Abstract
Candida palmioleophila has previously been misidentified as C. famata or C. guilliermondii. We have investigated traditional and modern identification methods for the identification of this and related species. Forty-one clinical isolates previously identified as C. famata or C. guilliermondii and 8 reference strains were included. Color development on CHROMagar, growth temperature ranges, micromorphologies, carbon assimilation (ID32C), matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) profiles, and susceptibility profiles (mica- and anidulafungin and itra-, vori-, posa-, and fluconazole MICs were determined by EUCAST method EDef 7.1, and caspofungin MICs were determined by Etest) were determined, and results were compared to those of molecular identification (ITS1 and ITS2 sequencing). The following five different species were identified among the clinical isolates by sequencing, but no C. famata isolates were found: C. guilliermondii (22 isolates), C. palmioleophila (8 isolates), C. fermentati (6 isolates), C. lusitaniae (3 isolates), and C. intermedia (2 isolates). C. palmioleophila developed a distinct scintillating color of turquoise to rose, grew at 40°C, and failed to produce pseudohyphae within 14 days. The ID32C profile for 7/9 C. palmioleophila isolates was 5367352315, and all were unable to hydrolyze esculin (Esc). The six related species were well discriminated by MALDI-TOF MS. The susceptibility pattern for C. palmioleophila was unique, as the echinocandin MICs were low (range, 0.008 to 0.125 μg/ml) and fluconazole MICs were high (range, 8 to >16 μg/ml). Correct identification of C. palmioleophila is important due to its unique susceptibility profile. Identification is possible yet laborious with conventional techniques, whereas MALDI-TOF MS easily separated the related species.
Modern molecular techniques and growing databases on fungal genome sequences have enabled reliable identification to the species level for species with indistinguishable phenotypic characteristics (11, 23, 24, 26, 34). One example is the discovery of Candida dubliniensis among C. albicans isolates in 1995 (41). Likewise, molecular taxonomic studies have led to the identification of C. nivariensis (1) and C. bracarensis (12), within the C. glabrata complex, and of C. metapsilosis, C. orthopsilosis, and Lodderomyces elongisporus (20, 43), within the C. parapsilosis complex. Due to unique susceptibility profiles of some species, accurate identification is important because treatment strategies are often guided by the species identification (5, 8, 16, 30–32). Thus, misidentification may lead to inappropriate treatment, particularly if accurate susceptibility testing is not performed (7, 8, 28, 35). We show here that this may be the case for C. palmioleophila, which appears to be an emerging species in Denmark (2). C. palmioleophila was first described by Nakase et al. in 1988 (25) and subsequently reported in 1999 as an opportunistic pathogen causing intravenous catheter-associated fungemia (39). This species has notoriously been misidentified as C. famata (Debaryomyces hansenii) (14, 29), which again is phenotypically indistinguishable from C. guilliermondii (Pichia guilliermondii) (27, 44, 49). On this background, we investigated the prevalence of C. palmioleophila in a Danish collection of clinical isolates previously identified as C. famata or C. guilliermondii, using internal transcribed spacer (ITS) sequencing, and subsequently assessed susceptibility profiles and identification results by using routine identification methods and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (33).
(Part of this study was presented at the Interscience Conference on Antimicrobial Agents and Chemotherapy [ICAAC], Boston, MA, September 2010.)
MATERIALS AND METHODS
Strains.
Forty-one clinical isolates (including 31 blood culture isolates) from 37 patients, identified as either C. famata or C. guilliermondii at the mycology reference laboratory at Statens Serum Institut during the period 1949–2009, were included (Table 1). Five strains obtained from the Centraalbureau voor Schimmelcultures (CBS; Utrecht, Netherlands) (C. palmioleophila CBS 7418, C. guilliermondii CBS 6021, C. fermentati [Pichia caribbica] CBS 9966, C. intermedia CBS 572, and C. famata CBS 796) and three isolates received as external quality assessment strains from the United Kingdom National External Quality Assessment Service (UKNEQAS) (P. guilliermondii UKNEQAS 4447, P. guilliermondii UKNEQAS 8112, and C. lusitaniae [Clavispora lusitaniae] UKNEQAS 4620) were included as reference strains. All isolates had been stored in 10% glycerol broth at −80°C.
Table 1.
