Abstract
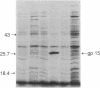
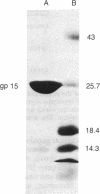
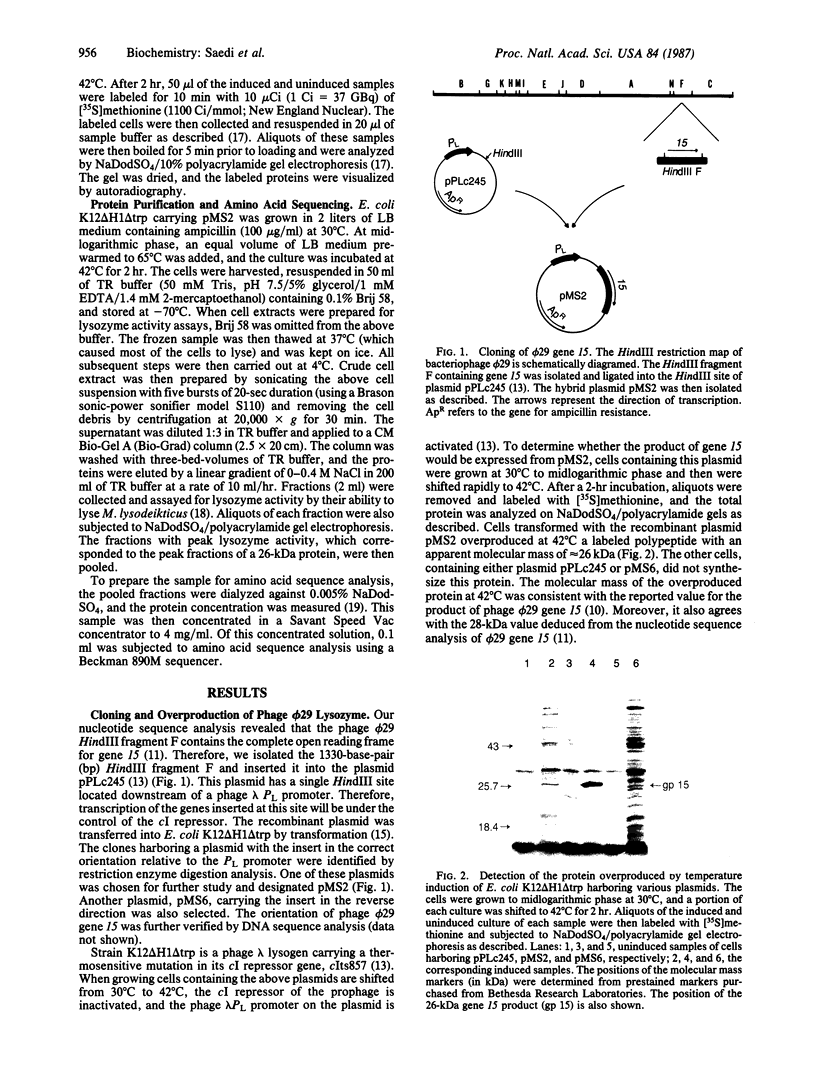
A DNA fragment of the bacteriophage phi 29 chromosome, encoding the entire sequence of phi 29 gene 15, has been cloned into the Escherichia coli expression vector pPLc245 under the control of the phage lambda major leftward promoter, PL. Upon heat induction, a protein with an apparent molecular mass of 26 kDa was overproduced. The molecular mass of this protein corresponds to the 28 kDa predicted for the product of gene 15 from its nucleotide sequence. The overproduced protein has been purified to near homogeneity and confirmed to be the product of gene 15 by amino acid sequence analysis of its N terminus. The purified product of gene 15 has a lysozyme activity similar to other phage-type lysozymes: products of phage T4 gene e and of phage P22 gene 19. However, to our knowledge phi 29 lysozyme is structurally unique among the phage-type lysozymes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bieńkowska-Szewczyk K., Taylor A. Murein transglycosylase from phage lambda lysate. Purification and properties. Biochim Biophys Acta. 1980 Oct;615(2):489–496. doi: 10.1016/0005-2744(80)90515-x. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Carrascosa J. L., Camacho A., Moreno F., Jiménez F., Mellado R. P., Viñuela E., Salas M. Bacillus subtilis phage phi29. Characterization of gene products and functions. Eur J Biochem. 1976 Jul 1;66(2):229–241. doi: 10.1111/j.1432-1033.1976.tb10512.x. [DOI] [PubMed] [Google Scholar]
- Grütter M. G., Matthews B. W. Amino acid substitutions far from the active site of bacteriophage T4 lysozyme reduce catalytic activity and suggest that the C-terminal lobe of the enzyme participates in substrate binding. J Mol Biol. 1982 Jan 25;154(3):525–535. doi: 10.1016/s0022-2836(82)80011-9. [DOI] [PubMed] [Google Scholar]
- Hagen E. W., Reilly B. E., Tosi M. E., Anderson D. L. Analysis of gene function of bacteriophage phi 29 of Bacillus subtilis: identification of cistrons essential for viral assembly. J Virol. 1976 Aug;19(2):501–517. doi: 10.1128/jvi.19.2.501-517.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M., Arnheim N., Sternglanz R. Bacteriophage T7 lysozyme is an N-acetylmuramyl-L-alanine amidase. J Biol Chem. 1973 Oct 25;248(20):7247–7252. [PubMed] [Google Scholar]
