Abstract
With improved measles virus (MV) control, the genetic variability of the MV-nucleoprotein hypervariable region (NP-HVR) decreases. Thus, it becomes increasingly difficult to determine the origin of a virus using only this part of the genome. During outbreaks in Europe and Africa, we found MV strains with identical NP-HVR sequences. However, these strains showed considerable diversity within a larger sequencing window based on concatenated MV phosphoprotein and hemagglutinin genes (P/H pseudogenes). In Belarus, Germany, Russia, and the Democratic Republic of Congo, the P/H pseudogenes provided insights into chains of transmission, whereas identical NP-HVR provided none. In Russia, for instance, the P/H pseudogene identified temporal clusters rather than geographical clusters, demonstrating the circulation and importation of independent variants rather than large local outbreaks lasting for several years, as suggested by NP-HVR. Thus, by extending the sequencing window for molecular epidemiology, a more refined picture of MV circulation was obtained with more clearly defined links between outbreaks and transmission chains. Our results also suggested that in contrast to the P gene, the H gene acquired fixed substitutions that continued to be found in subsequent outbreaks, possibly with consequences for its antigenicity. Thus, a longer sequencing window has true benefits both for the epidemiological surveillance of measles and for the better monitoring of viral evolution.
Since the introduction of measles vaccination, the global burden of measles disease has continuously decreased (13). Significant progress has been made during the last decade, with the elimination of measles from the Americas and the dramatic reduction in measles mortality worldwide (8). However, measles continues to be endemic in many developing countries and some industrialized countries (6). The molecular epidemiology of measles virus (MV) has proven to be a very useful tool for monitoring the progress in measles control (7). The negative-sense RNA genome (15,894 nucleotides [nt]) contains six genes, encoding the nucleoprotein (NP), the phosphoprotein (P), and the matrix (M), fusion (F), hemagglutinin (H), and large (L) proteins (5). Although MV is serologically a monotypic virus, genetic characterization so far has identified eight clades (A to H), subdivided into 24 genotypes (A, B1 to B3, C1 and C2, D1 to D11, E, F, G1 to G3, and H1 and H2) (8, 16). Since 1998, the World Health Organization (WHO) recommends the sequencing of the hypervariable region of 450 nt encoding the C-terminal 150 amino acids of the NP (NP-HVR) and the use of it as the minimal data for MV genotyping (12). Additionally, the complete H sequence should be obtained if a new genotype is suspected (8). However, only the sequences of the NP-HVR are available for most strains obtained from clinical cases.
MV genotyping is an important tool of measles surveillance to document chains of transmission, discriminate between imported or indigenous viruses, and monitor elimination programs. However, identical NP-HVR sequences have been found for several years in Europe and beyond (8). For instance, two main variants of genotype D6, differing by a single nucleotide in their NP-HVR, were widely distributed in the WHO European region in 2005 and 2006 (3). Thus, it is difficult to determine the origin of a virus using molecular tools based on the NP-HVR alone.
In this study, the sequence variability of P and H genes of strains with identical or very similar NP-HVR sequences was investigated. We showed for four different outbreaks in Europe and Africa that the phylogenetic analysis of the P/H pseudogene sequences provides a more refined picture of MV circulation.
MATERIALS AND METHODS
Clinical specimens.
A total of 73 strains from four epidemiological settings in Europe and Africa were analyzed (see Table S1 in the supplemental material). The strains were selected for the similarity or identity of their NP-HVR sequences. Clinical specimens from 13 patients (D6b) were collected between April and June 2006 from 11 different locations in North Rhine Westphalia (NRW; Germany). Samples from 10 patients were collected between March and September 2006 from three different areas in Belarus: Minsk city (n = 1), Minsk region (n = 4), and Grodno region (n = 5). Thirty-one clinical samples related only to outbreaks of genotype D6a were collected throughout the Russian Federation, Uzbekistan, Kazakhstan, and Kyrgyzstan during March 2003 to May 2007. Nineteen samples were collected between December 2004 and February 2006 in three different regions of the Democratic Republic of Congo (DR-Congo): Bas-Congo (n = 4), Kinshasa (n = 12), and Kasai-Oriental (n = 3). Most cases also were confirmed serologically by measles-specific IgM using a commercial enzyme-linked immunosorbent assay (ELISA) kit (Enzygnost, Siemens, Germany).