Phenotypic characteristics of 8 reference strains and 41 clinical isolates originally identified as either C. guilliermondii or C. famataa
| Strain (accession no.) | Original species identification | Growth at 37/40/42°Cb | Formation of pseudohyphae on CMAc | Color on CHROMagar | ATB strip (ID32C) profile | Species identified by ITS sequencing |
|---|---|---|---|---|---|---|
| CBS 7418 | C. palmioleophila | +/+/− | B | Turquoise | 5367352315E− | C. palmioleophila |
| W38018-09 | C. famata | +/+/− | B | Turquoise/rose | 5367352315E− | C. palmioleophila |
| W27955-07 | C. guilliermondii | +/+/− | B | Turquoise/rose | 5367352315E− | C. palmioleophila |
| T47982-09 | C. famata | +/+/− | B | Turquoise/rose | 5367352315E− | C. palmioleophila |
| M45383-09 | C. famata | +/+/− | B | Turquoise/rose | 5367352315E− | C. palmioleophila |
| W29590-09 | C. famata | +/+/− | B | Turquoise/rose | 5367352315E− | C. palmioleophila |
| M62963-09 | C. guilliermondii | +/+/− | B | Turquoise/rose | 5367352315E− | C. palmioleophila |
| M67303-09 | C. famata | +/+/− | B | Turquoise/rose | 367352315E−d | C. palmioleophila |
| F27811-07 | C. guilliermondii | +/+/− | − | Turquoise/rose | 5367352215E−d | C. palmioleophila |
| UKNEQAS 4447 | C. guilliermondii | +/+/− | − | Dark purple | 7577755715E+ | C. guilliermondii |
| UKNEQAS 8112 | C. guilliermondii | +/+/− | − | Light purple | 7577752517E+ | C. guilliermondii |
| CBS 6021 | C. guilliermondii | +/+/− | + | Purple | 7577377315E+ | C. guilliermondii |
| A3970-87 | C. guilliermondii | +/+/− | − | Purple | 5577752717E+ | C. guilliermondii |
| F15415-06 | C. guilliermondii | +/+/− | + | Dark purple | 7177352117E− | C. guilliermondii |
| W63031-08 | C. guilliermondii | +/+/− | − | Purple | 7577352117E+ | C. guilliermondii |
| T51653-07 | C. famata | +/+/− | − | Light purple | 7177352117E− | C. guilliermondii |
| F38277-07 | C. guilliermondii | +/+/− | − | Light purple | 7577352117E+ | C. guilliermondii |
| A173-94 | C. guilliermondii | +/+/w | + | Light purple | 7577352117E+ | C. guilliermondii |
| A336-99 | C. guilliermondii | +/+/− | − | Light purple | 7577357317E+ | C. guilliermondii |
| A331-75 | C. guilliermondii | +/+/w | + | Light purple | 7577752113E+ | C. guilliermondii |
| A194-53 | C. guilliermondii | +/+/w | − | Purple | 7577752117E+ | C. guilliermondii |
| A2189-86 | C. guilliermondii | +/+/− | − | Purple | 7577350515E+ | C. guilliermondii |
| A2551-86 | C. guilliermondii | +/+/− | − | Purple | 5577350117E+ | C. guilliermondii |
| W56260-07 | C. guilliermondii | +/+/− | − | Light purple | 7177352117E− | C. guilliermondii |
| A221-73 | C. guilliermondii | +/+/− | − | Purple | 7577352117E+ | C. guilliermondii |
| PF6934-86 | C. guilliermondii | +/+/− | − | Purple | 7577352117E+ | C. guilliermondii |
| A149-49 | C. guilliermondii | +/+/− | − | Purple | 7577750115E+ | C. guilliermondii |
| A2745-86 | C. guilliermondii | +/+/− | − | Purple | 7577752115E+ | C. guilliermondii |
| F44185-06 | C. guilliermondii | +/+/− | − | Purple | 7577752117E+ | C. guilliermondii |
| A1499-76 | C. guilliermondii | +/+/− | + | Purple | 7577752317E+ | C. guilliermondii |
| A1469-70 | C. guilliermondii | +/+/− | + | Purple | 7577750315E+ | C. guilliermondii |
| M8464-08 | C. guilliermondii | +/+/− | − | Light purple | 7577750315E+ | C. guilliermondii |
| H31147-09 | C. guilliermondii | +/+/− | − | Purple | 7577352117E+ | C. guilliermondii |
| T38215-06 | C. guilliermondii | +/+/− | − | Purple | 7577350117E− | C. guilliermondii |
| CBS 9966 | C. fermentati | +/+/− | − | Purple | 5577350115E+ | C. fermentati |
| F49572-08 | C. guilliermondii | +/+/− | − | Purple | 7577350117E+ | C. fermentati |
| M10611-04 | C. guilliermondii | +/+/− | − | Purple | 7577350117E+ | C. fermentati |
| W63245-01 | C. guilliermondii | +/+/− | + | Purple | 7577750717E+ | C. fermentati |
| T38768-04 | C. guilliermondii | +/+/w | − | Purple | 7577350117E+ | C. fermentati |
| T32779-07 | C. guilliermondii | +/+/w | − | Purple | 7577750117E+ | C. fermentati |
| M39632-09 | C. guilliermondii | +/+/w | − | Dark purple | 7577752317E− | C. fermentati |
| UKNEQAS 4620 | C. lusitaniae | +/+/+ | − | Purple | 5157370317E+ | C. lusitaniae |
| H45593-09 | C. famata | +/+/+ | + | Purple | 5377370317E+ | C. lusitaniae |
| H27507-04 | C. famata | +/+/+ | − | Light red | 5777770317E+ | C. lusitaniae |
| F47819-04 | C. famata | +/+/+ | − | Light red | 5757770317E+ | C. lusitaniae |
| CBS 572 | C. intermedia | −/−/− | + | Purple | 5377340733E+ | C. intermedia |
| A933-78 | C. famata | −/−/− | + | Light purple | 5177360137E+ | C. intermedia |
| F61108-07 | C. famata | −/−/− | + | Dark purple | 5177360137E+ | C. intermedia |
| CBS 796 | C. famata | −/−/− | − | Light red | 5577755117E+ | C. famata |
Growth at 37°C, 40°C, and 42°C, the presence of pseudohyphae on corn meal agar, and the colony color on CHROMagar were analyzed. All isolates were analyzed biochemically by 2nd day ID32C reads, and the ITS1/ITS2 region was sequenced.
w, weak growth.
B, budding present, but no pseudohyphae present.
Unacceptable ATB strip profile. Digits in gray differ from the consensus C. palmioleophila ID32C profile.
Phenotypic characterization.
All isolates were inoculated on corn meal agar (CMA; SSI Diagnostika, Hillerød, Denmark) and incubated at 25°C, and micromorphology was evaluated on days 2, 7, and 14. The ability to grow at 37°C, 40°C, and 42°C was observed on day 2 after inoculation on preheated Sabouraud agar (pH 4; SSI Diagnostika, Hillerød, Denmark), and growth was categorized as either present (+), weak (w), or absent (−). Colony color on CHROMagar (SSI Diagnostika, Hillerød, Denmark) was examined on days 1, 2, and 3 after incubation at 37°C. Carbon assimilation patterns were obtained using commercially available ATB strips (ID32C; bioMérieux, Marcy l'Etoile, France), which were read on day 2.
Susceptibility testing.
