to compare the effects of the media on the expression of biomarkers, cells cultured in BGJb medium were compared to their equivalent biomarker expressing cells of the same passage in OIM. At P1, ALP+ and STRO-1+ cells stained equally low in both media. The P7 cells expressing ALP markers stained considerably more in OIM compared to BGJb medium, but the STRO-1+ cells at P7 did not. The immunostaining of these cells were more concentrated on the surface of the cells. On the contrary, P1 Runx-2+ cells stained more visibly in OIM in comparison to BGJb medium. The expression of the markers was about the same for the P7 cells in both media, although the staining of cells in OIM cultures was well defined (Figure 1).
For further identification of cellular developmental stages, morphological differences of cells were quantified using fractal analysis technique. The fractal dimension is a measure of the cellular shape complexity, and was used to quantify the complex morphology of the cultures, as we had reported previously [17].
In this experiment, decreased value of fractal dimension correlated to less complexity reflecting a more differentiated, and specialized cell. The results of morphometric analysis of immunostained cells at P1 compared to P7 cultured in BGJb medium showed an overall decrease in fractal dimension value for all cultured cells correlating to less complexity and more differentiated cells. The effect of BGJb medium on cell differentiation was studied and the results showed no significant effect on differentiation of ALP+ and STRO-1+ cells from P1 to P7. By contrast, Runx-2+ and the unmarked cells were affected significantly (Figure 1).
The results obtained for the effect of OIM on cell differentiation from P1 to P7 showed no significant effect on ALP+ and Runx-2+ cells; whereas, the STRO-1+ and the unmarked cells were significantly affected. The mean fractal dimension values for cultured cells from P1 to P7 showed a decrease for ALP+, STRO-1+, and unmarked cells indicating less complexity and more differentiation of cells in both media. The Runx-2+ cells showed an increase in the mean fractal dimension value in OIM. The results for Runx-2+ and ALP+ cells could be affected due to the heterogeneity of the samples.
The effect of the two media on marked cells cultured in BGJb medium and OIM was tested the result indicated a significant effect for ALP+ and STRO-1+ cells in BGJb medium, and for Runx-2+ cells in OIM (Figure 1).
Conclusion
In conclusion, the results of our study allowed us to conclude that according to our fractal analysis profiling the Runx-2+ cell population was the most differentiated cell population followed by the ALP+ and STRO- 1+ cell populations. This is in agreement with our results of the immunocytochemical staining study. The Runx+2 cells were stained more prominently compared to other marked cells and there was no increase in their cell size, they reached a plateau in which cell differentiation was at rest. Also, according to the results of our cell media study, OIM affected the rate of cell differentiation driving the cells to become a more osteogenically differentiated and specialized cells, as the data summarized in Figure 1 suggest.
We propose the hypothesis that the model research approach we have described here lays the foundation for future research studies relating osteoimmunological bone disorders and dysfunctions such as osteoarthritis, and temporomandibular joint disorders (TMD) correlating proteomic profiling using biomarkers and innovative instruments to assess and detect prognostic changes as well as provide progressive treatments.
Acknowledgments
The authors thank Professor George Bernard, D.D.S., Ph.D. for his indefatigable guidance, support and constructive suggestions.
Footnotes
Citation:Barkhordarian et al, Bioinformation 5(7): 278-281 (2011)
References
- 1.Pittenger MF, Martin BJ. Circ. Res. 2004;95:9. doi: 10.1161/01.RES.0000135902.99383.6f. [DOI] [PubMed] [Google Scholar]
- 2.Breen EC, et al. J. Cell. Physiol. 1994;160:323. doi: 10.1002/jcp.1041600214. [DOI] [PubMed] [Google Scholar]
- 3.Dahir GA, et al. Clin. Orthop. Relat. Res. 2000;379:S134. doi: 10.1097/00003086-200010001-00018. [DOI] [PubMed] [Google Scholar]
- 4.Fried A, et al. J. Cell. Physiol. 1993;155:472. doi: 10.1002/jcp.1041550306. [DOI] [PubMed] [Google Scholar]
- 5.Gronthos S, et al. Blood. 1994;84:4164. [PubMed] [Google Scholar]
- 6.Olivier E, et al. Stem Cells. 2006;24:1914. doi: 10.1634/stemcells.2005-0648. [DOI] [PubMed] [Google Scholar]
- 7.Boyan BD, et al. Clin. Orthp. Relat. Res. 1992;277:266. [PubMed] [Google Scholar]
- 8.Matsuzaka K, et al. Clin. Oral. Implants. Res. 2000;11:325. doi: 10.1034/j.1600-0501.2000.011004325.x. [DOI] [PubMed] [Google Scholar]
- 9.Petite H, et al. Nat Biotechnol. 2000;18:959. doi: 10.1038/79449. [DOI] [PubMed] [Google Scholar]
- 10.Bruder sp, et al. Bone Miner. Res. 1998;13:655. doi: 10.1359/jbmr.1998.13.4.655. [DOI] [PubMed] [Google Scholar]
- 11.Devline H, Sloan P. Int. J. Oral. Maxillofac. Surg. 2002;31:641. doi: 10.1054/ijom.2002.0292. [DOI] [PubMed] [Google Scholar]
- 12.Herbertson A, Aubin JE. Bone. 1997;21:491. doi: 10.1016/s8756-3282(97)00197-x. [DOI] [PubMed] [Google Scholar]
- 13.Karsenty G, et al. Bone. 1999;25:107. doi: 10.1016/s8756-3282(99)00111-8. [DOI] [PubMed] [Google Scholar]
- 14.Maniapoluos C, et al. Cell Tissue Res. 1988;254:317. [Google Scholar]
- 15.Triffit JT, et al. J. Biomed. Mater. Res. 2002;63:384. doi: 10.1002/jbm.10260. [DOI] [PubMed] [Google Scholar]
- 16.Turken K, Aubin JE. J. Cell. Biol. 1991;114:373. doi: 10.1083/jcb.114.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiappelli F, et al. Front Biosci. 2005;10:3034. doi: 10.2741/1760. [DOI] [PubMed] [Google Scholar]
- 18.Ristanovic D, et al. Biol. Cybern. 2002;87:278. doi: 10.1007/s00422-002-0342-1. [DOI] [PubMed] [Google Scholar]
- 19.Bruder sp, et al. Bone. 1997;21:225. doi: 10.1016/s8756-3282(97)00127-0. [DOI] [PubMed] [Google Scholar]
- 20.Doherty MJ, et al. J. Bone Miner Res. 1998;13:828. doi: 10.1359/jbmr.1998.13.5.828. [DOI] [PubMed] [Google Scholar]
- 21.Friedenstein AJ, et al. Cell Tissue Kinet. 1987;20:263. doi: 10.1111/j.1365-2184.1987.tb01309.x. [DOI] [PubMed] [Google Scholar]