MV sequencing.
Viral RNA purification from clinical specimens was performed using a QIAamp viral RNA mini kit (Qiagen, The Netherlands) or MagMAX-96 AI/ND viral RNA isolation kit (Ambion, The Netherlands) according to the manufacturer's instructions. RNA was eluted in a final volume of 35 μl. Purified viral RNA was reverse transcribed for 80 min at 50°C using random primers (Invitrogen, Belgium) and SuperScript III reverse transcriptase (Invitrogen, Belgium).
Complete P and H genes as well as the 450 terminal nucleotides of the NP-HVR of all clinical specimens used in this study were sequenced and are available under the following NCBI accession numbers: NP-HVR, HM802038 to HM802117; P, HM801884 to HM802016; and H, HM801904 to HM802037. PCR products for NP-HVR and P were generated by nested PCRs as described before (1, 4). H genes were amplified using a seminested PCR strategy, generating two overlapping fragments in a first round using primers H1 and H6, as well as primers H5 and H2. Nested H PCRs were performed using primers H1 and H4, primers H3 and H6, primers H5 and H8, and primers H7 and H2 (Table 1). PCRs were done in a total volume of 25 μl, including 1 μl of cDNA or 5 μl of first-round product (50 times diluted in water), 2 mM MgCl2, 1× PCR buffer, 0.2 mM deoxynucleoside triphosphates (dNTPs), 0.5 U platinum Taq (Invitrogen, Belgium), 0.5 μM primer (first-round PCR), and 1 μM primer (nested PCR). Cycling conditions were denaturation at 94°C for 1 min 30 s, followed by 35 cycles of 94°C for 30 s, 1 min of annealing at 53°C (first round) or 58°C (nested), and 72°C for 1 min. PCRs were performed on Opticon 2 (Bio-Rad, Belgium) thermocyclers. PCR products were purified and sequenced as described in reference 3. Sequences were aligned using ClustalW (11) and further analyzed using SeqScape 2.5 and MEGA 4.0 (10). Phylogenetic trees were calculated by the neighbor-joining method using a number of nucleotide differences with 1,000 bootstrap replicates and pairwise deletion.
Table 1.
Primer used for the H gene PCR
| Name | Sequence (5′–3′) | MV genome position (nt) | PCR fragment size (nt) |
|---|---|---|---|
| H1 | TTAAAACTTAGGGTGCAAGATCATCCACA | 7242–7270 | H1-H6, 878 |
| H6 | GACCATTACTGACTGGTTGCTCAA | 8097–8120 | |
| H5 | GTACCGAGTGTTTGAAGTAGGTGTTA | 8023–8048 | H5-H2, 1134 |
| H2 | GGGTGACATCATGTGATTGGTTCA | 9134–9157 | |
| H4 | TATCCCTCATGCTGAAGTCTCTAG | 7653–7676 | H1-H4, 434 |
| H3 | CACCTCAGAGATTCACTGACCTAGT | 7590–7614 | H3-H6, 530 |
| H8 | TAACTAGTGTGTGCCGAGTCC | 8569–8589 | H5-H8, 541 |
| H7 | GATCTGAGTCTGACAGTTGAGCTTA | 8516–8540 | H7-H2, 641 |
Nucleotide sequence accession numbers.
The sequences determined in the course of this work are available under the following NCBI accession numbers: NP-HVR, HM802038 to HM802117; P, HM801884 to HM802016; and H, HM801904 to HM802037.
RESULTS
Genotype D6: Germany.