Susceptibilities to anidulafungin (MIC range, 0.008 to 4 μg/ml), caspofungin (two batches [TEK0010 and VEK0090]; MIC range, 0.008 to 4 μg/ml), micafungin (MIC range, 0.008 to 4 μg/ml), fluconazole (MIC range, 0.125 to 16 μg/ml), itraconazole (MIC range, 0.03 to 4 μg/ml), posaconazole (MIC range, 0.03 to 4 μg/ml), and voriconazole (MIC range, 0.03 to 4 μg/ml) were determined using the EUCAST microdilution method (13). MICs for caspofungin were also determined using Etest with RPMI–2% glucose medium (AB bioMérieux, Solna, Sweden) following the manufacturer's instructions.
MALDI-TOF MS.
All clinical isolates were subjected to MALDI-TOF MS by following the instructions of the manufacturer and following previously described guidelines for yeast identification (22, 45). Prior to sample preparation, all yeasts were grown for 2 days at 37°C (or 25°C if growth was absent at 37°C) on CHROMagar, subjected to 70% ethanol fixation, and either submitted to MALDI-TOF preparation or stored at −18°C until used. Measurements were performed with a Microflex mass spectrometer (Bruker Daltonics, Germany) using Flexcontrol, version 3.0, and spectra were imported and analyzed using Maldi Biotyper (version 2.0; Bruker Daltonics, Germany). Spectra were calibrated using Escherichia coli ribosomal proteins and evaluated against the Biotyper spectrum database by the default pattern-matching algorithm. Results are expressed as log values ranging from 0 to 3, where values of >1.7 are generally used for reliable genus identification and score values of >2.0 are used for reliable species identification (38). The original database did not contain reference spectra for C. palmioleophila or C. fermentati, so the MALDI-TOF profiles of our reference isolates were added manually to the library for a reevaluation of all obtained spectra (38). A MALDI-TOF score-oriented dendrogram was created using default settings in Biotyper (with distances measured by correlation with average linkages).
Molecular identification.
Single yeast colonies were transferred to sterile 1.5-ml Eppendorf tubes. DNA was extracted by a 2-step buffer extraction approach as previously described (9, 10), using colony material as a replacement for clinical specimens. The universal fungal primers ITS1 (CGTAGGTGAACCTGCGG) and ITS4 (TCCTCCGCTTATTGATATGC) (48) were employed to amplify the ribosomal ITS1, 5.8S, and ITS2 regions by conventional PCR, using 5 μl extracted DNA in 25-μl reaction mixtures, applying REDExtract-N-Amp PCR Readymix (R4775; Sigma-Aldrich, Denmark) containing deoxynucleoside triphosphates (dNTPs), MgCl2, and a hot start polymerase. The PCRs were performed in a Primus HT thermal cycler (MWG Biotech) with the following program: 5 min at 95°C, 10 touchdown cycles (94°C for 30 s, 58°C for 15 s [with a decrease of 1°C/cycle], and 75°C for 90 s), 30 cycles with a constant annealing temperature of 48°C, and a final elongation step at 75°C for 5 min. PCR products were purified on Qiagen spin columns (QIAquick PCR purification kit; Qiagen, Denmark) and sequenced by Macrogen, South Korea, applying the ITS1 and ITS4 primers for sequencing of both strands. Sequence analysis, alignments, and phylogenetics were performed with the bioinformatic software CLC DNA Workbench (CLC Bio, Denmark). Sequences were used for BLAST searches of sequence databases available through NCBI for species identifications, aligned to typed reference strains, and compared by phylogenetic analysis using the neighbor-joining algorithm (default) of Saitou and Nei (36), with 100 replicate bootstraps, based on alignment of the obtained ITS sequences.
Nucleotide sequence accession numbers.
The sequences of the clinical isolates have been deposited in GenBank under the following accession numbers: for strain F27811-07, HQ693769; M45383-09, HQ693770; M62963-09, HQ693771; M67303-09, HQ693772; T47982-09, HQ693773; W27955-07, HQ693774; W29590-09, HQ693775; W38018-09, HQ693776; F49572-08, HQ693777; M10611-04, HQ693778; M39632-09, HQ693779; T32779-07, HQ693780; T38768-04, HQ693781; W63245-01, HQ693782; A933-78, HQ693783; F61108-07, HQ693784; F47819-04, HQ693785; H27507-04, HQ693786; H45593-09, HQ693787; A1469-70, HQ693788; A149-49, HQ693789; A1499-76, HQ693790; A173-94, HQ693791; A194-53, HQ693792; A2189-86, HQ693793; A221-73, HQ693794; A2551-86, HQ693795; A2745-86, HQ693796; A331-75, HQ693797; A336-99, HQ693798; A3970-87, HQ693799; F15415-06, HQ693800; F38277-07, HQ693801; F44185-06, HQ693802; H31147-09, HQ693803; M8464-08, HQ693804; PF6934-86, HQ693805; T38215-06, HQ693806; T51653-07, HQ693807; W56260-07, HQ693808; and W63031-08, HQ693809.
RESULTS
Candida palmioleophila discovered by molecular identification.
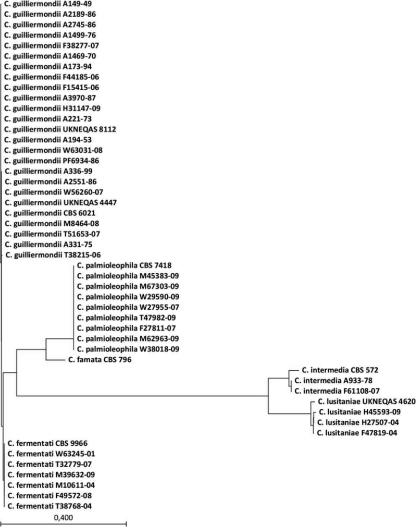
Sequencing of the rRNA ITS regions revealed that a total of 20 of 41 clinical isolates (49%) (Table 1) previously identified as C. guilliermondii or C. famata were misidentified by conventional methods. Among these, eight C. palmioleophila isolates (19.5%) were found (all from blood; two from 2007 and six from 2009). None of 11 isolates previously identified as C. famata were confirmed as this species, since 5 isolates were reidentified as C. palmioleophila, 3 as C. lusitaniae, 2 as C. intermedia, and 1 as C. guilliermondii. For comparison, among the 30 isolates initially identified as C. guilliermondii, 21 were confirmed by sequencing, whereas 6 were reidentified as C. fermentati (Pichia caribbica) and 3 were reidentified as C. palmioleophila. Phylogenetic analysis (Fig. 1) illustrated a genetic relatedness between C. guilliermondii and C. fermentati, while C. palmioleophila and the remaining three species clustered in separate groups.
