Abstract
Human saliva is a biological fluid with enormous diagnostic potential. Because saliva can be non-invasively collected, it provides an attractive alternative for blood, serum or plasma. It has been postulated that the blood concentrations of many components are reflected in saliva. Saliva harbors a wide array of proteins, which can be informative for the detection of diseases. Profiling the proteins in saliva over the course of disease progression could reveal potential biomarkers indicative of different stages of diseases, which may be useful in medical diagnostics. With advanced instrumentation and developed refined analytical techniques, proteomics is widely envisioned as a useful and powerful approach for salivary proteomic biomarker discovery. As proteomic technologies continue to mature, salivary proteomics have great potential for biomarker research and clinical applications. The progress and current status of salivary proteomics and its application in the biomarker discovery of oral and systematic diseases will be reviewed. The scientific and clinical challenges underlying this approach will also be discussed.
Keywords: Saliva, Biomarkers, Proteomics
Background
Human saliva is secreted from three pairs of major salivary glands namely parotid gland, submandibular gland and sublingual gland lying at the vicinity of oral cavity, and numerous minor salivary glands lying beneath the oral mucosa. It contains a large array of proteins, many of which can be informative for the detection of oral and systemic diseases. Saliva is an attractive diagnostic fluid because it has several key advantages for disease diagnosis and prognosis, for example low invasiveness, minimum cost, and easy sample collection and processing [1].
Relaying on the significant advances in mass spectrometry technologies, proteomics is widely envisioned as a useful and powerful approach for proteome research. Of particular note is the salivary proteomic biomarker discovery and validation, which involved high throughput proteomic approaches [1, 2]. Recently, salivary proteomics has demonstrated a great potential for biomarker discovery and validation for various oral and systemic diseases. In this review, the progress and prospects of salivary proteomics and its implications in oral and systematic diseases will be reviewed. In addition, the scientific and clinical challenges pertaining to the approach will be discussed.
Salivary Proteomics
Different proteomic technologies have been used to analyze the protein composition of saliva qualitatively and quantitatively. Based on advanced mass spectrometry technologies, the capability of saliva protein identification has been greatly enhanced by combining different protein separation technologies. As for example, free-flow electrophoresis has been coupled with linear ion-trap tandem mass spectrometry to identify salivary proteins. Multidimensional separation platform based on capillary isoelectric focusing and nano-reversed phase liquid chromatography has also been successfully utilized for salivary protein separation. Two dimensional gel electrophoresis (2-DE) coupled with matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) or liquid chromatography-mass spectrometry (LC-MS/MS) has been used as a routine approach for salivary protein separation, quantitation and identification [3]. Besides, non-gel based labeling methods such as iTRAQ [isobaric tag for relative and absolute quantitation] and label-free quantitation have been used for quantitative salivary proteome analysis. According to a recent consortium study, 1,166 proteins were identified in saliva (914 from the parotid fluid and 917 from the combined submandibular and sublingual fluids) and a high portion of these proteins was also found in serum [4].
Applications
Oral and systemic diseases may affect salivary glands directly or indirectly, and may influence the quantity of saliva that is produced, as well as the composition of the fluid [5]. These phenomena have significant impact in biomarker discovery for disease by utilizing proteomic methodologies. A schematic diagram regarding the rationale for human salivary proteomics has been presented in Figure 1.
Figure 1.
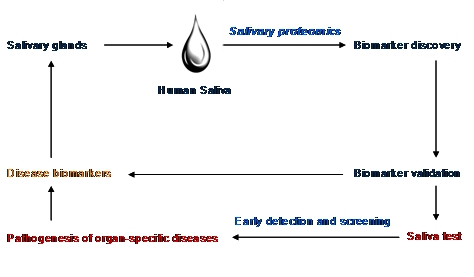
The rationale for salivary proteomic biomarkers.
