Abstract
A PCR targeting sau1-hsdS1 was developed for rapid detection of Staphylococcus aureus clonal complex 398 (CC398). High sensitivity (100%) and specificity (100%) were shown by evaluating the test on a large strain collection (n = 1,307). We recommend this test for accurate, rapid, and inexpensive diagnosis of methicillin-resistant S. aureus (MRSA) CC398 in hospitals and on farms.
Methicillin-resistant Staphylococcus aureus (MRSA) belonging to clonal complex 398 (CC398) has emerged in livestock worldwide and is presently regarded as an important zoonotic agent (3, 8, 10, 11). CC398 strains are not typeable by standard SmaI pulsed-field gel electrophoresis (PFGE) analysis (1) and are currently identified at the clonal level by spa typing or multilocus sequence typing (MLST). In this study, a lineage-specific PCR was developed for rapid detection of S. aureus CC398 based on the principle that clonal differences within S. aureus are reflected in the sequence of sau1-hsdS1, a gene responsible for the restriction modification specificity of this bacterial species (12). As the sequence of sau1-hsdS1 in CC398 was unknown, as the first step we studied the sequence variability of this gene in CC398. A universal reverse primer (5′-CAATTTGTCGGTCGAGTTTGCTG-3′) was designed for amplification of an approximately 530-bp region in sau1-hsdS1 by aligning publically available sau1-hsdS1 sequences (GenBank accession numbers DQ309449 to DQ309455) and used with the forward universal sau1-hsdS1 AF primer described by Cockfield et al. (2) (5′-AGGGTTTGAAGGCGAATGGG-3′). The amplicon sequences obtained from eight CC398 isolates displaying distinct spa types (t011, t034, t108, t567, t571, t1255, t1793, and t2876) were sequenced (TAG Copenhagen, Copenhagen, Denmark) and showed 100% identity and no homology to any publically available bacterial sequences (www.ncbi.nlm.nih.gov/blast). A CC398-specific reverse primer (CC398r1 [5′-CAGTATAAAGAGGTGACATGACCCCT-3′]) was designed to amplify a 296-bp fragment of the sau1-hsdS1 gene in combination with the AF primer (final primer concentration of 0.2 μM) using AmpliTaq Gold (Applied Biosystems). The following conditions were used: 12 min at 95°C, followed by 35 cycles of 30 s at 95°C, 30 s at 61°C, and 1 min at 72°C, and a final extension at 72°C for 10 min. The PCR test was evaluated using a large collection of strains (n = 1,307) comprising over 10 clonal complexes (Table 1). The collection included 40 human and 1,072 animal CC398 isolates representative of 13 spa types (t011, t034, t108, t567, t571, t899, t1255, t1344, t1456, t1793, t2330, t2876, and t4652) and 75 human and 160 animal non-CC398 isolates provided by the four laboratories involved in the study. All isolates had been previously characterized by spa typing (5), and clonal complex associations were determined using http://spaserver.ridom.de and by comparing MLST mapping of previous isolates with similar or related spa types with subsequent clustering by using eBURST v3 (4, 7). The results of the PCR validation showed 100% specificity (235/235) and 100% sensitivity (1,072/1,072). A multiplex PCR version allowing differentiation between MRSA and non-MRSA isolates belonging to CC398 was obtained by coupling the primers targeting sau1-hsdS1 with mecA-specific primers (6) (mecup1 [5′-GGGATCATAGCGTCATTATTC-3′] and mecup2 [5′-AACGATTGTGACACGATAGCC-3′]) (see Fig. 1). The size of the CC398-specific amplicon (527 bp) is easily distinguishable from the predicted product sizes of hospital-acquired S. aureus lineages by the use of the existing restriction-modification typing scheme of Cockfield et al. (2). This allows a potential expansion of the current typing scheme for rapid identification of livestock-associated MRSA CC398 in hospital settings.
Table 1.