Fig. 1.
Unrooted phylogenetic tree based on ITS sequence alignment, using the neighbor-joining algorithm (default) of Saitou and Nei (36), with 100 replicate bootstraps (CLC DNA Workbench). The scale bar represents the bootstrap distance. All isolates were named based on the individual BLAST result, followed by accession number and the year of isolation.
Phenotypic characteristics of C. palmioleophila are distinct.
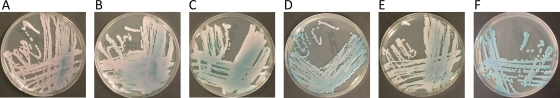
The nine C. palmioleophila isolates (reference strain included) showed uniform growth patterns (Table 1), a maximum growth temperature of 40°C, intense budding but no pseudohyphae after 14 days, and color development on CHROMagar from turquoise (reference isolate only) to a distinct scintillating turquoise to rose (all clinical isolates) (Fig. 2). For comparison, only C. lusitaniae isolates were consistently able to grow well at 42°C, C. famata and C. intermedia were unable to grow at 37°C, and C. intermedia was the only species producing pseudohyphae on day 2. Moreover, all species other than C. palmioleophila developed various but uniform shades of purple or red.
Fig. 2.
Mosaic of six clinical isolates and the CBS reference strain of C. palmioleophila photographed on day 3 after incubation at 37°C on CHROMagar. (A) M45383-09; (B) W27955-07; (C) W38018-09; (D) F27811-07; (E) T47982-09; (F) CBS 7418. The clinical isolates scintillate from turquoise to rose, whereas the reference isolate develops a uniform turquoise color.
Apart from a single isolate (M67303-09) tolerating cycloheximide (ACT) and one isolate (F27811-07) unable to assimilate d-melezitose (MLZ), the nine C. palmioleophila isolates had identical ID32C profiles and were all unable to hydrolyze esculin (Table 1). Consequently, ATB strip reads for C. palmioleophila gave either a 91.7% match for C. famata or an unacceptable profile. ID32C profiles for the other species were generally less uniform, but all were able to assimilate d-cellobiose, in contrast to C. palmioleophila.
MALDI-TOF MS accurately discriminates the six related species.
The obtained MALDI-TOF mass spectra were evaluated against the original spectrum database and against the updated version, which included the additional reference spectra. Evaluation against the original database gave 27 isolates with a correct best match, including 21 with a score of >2 and 6 with a score of 1.7 to <2.0. Unreliable species identifications were clearly noticed by low score values (<1.5), indicating the absence of appropriate reference spectra (8/8 C. palmioleophila isolates and 6/6 C. fermentati isolates). Evaluation against the updated database yielded 41 of 41 isolates with a correct best match and 37 (90%) isolates with a spectral score value of >2. Two isolates of C. palmioleophila scored 1.99, and a third isolate scored 1.90, whereas a single C. guilliermondii isolate scored 1.93.
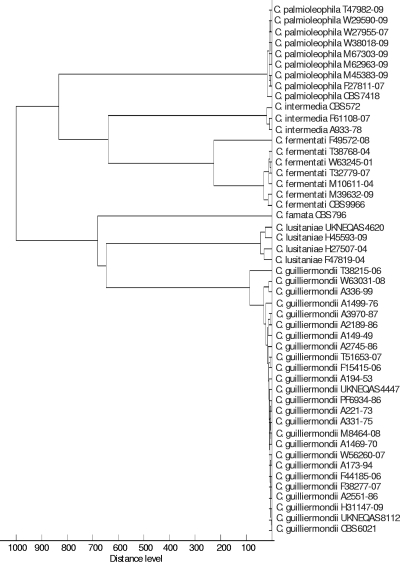
A score-oriented dendrogram was created based on the spectra (Fig. 3), and it shows all isolates clustering with the corresponding reference strains, in agreement with ITS sequence identification.
Fig. 3.
Score-oriented dendrogram using Euclidian squared distances and average linkages to cluster the MALDI-TOF mass spectra obtained for all included isolates. All samples were named based on molecular identification, illustrating the complete agreement between ITS sequencing and the MALDI-TOF spectra.
C. palmioleophila displays a unique susceptibility profile.
The clinical isolates of C. palmioleophila were highly susceptible to echinocandins (anidulafungin and micafungin MICs of ≤0.03 μg/ml and caspofungin Etest MICs of ≤0.125 μg/ml), less susceptible to itraconazole (MIC range, 0.125 to 1 μg/ml), posaconazole (MIC range, 0.06 to 0.25 μg/ml), and voriconazole (MIC range, 0.125 to 1 μg/ml), and resistant to fluconazole (MICs of ≥16 μg/ml). For comparison, echinocandin MICs were considerably higher for C. guilliermondii and C. fermentati isolates (e.g., anidulafungin MIC50s [ranges] of 2 μg/ml [0.125 to 4 μg/ml] and 1 μg/ml [0.5 to 2 μg/ml], respectively), and azole MICs were notably lower for C. lusitaniae, C. intermedia, and C. famata (Table 2). The MIC values for caspofungin determined by the EUCAST method were not in agreement with the Etest MICs. Thus, given by species and as MIC50s (MIC ranges), the values were as follows for both caspofungin lots: for C. palmioleophila, 0.5 (0.5 to 1); for C. guilliermondii, 1 (0.5 to 2); for C. fermentati, 1 (0.5 to 1); for C. lusitaniae, 1 (1); for C. intermedia, 0.5 (0.5 to 1); and for C. famata, 0.25 (0.25).