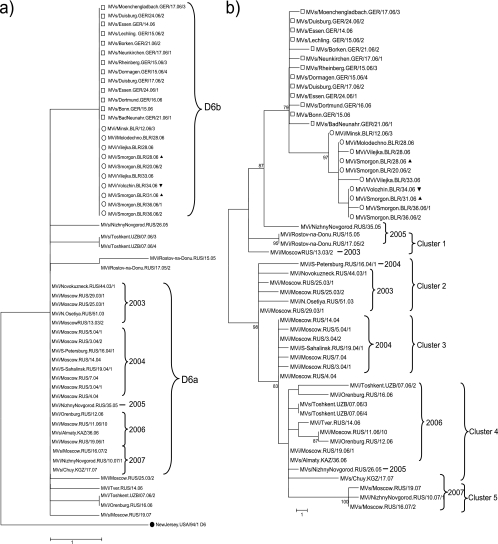
Between January and July 2006, 1,710 measles cases were reported during a measles outbreak in NRW (http://www3.rki.de/SurvStat), which was associated with MVi/Moenchengladbach.DEU/19.06. This D6 variant was identical to the D6 virus Mvs/Kyiv.UKR/03.06/1, which was associated with a large outbreak in the Ukraine, with more than 50,000 cases during 2005 to 2006. Eleven NP-HVR variants differing by up to 3 nt were found among the 125 MV strains sequenced in the context of this outbreak (14). Among these variants, we investigated the genetic diversity of P and H genes of 13 of these strains with identical NP-HVR sequences (Fig. 1 and 2). Nine of them were collected in April 2006 at the peak of the epidemic and the other four 1 or 2 months later. Six strains that were identical in their NP-HVR sequences each had a unique P/H pseudogene sequence, differing by 1 or 2 nt from the most common variant (Fig. 1b). The highest genetic distance of 4 nt was found between two strains (MVs/BadNeuenahr.GER/21.06/1 and MVs/Borken.GER/21.06/2) collected toward the end of the outbreak. In the H gene all 13 strains were identical, except for one (MVs/Neunkirchen.GER/17.06/1), with a single point mutation (Fig. 2b). Thus, the diversity of P/H pseudogene sequences was due mainly to random mutations in the P gene (Fig. 2a).
Fig. 1.
Phylogenetic trees of D6a strains (collected in the Russian Federation from 2003 to 2007) and D6b strains (collected in Germany (□; April to June 2006) or Belarus (○; March to September 2006). D6b strains from Russia (2005 to 2007) are not included in this study. The calculation was based on (a) the 450 nt of the NP-HVR and (b) the 3,377 nt of the P/H pseudogene. ●, WHO reference strain; ▴, strains with epidemiological link to the Ukraine; ▿, strain imported from the Grodno region to the Minsk region. Genetic distances are represented as the number of nucleotides of difference between strains.
Fig. 2.
Phylogenetic trees of D6a strains (collected in the Russian Federation from 2003 to 2007) and D6b strains (collected in Germany (□; April to June 2006) or Belarus (○; March to September 2006). The calculation was based on (a) the 1,524 nt of the P gene and (b) the 1,854 nt of the H gene. ▴, strains with an epidemiological link to the Ukraine; ▿, strain imported from the Grodno region to the Minsk region. Genetic distances are represented as the number of nucleotides of difference between strains.
Genotype D6: Belarus.
Between January and September 2006, 149 measles cases were reported from five different regions in Belarus (http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=3011 and a personal communication from G. Semeiko). A total of five NP-HVR variants, differing by 1 to 3 nt, were obtained from 47 MV strains, which were sequenced in the context of this outbreak (3). Ten of these strains with identical NP-HVR sequences were analyzed here for their P and H gene diversity (Fig. 1 and 2). The latter were collected between March and September 2006 in three different regions of Belarus (see Table S1 in the supplemental material). Among the 10 strains with identical NP-HVR sequences, four and three different P and H gene sequences, respectively, were found (Fig. 2a and b). Six different P/H pseudogene variants could be distinguished (Fig. 1b).
The strains imported independently from two very distant locations within the Ukraine (MVi/Smorgon.BLR/28.06, epidemiologically linked to Khmelnitsky, and MVi/Smorgon.BLR/31.06, from a patient returning from the Crimea 4 days before the onset of disease) had identical P gene sequences and differed by a single fixed nucleotide in the H gene (Fig. 2a and b). The latter nucleotide was found in several other strains from Smorgon (Grodno region) and Volozhin (Minsk region) collected several weeks later. Similarly, all four strains from Minsk region (Molodechno, Vilejka, and Volozhin; see Table S1 in the supplemental material) differed from each other by the same fixed H nucleotides as well as some random mutations in the P gene, suggesting several independent importations into both regions. Indeed, MVi/Volozhin.BLR/34.06 (Minsk Region) was epidemiologically linked to Grodno region.
In conclusion, six sequence variants could be distinguished among the 10 MV strains from Belarus (with identical NP-HVRs) on the basis of their P/H pseudogene sequences. Although the overall sequence diversity remained low, the fixed H nucleotides accurately reflected the known epidemiological links, while the random mutations of the P gene were less informative.