S. aureus strain collection used for validation of the CC398-specific PCR
| Origin (no. of isolates) | Clonal complexb | spa type(s) (no. of isolates where ≥10) | No. of isolates of indicated clonal complex type | No. of sau1-hsdS1 PCR-positive results |
|---|---|---|---|---|
| Cattle (8) | CC80 | t527 | 1 | 0 |
| CC9 | t2839 | 1 | 0 | |
| NDa | t524, 2873, t3046 | 6 | 0 | |
| Dog (9) | CC15 | t084, t774 | 2 | 0 |
| CC20 | t091 | 1 | 0 | |
| CC25 | t227 | 1 | 0 | |
| CC30 | t3055 | 1 | 0 | |
| CC5 | t548 | 1 | 0 | |
| CC8 | t030 | 1 | 0 | |
| ND | t1335, t1651 | 2 | 0 | |
| Goat (1) | ND | t1166 | 1 | 0 |
| Horse (13) | CC15 | t084 | 1 | 0 |
| ND | t1166, t1294, t2112, t2484, t3043, t3044 | 12 | 0 | |
| Human (115) | CC22 | t005, t022, t032, t223, t541 | 10 | 0 |
| CC30 | t012, t016, t019, t318 | 10 | 0 | |
| CC398 | t011, t034 (35), t108, t567, t571 | 40 | 40 | |
| CC45 | t015, t065, t230 | 7 | 0 | |
| CC5 | t001, t002, t003, t041, t045 | 20 | 0 | |
| CC8 | t008, t024, t037, t064 | 21 | 0 | |
| CC80 | t044, t376 | 5 | 0 | |
| ST152/377 | t355 | 2 | 0 | |
| Poultry (114) | CC5 | t002 (64), t306, t2049 | 66 | 0 |
| CC8 | t304 | 1 | 0 | |
| CC9 | t1430 | 19 | 0 | |
| CC80 | t203 | 1 | 0 | |
| CC398 | t011 (11), t034, t108, t567, t1456, t4652 | 25 | 25 | |
| ND | t324, t2038 | 2 | 0 | |
| Sheep (5) | ND | t2678, t3042, t3045, t3047 | 5 | 0 |
| Swine (1,042) | CC1 | t127 | 2 | 0 |
| CC5 | t151, t2164 | 2 | 0 | |
| CC30 | t1333, t2840 | 8 | 0 | |
| CC398 | t011 (404), t034 (13), t108 (575), t571, t899, t1255, t1344, t1793, t2330, t2876 | 1,007 | 1,007 | |
| CC9 | t337 (21), t899c, t2839 | 23 | 0 |
ND, not determined (but non-CC398).
Clonal complex associations were determined using http://spaserver.ridom.de and by comparing the results of MLST mapping of previous isolates with similar or related spa types with subsequent clustering by the use of eBURST v3 (http://eburst.mlst.net).
MLST typed.
Fig. 1.
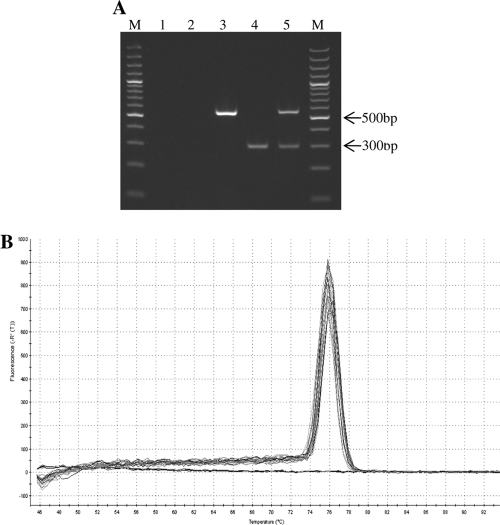
(A) Duplex PCR for CC398 sau1-hsdS1 and mecA detection. Lane M, 100-bp DNA ladder; lane 1, negative control (water); lane 2, non-CC398 methicillin-susceptible S. aureus (strain ATCC 6538); lane 3, non-CC398 methicillin-resistant S. aureus (strain ATCC 33591); lane 4, CC398 methicillin-sensitive S. aureus (SSI 52615); lane 5, CC398 methicillin-resistant S. aureus (KVL 288). (B) Real-time PCR melting curve analysis showing the melting temperature (Tm) of the amplicons. (−R′(T)), negative derivative of fluorescence with respect to temperature.
For a quicker turnaround time, the conventional PCR was converted to a real-time platform. Performance of the real-time PCR was validated using a separate strain collection consisting of 77 CC398 and 18 non-CC398 isolates (CC5, -8, -9, -22, -30, -45, -80, and -121) of human and animal origin, confirming the excellent results obtained by the conventional PCR (100% sensitivity and specificity). Real-time PCR (RT-PCR) was performed using 96-well plates on an Mx3000P platform (Stratagene) and Maxima SYBR green-ROX quantitative PCR (qPCR) master mix (Fermentas) with a 0.5 μM concentration of each primer in a final volume of 12.5 μl. PCR conditions were 5 min at 94°C followed by 35 cycles of 30 s at 94°C, 30 s at 57°C, and 1 min at 72°C. The specificity of the amplification products was confirmed by gel electrophoresis and melting curve analysis (Fig. 1).