Table 2.
Selected susceptibility results by species for the 41 clinical isolates and 8 reference strainsa
| Strain (accession no.) | Species | MIC (μg/ml) |
||||
|---|---|---|---|---|---|---|
| Fluconazole | Voriconazole | Anidulafungin | Micafungin | Caspofungin | ||
| CBS 7418 | C. palmioleophila | 8 | 0.06 | 0.008 | 0.008 | 0.032 |
| W38018-09 | C. palmioleophila | 16 | 0.125 | 0.016 | 0.016 | 0.125 |
| W27955-07 | C. palmioleophila | 16 | 0.25 | 0.03 | 0.03 | 0.125 |
| T47982-09 | C. palmioleophila | 16 | 0.125 | 0.03 | 0.016 | 0.125 |
| M45383-09 | C. palmioleophila | >16 | 1 | 0.016 | 0.016 | 0.125 |
| W29590-09 | C. palmioleophila | >16 | 0.5 | 0.03 | 0.016 | 0.125 |
| M62963-09 | C. palmioleophila | >16 | 0.125 | 0.03 | 0.016 | 0.064 |
| M67303-09 | C. palmioleophila | >16 | 0.25 | 0.03 | 0.016 | 0.064 |
| F27811-07 | C. palmioleophila | >16 | 1 | 0.06 | 0.016 | 0.125 |
| UKNEQAS 4447 | C. guilliermondii | 2 | ≤0.03 | 0.5 | 0.25 | 0.25 |
| UKNEQAS 8112 | C. guilliermondii | 4 | 0.06 | 1 | 0.25 | 0.5 |
| CBS 6021 | C. guilliermondii | 4 | ≤0.03 | 0.5 | 0.25 | 0.5 |
| A3970-87 | C. guilliermondii | 2 | 0.06 | 0.125 | 0.06 | 1 |
| F15415-06 | C. guilliermondii | 2 | ≤0.03 | 2 | 0.25 | 0.25 |
| W63031-08 | C. guilliermondii | 2 | 0.06 | 0.5 | 0.125 | 0.5 |
| T51653-07 | C. guilliermondii | 4 | 0.125 | 2 | 0.5 | 0.5 |
| F38277-07 | C. guilliermondii | 4 | 0.06 | 2 | 0.5 | 0.5 |
| A173-94 | C. guilliermondii | 4 | ≤0.03 | 2 | 0.25 | 0.5 |
| A336-99 | C. guilliermondii | 4 | 0.06 | 2 | 0.25 | 0.25 |
| A331-75 | C. guilliermondii | 4 | 0.125 | 1 | 0.25 | 0.5 |
| A194-53 | C. guilliermondii | 4 | 0.06 | 2 | 0.25 | 2 |
| A2189-86 | C. guilliermondii | 8 | 0.06 | 0.5 | 0.125 | 0.5 |
| A2551-86 | C. guilliermondii | 8 | 0.06 | 0.5 | 0.25 | 1 |
| W56260-07 | C. guilliermondii | 8 | 0.125 | 2 | 0.5 | 0.5 |
| A221-73 | C. guilliermondii | 8 | 0.125 | 2 | 0.25 | 0.5 |
| PF6934-86 | C. guilliermondii | 8 | 0.125 | 1 | 0.25 | 0.5 |
| A149-49 | C. guilliermondii | 8 | ≤0.03 | 2 | 0.25 | 0.5 |
| A2745-86 | C. guilliermondii | 8 | 0.06 | 2 | 0.25 | 0.5 |
| F44185-06 | C. guilliermondii | 8 | 0.125 | 2 | 0.5 | 0.5 |
| A1499-76 | C. guilliermondii | 8 | 0.06 | 4 | 0.25 | 0.5 |
| A1469-70 | C. guilliermondii | >16 | 0.5 | 1 | 1 | 1 |
| M8464-08 | C. guilliermondii | >16 | 0.5 | 1 | 0.25 | 0.25 |
| H31147-09 | C. guilliermondii | >16 | 0.5 | 1 | 0.25 | 1 |
| T38215-06 | C. guilliermondii | >16 | 1 | 2 | 1 | 0.5 |
| CBS 9966 | C. fermentati | 2 | ≤0.03 | 2 | 0.25 | 0.5 |
| F49572-08 | C. fermentati | >16 | 2 | 2 | 0.5 | 0.5 |
| M10611-04 | C. fermentati | 1 | ≤0.03 | 1 | 0.125 | 0.5 |
| W63245-01 | C. fermentati | 1 | 0.06 | 0.5 | 0.125 | 0.25 |
| T38768-04 | C. fermentati | 1 | ≤0.03 | 0.5 | 0.125 | 0.25 |
| T32779-07 | C. fermentati | 1 | 0.06 | 1 | 0.25 | 0.25 |
| M39632-09 | C. fermentati | 4 | 0.125 | 2 | 0.25 | 0.25 |
| UKNEQAS 4620 | C. lusitaniae | 0.25 | ≤0.03 | 0.03 | 0.016 | 0.5 |
| H45593-09 | C. lusitaniae | ≤0.125 | ≤0.03 | 0.06 | 0.06 | 0.25 |
| H27507-04 | C. lusitaniae | 0.25 | ≤0.03 | 0.03 | 0.03 | 0.5 |
| F47819-04 | C. lusitaniae | 0.5 | ≤0.03 | 0.06 | 0.06 | 0.25 |
| CBS 572 | C. intermedia | ≤0.125 | ≤0.03 | 0.06 | 0.008 | 0.25 |
| A933-78 | C. intermedia | 1 | ≤0.03 | 0.03 | 0.008 | 0.125 |
| F61108-07 | C. intermedia | 2 | ≤0.03 | 0.03 | 0.008 | 0.125 |
| CBS 796 | C. famata | ≤0.125 | ≤0.03 | 0.03 | 0.008 | 0.032 |
MICs for fluconazole, voriconazole, anidulafungin, and micafungin were determined by EUCAST methodology, and those for caspofungin were determined using Etest.