Genotype D6: Russia.
To monitor the evolution of the P and H gene of MV strains with identical or very similar NP-HVR sequences during a prolonged period, 31 genotype D6 viruses collected from March 2003 to May 2007 in the Russian Federation and the Newly Independent States (NIS) Uzbekistan, Kazakhstan, and Kyrgyzstan were analyzed. Most of the strains (n = 21) collected during these 5 years had the same NP-HVR sequence (variant D6a). Ten strains differed by 1 or 2 nt from variant D6a and by up to 3 nt (0.67%) from each other (Fig. 1a). The maximal overall genetic distance between all MV sequences reported during the same period from the Russian Federation was 4 nt (0.89%) in the NP-HVR (9).
In our study, the P/H pseudogene revealed 18 variants among the 21 D6a strains (maximal genetic distance, 20 nt [0.59%]) (Fig. 1b). Using H gene sequences only of the same strains, 13 variants (maximal genetic distance, 10 nt [0.54%]) could be distinguished. Similarly, 13 different P gene variants were found among the 21 D6a strains (maximal genetic distance, 10 nt [0.66%]) (Fig. 2a and b). The maximal genetic distance increased to 11 nt (0.72%) in the P gene when the 11 other MV variants with slightly different NP-HVR sequences were included in the analysis, but it did not increase for the H gene.
Thus, phylogenetic dendrograms of P and/or H genes were in contrast to the clearly structured NP-HVR tree. The fused P and H genes formed at least five clusters (Fig. 1b). Cluster 1 included strains identified in the Russian Federation during 2003 until 2005; cluster 2 included strains mainly from throughout Russia in 2003, including Moscow; cluster 3 strains were mostly from Moscow in 2004; cluster 4 had mostly 2006 strains from various regions throughout Russia and the NIS; and finally, cluster 5 was comprised of strains collected throughout Russia in 2007. These clusters resulted mainly, but not exclusively, from accumulating fixed nucleotides in the H gene.
The three MV strains from Nizhny Novgorod (MVs/NizhnyNovgorod.RUS/26.05, MVi/NizhnyNovgorod.RUS/10.07/1, and MVi/NizhnyNovgorod.RUS/35.05) had highly diverse P and H gene sequences. Interestingly, the MVi/NizhnyNovgorod.RUS/35.05 strain was relatively closely related to the strain that spread from the Ukrainian outbreak 1 year later. This strain also showed the highest genetic distance of all strains represented here to the 2007 strain from the same city, suggesting the repeated independent importation of MV rather than continued circulation in Nizhny Novgorod. Similarly, the P/H pseudogene variability clearly demonstrated that the strains from Moscow were not directly related to or part of a larger outbreak.
Considering the large differences in sequence diversity (Table 2), strains with identical P and/or H sequences were much more likely to be epidemiologically linked than strains with identical NP-HVR sequences. For instance, two strains from Rostov-na-Donu (MVi/Rostov-na-Donu.RUS/15.05 and MVi/Rostov-na-Donu.RUS/17.05/2) differed by 1 nt in their NP-HVR but had identical P and H sequences. MVs/Almaty.KAZ/36.06 and MVi/Moscow.RUS/19.06/1 had identical sequences in all genes, suggesting a direct epidemiological link between measles cases in Moscow and Kazakhstan in 2006. Similarly, two of the three MV strains obtained in Tashkent (Uzbekistan) during the same week of 2006 had identical sequences in all genes, whereas the third one seems to represent a different chain of transmission.
Table 2.
Comparison of the overall genetic diversity of the MV genes used
| Source | NP-HVR | P gene | H gene | P/H pseudogene |
|---|---|---|---|---|
| D6b (Germany) | 0 | 0.26 | 0.05 | 0.12 |
| D6b (Belarus) | 0 | 0.20 | 0.11 | 0.09 |
| D6a (Russia) | 0 | 0.66 | 0.54 | 0.59 |
| B2KIN-A (DR-Congo) | 0 | 0.39 | 0.05 | 0.21 |
| B2KIN-C (DR-Congo) | 0 | 0.72 | 0.22 | 0.41 |
In conclusion, P and H gene sequences further structured chains of transmission of strains with virtually identical NP-HVR sequences.
Genotype B2: Democratic Republic of Congo.