Oral diseases biomarker discovery Salivary proteomics has been extensively used for oral diseases biomarker discovery, such as oral cancer and Sjögren's syndrome. Recently, 5 protein biomarkers (Table 1 see Table 1) have been discovered by 2-DE/MS and validated by immunoassays for oral cancer detection [6]. As for primary Sjögren's syndrome, dozens of salivary proteins were found to be downregulated or up-regulated in patients when comparing with matched healthy control subjects (Table 1 see Table 1). These proteins reflected the damage of glandular cells and inflammation of the oral cavity system in primary Sjögren’s syndrome [7].
Systematic diseases biomarker discovery Salivary proteomics has also been used for systematic diseases biomarker discovery, for example breast cancer. The soluble fragment of the c-erbB-2 oncogene was found to be significantly higher in saliva collected from patients with breast cancer compared to patients with benign tumors and healthy controls. A pilot study has indicated that the saliva test for this oncogene is sensitive, reliable, and potentially useful in initial detection of and/or follow-up screening for breast cancer [8]. Efforts are being made to apply salivary proteomics for disease-specific biomarker discovery, such as lung, gastric and pancreatic cancer.
Challenges
Although there is increasing interest in salivary proteome analysis, there are still some bottlenecks for salivary proteomic biomarker discovery. First, there are some high abundant proteins in saliva, such as amylase, albumin, and immunoglobulins, which become the hurdles for the detection and quantitation of low abundant proteins. Depletion of these interfering proteins prior to definitive analyses should improve the resolution and sensitivity of salivary proteome analysis [1]. Second, high throughput and quantitative validation methods are urgently needed for the confirmation of potential biomarkers that lack available antibodies. The development of new technologies will accelerate the biomarker research in saliva. Third, and probably not the least, the mechanism behind the disease biomarker distribution in saliva should be clarified for further clinical screening and detection. A recent study has shown that biomarkers can be secreted from lung cancer tumor to mouse saliva [9], which suggests that the salivary glands may be regulated by mediators released from remote tumors.
Conclusion
The proteomes in saliva have been characterized by different proteomic technologies, including qualitative and quantitative methods. Based on these platforms, salivary proteomic biomarker discovery has been carried out in selective oral and systematic diseases. Most importantly, when a saliva test could be easily conducted in clinic for early detection of diseases, the life quality of patients will be greatly improved. Although there are technical and mechanistic challenges, one can anticipate that the demands for clinical applications will be the great impetus for the salivary proteomics development.
Supplementary material
Acknowledgments
We thank Dr. Jieping Yang and Dr. Abu Nazmul-Hossain for critical review of the manuscript. This work was supported by NIH ⁄ NIDCR Grant U01-DE16275, U01-DE17790, R01-DE17593 and R01-DE17170 to DTW.
Footnotes
Citation:Xiao et al, Bioinformation 5(7): 294-296 (2011)
References
- 1.Hu S, et al. Proteomics. 2006;6:6326. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L, et al. Mol Diagn Ther. 2009;13:245. doi: 10.1007/BF03256330. [DOI] [PubMed] [Google Scholar]
- 3.Hu S, et al. Proteomics. 2005;5:1714. doi: 10.1002/pmic.200401037. [DOI] [PubMed] [Google Scholar]
- 4.Denny P, et al. J Proteome Res. 2008;7:1994. doi: 10.1021/pr700764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaufman E, et al. Crit. Rev. Oral Biol. Med. 2002;13:197. doi: 10.1177/154411130201300209. [DOI] [PubMed] [Google Scholar]
- 6.Hu S, et al. Clin Cancer Res. 2008;14:6246. doi: 10.1158/1078-0432.CCR-07-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu S, et al. Arthritis Rheum. 2007;56:3588. doi: 10.1002/art.22954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Streckfus C, et al. Clin Cancer Res. 2000;6:2363. [PubMed] [Google Scholar]
- 9.Gao K, et al. Plos One. 2009;4:e5875. doi: 10.1371/journal.pone.0005875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