DISCUSSION
In this work, we report on Candida palmioleophila as a previously overlooked fungal pathogen, characterize its unique susceptibility profile, and evaluate classical and new identification tools for correct identification of this species.
Within the collection of clinical isolates previously identified as C. famata or C. guilliermondii, four species were discovered: C. palmioleophila was discovered among both sets, C. fermentati among “C. guilliermondii” isolates only, and C. lusitaniae and C. intermedia solely among “C. famata” isolates. Overall, almost half of the isolates were originally misidentified by conventional diagnostics, even though the tests were performed at a specialized laboratory, and notably, no clinical isolates were confirmed to be C. famata. This suggests that C. famata may be an even more uncommon human pathogen than initially thought, if not totally absent. Our findings are in agreement with the work of Desnos-Ollivier et al. (14), who were able to confirm only 3 of 26 clinical isolates as C. famata by molecular methods, with only 1 confirmed as an invasive isolate. Interestingly, they also found a panel of misidentified species, including three clinical C. palmioleophila isolates misidentified as C. famata. However, in contrast to our findings, no C. intermedia isolates were found, but Pichia jadinii and C. haemulonii type II isolates were identified, again emphasizing the challenges for discriminating related species within the C. guilliermondii/C. famata group and the geographical variation in species distribution.
As shown in this study, correct identification could be obtained for C. palmioleophila, C. lusitaniae, C. intermedia, and C. famata by carefully combining traditional routine identification methods, whereas C. guilliermondii and C. fermentati remain notoriously inseparable (Table 3) (6, 17, 21, 37). Discriminating C. palmioleophila from C. guilliermondii is highly clinically relevant because the latter has reduced susceptibility to echinocandins due to a naturally occurring polymorphism at a locus that reduces susceptibility (Fks1p M642) (David Perlin, personal communication), whereas C. palmioleophila was shown to be highly sensitive. Thus, echinocandins would be an excellent choice for C. palmioleophila but less so for C. guilliermondii (21). C. lusitaniae is important to identify correctly because it is often misidentified (14, 42), but it should be regarded as a poor target for amphotericin B despite being classified as susceptible based on MIC determinations (5). Overall, compared to species complexes such as the C. parapsilosis complex or the C. glabrata complex, accurate identification within the C. guilliermondii/C. famata group has more clinical importance due to the remarkably diverse susceptibility profiles of the group members (18–20, 40). The challenges in discriminating C. fermentati and C. guilliermondii have been assessed previously, and the need for molecular methods for accurate identification was demonstrated (6, 17, 37, 46). Nevertheless, although interesting from an epidemiological perspective, the discrimination of these two species is clinically less crucial due to their equivalent susceptibility patterns.
Table 3.
Summary of key phenotypic and biochemical characteristics of included Candida isolates
| Species | Growth at 37/40/42°C | Formation of pseudohyphaea | Color on CHROMagar | Esculin hydrolysisa | d-Cellobiose assimilation | MIC range (μg/ml) |
||
|---|---|---|---|---|---|---|---|---|
| Anidulafungin | Fluconazole | Voriconazole | ||||||
| C. palmioleophila | +/+/− | − | Turquoise/rose | − | − | 0.008–0.06 | 8–>16 | 0.06–1 |
| C. guilliermondii | +/+/− | V | Purple | Vb | + | 0.125–4 | 2–>16 | ≤0.03–1 |
| C. fermentati | +/+/− | V | Purple | Vc | + | 0.5–2 | 1–>16 | ≤0.03–2 |
| C. lusitaniae | +/+/+ | V | Purple/red | + | + | 0.03–0.06 | ≤0.125–0.5 | ≤0.03 |
| C. intermedia | −/−/− | + | Purple | + | + | 0.03–0.06 | ≤0.125–2 | ≤0.03 |
| C. famata | −/−/− | − | Light red | + | + | 0.03 | ≤0.125 | ≤0.03 |
V, variable.
A total of 20/25 isolates were able to hydrolyze esculin.
A total of 6/7 isolates were able to hydrolyze esculin.
Evaluation of caspofungin MICs with the EUCAST method gave a uniform and narrow MIC range of 0.25 to 2 μg/ml across the included species and both caspofungin lots (TEK0010 and VEK0090). This finding contrasts with the diverse susceptibility patterns suggested by the anidulafungin and micafungin EUCAST MIC results and the caspofungin Etest end points. We previously reported on variability in caspofungin microdilution MIC values across microdilution methods, time, and country (3, 4). So far, no data suggest a variable susceptibility to the three echinocandins, and we believe that the elevated caspofungin EUCAST MIC ranges for the most anidulafungin- and micafungin-susceptible species in this study reflect an in vitro phenomenon rather than true differences in susceptibility.
Conventional identification methods often require several phenotypic and biochemical assays, which are time-consuming and still insufficient for precise discrimination of some species (28, 29, 42, 47). On the other hand, molecular methods are rapid and highly discriminative identification tools, yet they are often expensive and may require skilled technicians (15, 22, 44). MALDI-TOF has recently proved a useful and powerful identification tool for several yeast and mold species and has been introduced over recent years to many routine clinical microbiology laboratories (22, 33, 38, 45). We therefore examined the performance of this method with this group of closely related Candida species. As shown in this study, the accuracy of the MALDI-TOF analysis and spectrum evaluation against the updated database was significant, yielding only four spectra which should be considered in the borderline range for reliable species identification (spectral scores of 1.9 to <2.0). However, a correct best match was obtained for all clinical isolates. Moreover, phylogenetic analysis based on the obtained MALDI-TOF spectra depicted an excellent discriminatory power of this analysis, since all isolates were clustered with the corresponding reference strain and distinct from the other species. To our knowledge, this is the first report to describe the excellent performance of MALDI-TOF for the identification of C. palmioleophila and related cryptic species.