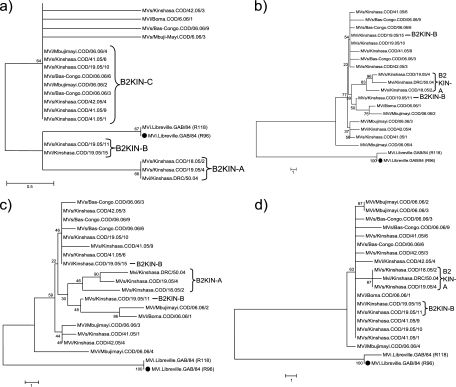
Between 2004 and 2006, a large measles epidemic with more than 36,000 cases occurred in the DR-Congo. Among 45 MV strains (genotype B2) sequenced from this outbreak, three main NP-HVR variants (B2KIN-A, B2KIN-B, and B2KIN-C) differing by up to 2 nt were detected (2). From 18 of these viruses, including three B2KIN-A, two B2KIN-B, and nine B2KIN-C strains collected between December 2004 and February 2006, the P and H genes were sequenced (Fig. 3). Four additional strains that differed by a single nucleotide from B2KIN-C also were included.
Fig. 3.
Phylogenetic trees showing B2 strains collected in the DR-Congo (December 2004 to February 2006). The calculation was based on (a) the 450 nt of the NP-HVR, (b) the 3,377 nt of the P/H pseudogene, (c) the 1,524 nt of the P gene, and (d) the 1,854 nt of the H gene. ●, WHO reference strain. Genetic distances are represented as the number of nucleotides of difference between strains.
All strains except two (MVi/Kinshasa.COD/19.05/15 and MVs/Kinshasa.COD/19.05/10) had unique P/H pseudogene sequences (Fig. 3b). However, only the three strains from variant B2KIN-A formed a separate cluster in their P/H pseudogenes, containing three distinct variants with a maximal genetic distance of 7 nt (0.21%). This difference was due mostly to the P gene, with a maximal genetic distance of 6 nt (0.39%), but also to two H gene variants differing by a single nucleotide (Fig. 3c and d). B2KIN-B and B2KIN-C strains did not form distinct clusters in the phylogenetic tree based on P/H pseudogene sequences, and genetic distances between B2KIN-C strains were due to substitutions in their P genes (maximal diversity, 11 nt [0.72%]) rather than their H genes (maximal diversity, 4 nt [0.22%]). On the other hand, four strains with different NP-HVR sequences were identical in their P genes (MVs/Kinshasa.COD/41.05/6, MVs/Kinshasa.COD/42.05/3, MVs/Kinshasa.COD/19.05/10, MVi/Kinshasa.COD/19.05/15, and MVs/Bas-Congo.COD/06.06/9).
The earliest available sequence of the 2004-2006 outbreak, a B2KIN-A variant (Mvi/Kinshasa.DRC/50.04), differed by only 2 nt (0.44%) in the NP-HVR from a B2 strain detected in Gabon 20 years earlier (MVi.Libreville.GAB/84 [R96] [U01994]). Also, B2KIN-B and B2KIN-C differed by only 1 or 2 nt from the Gabon strains. In contrast, in the P/H pseudogene the B2 strains from DR-Congo differed by 45 and 53 nt (1.33 and 1.57%) from the Gabon strains. Most of the latter mutations (29 to 34) were found in the H gene, whereas their P genes showed fewer differences (16 to 23).
The pseudogene of the B2KIN-A variants, which were detected only during the early phase of the epidemic in Kinshasa, were genetically more distant (minimal distance, 49 nt [1.45%]) from the Gabon strains from 1984 than B2KIN-B and B2KIN-C variants. Therefore, it is unlikely that B2KIN-B and B2KIN-C evolved from B2KIN-A during the 2004-2006 epidemic in Kinshasa but rather represent independent introductions of different genotype B2 variants during 2005.
The three strains collected during the same week in the province of Bas-Congo (MVs/Bas-Congo.COD/06.06/6, MVs/Bas-Congo.COD/06.06/9, and MVs/Bas-Congo.COD/06.06/3) differed by 3 to 5 nt from each other. Similarly, differences between 9 and 14 nt were found in the P/H pseudogene sequences of strains MVi/Mbujimayi.COD/06.06/4, MVi/Mbujimayi.COD/06.06/3, and MVi/Mbujimayi.COD/06.06/2, which were collected in the same week in the province of Kasai-Oriental. These sequence data, as well as their phylogeny (Fig. 3b), suggest that measles outbreaks in Bas-Congo and Kasai-Oriental, which occurred during the epidemic peak in Kinshasa, were linked by several independent chains of transmission with Kinshasa.