- Inouye M., Imada M., Tsugita A. The amino acid sequence of T4 phage lysozyme. IV. Dilute acid hydrolysis and the order of tryptic peptides. J Biol Chem. 1970 Jul 25;245(14):3479–3484. [PubMed] [Google Scholar]
- Ito J. Bacteriophage phi29 terminal protein: its association with the 5' termini of the phi29 genome. J Virol. 1978 Dec;28(3):895–904. doi: 10.1128/jvi.28.3.895-904.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez F., Camacho A., De La Torre J., Viñuela E., Salas M. Assembly of Bacillus subtilis phage phe29. 2. Mutants in the cistrons coding for the non-structural proteins. Eur J Biochem. 1977 Feb 15;73(1):57–72. doi: 10.1111/j.1432-1033.1977.tb11291.x. [DOI] [PubMed] [Google Scholar]
- Jollès P., Jollès J. What's new in lysozyme research? Always a model system, today as yesterday. Mol Cell Biochem. 1984 Sep;63(2):165–189. doi: 10.1007/BF00285225. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Morita T., Hara S., Matsushima Y. Purification and characterization of lysozyme produced by Streptomyces erythraeus. J Biochem. 1978 Mar;83(3):893–903. doi: 10.1093/oxfordjournals.jbchem.a131987. [DOI] [PubMed] [Google Scholar]
- Remaut E., Stanssens P., Fiers W. Inducible high level synthesis of mature human fibroblast interferon in Escherichia coli. Nucleic Acids Res. 1983 Jul 25;11(14):4677–4688. doi: 10.1093/nar/11.14.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennell D., Poteete A. R. Phage P22 lysis genes: nucleotide sequences and functional relationships with T4 and lambda genes. Virology. 1985 May;143(1):280–289. doi: 10.1016/0042-6822(85)90115-1. [DOI] [PubMed] [Google Scholar]
- Tsugita A., Inouye M. Complete primary structure of phage lysozyme from Escherichia coli T4. J Mol Biol. 1968 Oct 14;37(1):201–212. doi: 10.1016/0022-2836(68)90083-1. [DOI] [PubMed] [Google Scholar]
- Weaver L. H., Grütter M. G., Remington S. J., Gray T. M., Isaacs N. W., Matthews B. W. Comparison of goose-type, chicken-type, and phage-type lysozymes illustrates the changes that occur in both amino acid sequence and three-dimensional structure during evolution. J Mol Evol. 1984;21(2):97–111. doi: 10.1007/BF02100084. [DOI] [PubMed] [Google Scholar]
- Weaver L. H., Rennell D., Poteete A. R., Mathews B. W. Structure of phage P22 gene 19 lysozyme inferred from its homology with phage T4 lysozyme. Implications for lysozyme evolution. J Mol Biol. 1985 Aug 20;184(4):739–741. doi: 10.1016/0022-2836(85)90318-3. [DOI] [PubMed] [Google Scholar]