In conclusion, Candida palmioleophila is an emerging pathogen in Denmark and is often misidentified as C. guilliermondii or C. famata. Due to its unique susceptibility profile, correct identification has a high clinical importance. By conventional mycological methods, identification requires several tests and technician experience, yet MALDI-TOF MS and ITS sequencing may provide rapid and powerful alternatives to conventional identification techniques.
ACKNOWLEDGMENTS
We thank the technical staff from the Mycology Division, Department for Microbiological Diagnostics, Statens Serum Institute, especially Birgit Brandt and Stéphane Fabrice Butteux, for contributions to the experimental work. Moreover, we thank Rimtas Dargis for his expertise and assistance in the MALDI-TOF mass spectrometry analysis.
Footnotes
Published ahead of print on 8 December 2010.
REFERENCES
- 1. Alcoba-Florez J., et al. 2005. Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. J. Clin. Microbiol. 43:4107–4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arendrup M. C., et al. National surveillance of fungemia in Denmark (2004 to 2009). J. Clin. Microbiol. 49:325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arendrup M. C., et al. 2009. Breakthrough Aspergillus fumigatus and Candida albicans double infection during caspofungin treatment: laboratory characteristics and implication for susceptibility testing. Antimicrob. Agents Chemother. 53:1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arendrup M. C., et al. 2010. Echinocandin susceptibility testing of Candida species: comparison of EUCAST EDef 7.1, CLSI M27–A3, Etest, disk diffusion, and agar dilution methods with RPMI and Iso-Sensitest media. Antimicrob. Agents Chemother. 54:426–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Atkinson B. J., Lewis R. E., Kontoyiannis D. P. 2008. Candida lusitaniae fungemia in cancer patients: risk factors for amphotericin B failure and outcome. Med. Mycol. 46:541–546 [DOI] [PubMed] [Google Scholar]
- 6. Bai F. Y., Liang H. Y., Jia J. H. 2000. Taxonomic relationships among the taxa in the Candida guilliermondii complex, as revealed by comparative electrophoretic karyotyping. Int. J. Syst. Evol. Microbiol. 50:417–422 [DOI] [PubMed] [Google Scholar]
- 7. Bishop J. A., et al. 2008. Candida bracarensis detected among isolates of Candida glabrata by peptide nucleic acid fluorescence in situ hybridization: susceptibility data and documentation of presumed infection. J. Clin. Microbiol. 46:443–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Borman A. M., et al. 2008. Candida nivariensis, an emerging pathogenic fungus with multidrug resistance to antifungal agents. J. Clin. Microbiol. 46:933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brillowska-Dabrowska A. December 2006. DNA preparation from nail samples. Denmark patent WO2006133701
- 10. Brillowska-Dabrowska A., Saunte D. M., Arendrup M. C. 2007. Five-hour diagnosis of dermatophyte nail infections with specific detection of Trichophyton rubrum. J. Clin. Microbiol. 45:1200–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coignard C., et al. 2004. Resolution of discrepant results for Candida species identification by using DNA probes. J. Clin. Microbiol. 42:858–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Correia A., Sampaio P., James S., Pais C. 2006. Candida bracarensis sp. nov., a novel anamorphic yeast species phenotypically similar to Candida glabrata. Int. J. Syst. Evol. Microbiol. 56:313–317 [DOI] [PubMed] [Google Scholar]
- 13. Cuenca-Estrella M., et al. 2003. Multicenter evaluation of the reproducibility of the proposed antifungal susceptibility testing method for fermentative yeasts of the Antifungal Susceptibility Testing Subcommittee of the European Committee on Antimicrobial Susceptibility Testing (AFST-EUCAST). Clin. Microbiol. Infect. 9:467–474 [DOI] [PubMed] [Google Scholar]
- 14. Desnos-Ollivier M., et al. 2008. Debaryomyces hansenii (Candida famata), a rare human fungal pathogen often misidentified as Pichia guilliermondii (Candida guilliermondii). J. Clin. Microbiol. 46:3237–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fujita S., Hashimoto T. 2000. DNA fingerprinting patterns of Candida species using HinfI endonuclease. Int. J. Syst. Evol. Microbiol. 50:1381–1389 [DOI] [PubMed] [Google Scholar]
- 16. Garey K. W., et al. 2006. Time to initiation of fluconazole therapy impacts mortality in patients with candidemia: a multi-institutional study. Clin. Infect. Dis. 43:25–31 [DOI] [PubMed] [Google Scholar]
- 17. Lan L., Xu J. 2006. Multiple gene genealogical analyses suggest divergence and recent clonal dispersal in the opportunistic human pathogen Candida guilliermondii. Microbiology 152:1539–1549 [DOI] [PubMed] [Google Scholar]
- 18. Lockhart S. R., et al. 2009. Identification of Candida nivariensis and Candida bracarensis in a large global collection of Candida glabrata isolates: comparison to the literature. J. Clin. Microbiol. 47:1216–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lockhart S. R., Messer S. A., Pfaller M. A., Diekema D. J. 2008. Geographic distribution and antifungal susceptibility of the newly described species Candida orthopsilosis and Candida metapsilosis in comparison to the closely related species Candida parapsilosis. J. Clin. Microbiol. 46:2659–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lockhart S. R., Messer S. A., Pfaller M. A., Diekema D. J. 2008. Lodderomyces elongisporus masquerading as Candida parapsilosis as a cause of bloodstream infections. J. Clin. Microbiol. 46:374–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lockhart S. R., Messer S. A., Pfaller M. A., Diekema D. J. 2009. Identification and susceptibility profile of Candida fermentati from a worldwide collection of Candida guilliermondii clinical isolates. J. Clin. Microbiol. 47:242–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marklein G., et al. 2009. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 47:2912–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McMullan R., et al. 2008. A prospective clinical trial of a real-time polymerase chain reaction assay for the diagnosis of candidemia in nonneutropenic, critically ill adults. Clin. Infect. Dis. 46:890–896 [DOI] [PubMed] [Google Scholar]