DISCUSSION
By extending the sequencing window recommended by WHO for the molecular epidemiology of MV from the NP-HVR to P and H genes, links between outbreaks and transmission chains became more clearly defined. Without epidemiological data, identical NP-HVR sequences found in Belarus and Germany have suggested that these strains belong to the same transmission chain. However, their P/H pseudogene sequences formed distinct clusters supported by high bootstrap values that clearly identified the cases in both countries as part of two independent outbreaks. Interestingly, the outbreak in Germany showed only a single cluster in the P/H pseudogene, whereas the Belarusian strains from both Grodno and Minsk regions showed at least two clusters of pseudogenes, which corresponded to several independent introductions from the Ukraine. Thus, the strains from both regions do not correspond to an ongoing transmission, affirming a better measles control in Belarus than that suggested by the NP-HVR analysis. Unlike the German strains, the Belarusian pseudogenes showed an apparent evolution during the 24 weeks of observation, which was due mainly to the fixation of two specific mutations after week 12 and week 31 in the H gene, while the P gene added additional variability to the P/H pseudogene. In contrast, all German H genes except one were identical. Also, in this case the variability within the single cluster of the pseudogene was due to mutations in the P gene.
In the NP-HVR, the strains from Russia (D6a) and Belarus/Germany (D6b) differed by a single nucleotide with no intermixing. While this is suggestive of at least two parallel outbreaks in both regions, the NP-HVR provided no further insights into transmission pathways. In the P/H pseudogenes of the Belarusian/German strains, the tree structure suggested a common most recent ancestor with 2003 and 2005 strains from Russia. Most other strains from Russia formed a clearly separated cluster of their own with high bootstrap support. Interestingly, the analysis of the pseudogenes of the Russian strains revealed well-defined subclusters by calendar years, suggesting that closely related viruses circulated simultaneously in different locations throughout Russia and NIS as a single epidemiological space and continued to evolve. Thus, the P/H pseudogene permitted a temporal and geographic interpretation of MV circulation. In contrast, the NP-HVR sequences provided no insight into circulation patterns of MV and could be interpreted as belonging to a single transmission chain lasting for several years at the different locations. Strains collected in Moscow during 5 years all were identical in their NP-HVR, suggesting a continuous circulation of MV in the city. However, the P/H pseudogene showed that most of the strains in Moscow were more closely related to strains from several other locations during the same years than to earlier or later strains from Moscow. This suggests that every year virus strains have been reintroduced into Moscow from different regions within Russia and NIS with ongoing outbreaks. Interestingly, the phylogenetic structure reflects a molecular evolution of the H gene between 2003 and 2007, with the irreversible fixation of distinct mutations. This was much less evident for the P gene, but the phylogeny of the constructed P/H pseudogenes confirms this evolution.
Recently three distinct variants of NP-HVR genotype B2 (B2KIN-A/B/C) were found in DR-Congo (2). However, using their P/H pseudogenes, only strains from variant B2KIN-A formed a distinct cluster, supported by bootstrap values for the H gene. Thus, our findings suggest the circulation of variant B2KIN-A in Kinshasa and multiple independent importations of B2KIN-B and B2KIN-C into Kinshasa. The most surprising finding in DR-Congo was that strains collected during this outbreak showed only a very short genetic distance (maximum, 2 nt) in their NP-HVR compared to that of strains collected more than 20 years before in Gabon. In contrast, they displayed a very high genetic diversity in their P/H pseudogenes (>45 nt) compared to those of these strains. Interestingly, two-thirds of this variability was contributed by the H gene.
The NP-HVR provides the most bar-coding information per sequence length (12%) (15). Nevertheless, the P and H genes (and most other MV genes) also have considerable sequence diversity (5.5 and 6.1% [1]), which, as we showed here, can be exploited to monitor viruses with reduced genetic diversity. Thus, by extending the sequencing window for molecular epidemiology, a more refined picture of MV circulation was obtained with more clearly defined links between outbreaks and transmission chains.