The very high sensitivity and specificity of the singleplex and multiplex PCR methods presented in this study are explained by the observation that the sau1-hsdS1 sequence in CC398 is highly conserved and differs significantly from the homologous sequence in other S. aureus lineages. van Wamel et al. (9) recently proposed four PCRs to identify CC398 isolates. The two that showed 100% accuracy when tested with a collection of 133 isolates were A07, representing gene SAPIG2195, and C01, representing gene SAPIG2194. Both genes are carried on a transposon and are therefore unsafe for use as a stable marker of lineage. Our results confirm the general findings by Waldron et al. indicating that sau1-hsdS1 is highly clonal specific and exhibits very high sequence homology within lineages. The sequence variability of the gene in CC398 showed 100% nucleotide identity across eight different CC398-associated spa types and 100% homology to SAPIG0500 from the sequenced ST398 genome (GenBank accession number AM990992). Sequence analysis of the 64 publicly available S. aureus whole-genome sequencing projects did not show any similarity to existing sau1-hsdS sequences (data not shown). Accordingly, the gene appears to be a very conserved and discriminatory epidemiological marker for clonal identification of S. aureus CC398.
Molecular typing of MRSA is an important tool for epidemiological surveillance and for development of infection control measures aimed at preventing dissemination within hospitals as well as from the community to hospitals. In developing such control measures, it is necessary to identify genetic markers allowing rapid and reliable MRSA identification at the CC level and easy communication of results between laboratories. Due to the increasing public health concern associated with this livestock-associated MRSA strain, the need for rapid methods for MRSA identification at the CC level is no longer limited to human medicine but is now extended to MRSA surveillance of living animals, farm environments, and animal food products. Therefore, the PCR test presented here has important applications in both human and veterinary public health. We recommend the use of this test for rapid, accurate, and inexpensive identification of MRSA CC398 in human diagnostic specimens as well as in any human, animal, food, and environmental samples analyzed for surveillance purposes in hospitals or on farms.
Acknowledgments
The work was partly supported by the EU-HEALTH Project PILGRIM (223050) of the Seventh Framework Programme (FP7).
A Danish patent application based on the findings of this study was filed in June 2009 (PA 2009 00767).
Footnotes
Published ahead of print on 1 December 2010.
REFERENCES
- 1. Bens C. C., Voss A., Klaassen C. H. 2006. Presence of a novel DNA methylation enzyme in methicillin-resistant Staphylococcus aureus isolates associated with pig farming leads to uninterpretable results in standard pulsed-field gel electrophoresis analysis. J. Clin. Microbiol. 44:1875–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cockfield J. D., Pathak S., Edgeworth J. D., Lindsay J. A. 2007. Rapid determination of hospital-acquired meticillin-resistant Staphylococcus aureus lineages. J. Med. Microbiol. 56:614–619 [DOI] [PubMed] [Google Scholar]
- 3. European Food Safety Authority 2009. Analysis of the baseline survey on the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in holdings with breeding pigs, in the EU, 2008, part A: MRSA prevalence estimates. Eur. Food Saf. Authority 7:1376 doi:10.2903/j.efsa.2009.1376. http://www.efsa.europa.eu/en/scdocs/doc/1376.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Feil E. J., Li B. C., Aanensen D. M., Hanage W. P., Spratt B. G. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harmsen D., et al. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Poulsen A. B., Skov R., Pallesen L. V. 2003. Detection of methicillin resistance in coagulase-negative staphylococci and in staphylococci directly from simulated blood cultures using the EVIGENE MRSA Detection Kit. J. Antimicrob. Chemother. 51:419–421 [DOI] [PubMed] [Google Scholar]
- 7. Spratt B. G., Hanage W. P., Li B., Aanensen D. M., Feil E. J. 2004. Displaying the relatedness among isolates of bacterial species—the eBURST approach. FEMS Microbiol. Lett. 241:129–134 [DOI] [PubMed] [Google Scholar]
- 8. Springer B., et al. 2009. Methicillin-resistant Staphylococcus aureus: a new zoonotic agent? Wien. Klin. Wochenschr. 121:86–90 [DOI] [PubMed] [Google Scholar]
- 9. van Wamel W. J., et al. 2009. Short term micro-evolution and PCR-detection of methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398. Eur. J. Clin. Microbiol. Infect. Dis. 29:119–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Loo I., et al. 2007. Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg. Infect. Dis. 13:1834–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Voss A., Loeffen F., Bakker J., Klaassen C., Wulf M. 2005. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11:1965–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Waldron D. E., Lindsay J. A. 2006. SauI: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J. Bacteriol. 188:5578–5585 [DOI] [PMC free article] [PubMed] [Google Scholar]