- 24. Mirhendi H., et al. 2010. Molecular screening for Candida orthopsilosis and Candida metapsilosis among Danish Candida parapsilosis group blood culture isolates: proposal of a new RFLP profile for differentiation. J. Med. Microbiol. 59:414–420 [DOI] [PubMed] [Google Scholar]
- 25. Nakase T., Itoh M., Suzuki M., Komagata K., Kodama T. 1988. Candida palmioleophila sp. nov., a yeast capable of assimilating crude palm oil, formerly identified as Torulopsis candida. J. Gen. Appl. Microbiol. 34:493–498 [Google Scholar]
- 26. Nishikawa A., Sugita T., Shinoda T. 1997. Differentiation between Debaryomyces hansenii/Candida famata complex and Candida guilliermondii by polymerase chain reaction. FEMS Immunol. Med. Microbiol. 19:125–129 [DOI] [PubMed] [Google Scholar]
- 27. Nishikawa A., Sugita T., Shinoda T. 1999. Rapid identification of Debaryomyces hansenii/Candida famata by polymerase chain reaction. Med. Mycol. 37:101–104 [PubMed] [Google Scholar]
- 28. Page B. T., Kurtzman C. P. 2005. Rapid identification of Candida species and other clinically important yeast species by flow cytometry. J. Clin. Microbiol. 43:4507–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Page B. T., Shields C. E., Merz W. G., Kurtzman C. P. 2006. Rapid identification of ascomycetous yeasts from clinical specimens by a molecular method based on flow cytometry and comparison with identifications from phenotypic assays. J. Clin. Microbiol. 44:3167–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pfaller M. A., et al. 2010. Wild-type MIC distributions and epidemiological cutoff values for the echinocandins and Candida spp. J. Clin. Microbiol. 48:52–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pfaller M. A., et al. 2005. In vitro activities of anidulafungin against more than 2,500 clinical isolates of Candida spp., including 315 isolates resistant to fluconazole. J. Clin. Microbiol. 43:5425–5427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pfaller M. A., et al. 2010. Results from the Artemis Disk Global Antifungal Surveillance Study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J. Clin. Microbiol. 48:1366–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qian J., Cutler J. E., Cole R. B., Cai Y. 2008. MALDI-TOF mass signatures for differentiation of yeast species, strain grouping and monitoring of morphogenesis markers. Anal. Bioanal. Chem. 392:439–449 [DOI] [PubMed] [Google Scholar]
- 34. Romeo O., Scordino F., Pernice I., Lo P. C., Criseo G. 2009. A multiplex PCR protocol for rapid identification of Candida glabrata and its phylogenetically related species Candida nivariensis and Candida bracarensis. J. Microbiol. Methods 79:117–120 [DOI] [PubMed] [Google Scholar]
- 35. Rowen J. L., Tate J. M., Nordoff N., Passarell L., McGinnis M. R. 1999. Candida isolates from neonates: frequency of misidentification and reduced fluconazole susceptibility. J. Clin. Microbiol. 37:3735–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 37. San Millan R. M., Wu L. C., Salkin I. F., Lehmann P. F. 1997. Clinical isolates of Candida guilliermondii include Candida fermentati. Int. J. Syst. Bacteriol. 47:385–393 [DOI] [PubMed] [Google Scholar]
- 38. Stevenson L. G., Drake S. K., Shea Y. R., Zelazny A. M., Murray P. R. 2010. Evaluation of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF) for the identification of clinically important yeast species. J. Clin. Microbiol. 48:3482–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sugita T., et al. 1999. A clinical isolate of Candida palmioleophila formerly identified as Torulopsis candida. Nippon Ishinkin Gakkai Zasshi 40:21–25 [DOI] [PubMed] [Google Scholar]
- 40. Sullivan D., Coleman D. 1998. Candida dubliniensis: characteristics and identification. J. Clin. Microbiol. 36:329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sullivan D. J., Westerneng T. J., Haynes K. A., Bennett D. E., Coleman D. C. 1995. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology 141:1507–1521 [DOI] [PubMed] [Google Scholar]
- 42. Szabo Z., et al. 2008. Evaluation of the new Micronaut-Candida system compared to the API ID32C method for yeast identification. J. Clin. Microbiol. 46:1824–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tavanti A., Davidson A. D., Gow N. A., Maiden M. C., Odds F. C. 2005. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J. Clin. Microbiol. 43:284–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tsui C. K., Daniel H. M., Robert V., Meyer W. 2008. Re-examining the phylogeny of clinically relevant Candida species and allied genera based on multigene analyses. FEMS Yeast Res. 8:651–659 [DOI] [PubMed] [Google Scholar]
- 45. van Veen S. Q., Claas E. C., Kuijper E. J. 2010. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization–time of flight mass spectrometry in conventional medical microbiology laboratories. J. Clin. Microbiol. 48:900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vaughan-Martini A., Kurtzman C. P., Meyer S. A., O'Neill E. B. 2005. Two new species in the Pichia guilliermondii clade: Pichia caribbica sp. nov., the ascosporic state of Candida fermentati, and Candida carpophila comb. nov. FEMS Yeast Res. 5:463–469 [DOI] [PubMed] [Google Scholar]
- 47. Verweij P. E., Breuker I. M., Rijs A. J., Meis J. F. 1999. Comparative study of seven commercial yeast identification systems. J. Clin. Pathol. 52:271–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. White T. J., Bruns T., Lee S., Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315–322 In Innis M. A., Gelfand D. H., Sninsky J. J. (ed.), PCR protocols. A guide to methods and applications. Academic Press, San Diego, CA [Google Scholar]
- 49. Yamamura M., et al. 2009. Polymerase chain reaction assay for specific identification of Candida guilliermondii (Pichia guilliermondii). J. Infect. Chemother. 15:214–218 [DOI] [PubMed] [Google Scholar]