The examples described above suggest some striking differences between the molecular evolution of the P and H genes. While mutations in the H gene seem to have a high tendency to become fixed in the viral genome, mutations in the P gene seem to be more random and variable. As a result, mutations in the H gene tend to form distinct clusters, whereas P gene mutations only add variability to these clusters. This was very obvious in the Belarus and the Russian strains, where several more or less time-defined clusters evolved from each other, while during the short (4 weeks) observation period H genes of the German strains showed no fixed mutation and only a single cluster in the P/H pseudogene. The evolutionary pattern described was particularly obvious when a recent B2 strain from DR-Congo was compared to a 22-year-old sequence from Gabon, where the H gene and the P gene showed a stronger and a weaker evolutionary drift, respectively, while the NP-HVR showed none during the 22-year period. Our results suggest that the rate of accumulation of fixed mutations in the H gene in the four outbreaks described here is about one per year. A similar antigenetic drift so far has not been reported for the H gene. This may have important consequences for the antigenicity of the H protein, which is the main target of neutralizing antibodies. Thus, a longer sequencing window has true benefits both for the epidemiological surveillance of measles and for the better monitoring of viral evolution.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Sausy and E. Charpentier for perfect technical assistance. We also thank the WHO-Lab-Net for providing samples from measles patients.
This work represents part of the Ph.D. thesis of J. R. Kessler.
This work was supported by the Fonds National de la Recherche, Luxembourg.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 24 November 2010.
ADDENDUM
The recently identified genotype D11 in China (16) was not included in our calculations.
REFERENCES
- 1. Bankamp B., et al. 2008. Genetic variability and mRNA editing frequencies of the phosphoprotein genes of wild-type measles viruses. Virus Res. 135:298–306 [DOI] [PubMed] [Google Scholar]
- 2. Kremer J. R., et al. 2010. Measles virus strain diversity, Nigeria and the Democratic Republic of the Congo. Emerg. Infect. Dis. 16:1724–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kremer J. R., et al. 2008. High genetic diversity of measles virus, World Health Organization European Region, 2005–2006. Emerg. Infect. Dis. 14:107–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kremer J. R., et al. 2007. Genotyping of recent measles virus strains from Russia and Vietnam by nucleotide-specific multiplex PCR. J. Med. Virol. 79:987–994 [DOI] [PubMed] [Google Scholar]
- 5. Longhi S. 2009. Nucleocapsid structure and function. Curr. Top. Microbiol. Immunol. 329:103–128 [DOI] [PubMed] [Google Scholar]
- 6. Papania M. J., Orenstein W. A. 2004. Defining and assessing measles elimination goals. J. Infect. Dis. 189(Suppl. 1):S23–S26 [DOI] [PubMed] [Google Scholar]
- 7. Riddell M. A., Rota J. S., Rota P. A. 2005. Review of the temporal and geographical distribution of measles virus genotypes in the prevaccine and postvaccine eras. Virol. J. 2:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rota P. A., Featherstone D. A., Bellini W. J. 2009. Molecular epidemiology of measles virus. Curr. Top. Microbiol. Immunol. 330:129–150 [DOI] [PubMed] [Google Scholar]
- 9. Shulga S. V., et al. 2009. Genetic variability of wild-type measles viruses, circulating in the Russian Federation during the implementation of the National Measles Elimination Program, 2003–2007. Clin. Microbiol. Infect. 15:528–537 [DOI] [PubMed] [Google Scholar]
- 10. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 11. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. WHO 2007. Manual for the laboratory diagnosis of measles and rubella virus infection. WHO/IVB/07.01 WHO, Geneva, Switzerland [Google Scholar]
- 13. WHO 2006. Progress in reducing global measles deaths: global measles and rubella laboratory network–update. Wkly. Epidemiol. Rec. 81:90–9416671215 [Google Scholar]
- 14. Wichmann S., Seidler A., Sagebiel D., Hellenbrand W., Santibanez S., Mankertz A., Vogt G., Treeck U., Krause G. 2009. Further efforts needed to achieve measles elimination in Germany: results of an outbreak investigation. Bull. World Health Organ. 87:108–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu W., et al. 1998. New genetic group of measles virus isolated in the People's Republic of China. Virus Res. 54:147–156 [DOI] [PubMed] [Google Scholar]
- 16. Zhang Y., et al. 2010. New measles virus genotype associated with outbreak, China. Emerg. Infect. Dis. 16:943–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.