Abstract
Enterobacterial isolates expressing the carbapenemase NDM-1 are emerging worldwide. Twenty-seven NDM-1-positive isolates of worldwide origin were included in this study to identify these strains as not only pathogens but also colonizers of normal flora for infection control screening. Although susceptibility to carbapenems varied, a combined test (IMP/IMP + EDTA), the Etest MBL, and automated susceptibility testing by Vitek2 (bioMérieux) identified those NDM-1 producers as verified by PCR using specific primers. Screening for carriers of NDM-1 producers may be based on media such as the ChromID ESBL culture medium routinely used to screen for extended-spectrum β-lactamase producers, which gives excellent detection levels with low limits of detection ranging from 8 × 100 to 5 × 102 CFU/ml. The CHROMagar KPC culture medium had higher limits of detection (1 × 101 to 5 × 105 CFU/ml) and may be proposed for the follow-up of outbreaks of infections with NDM-1 producers. Colonies growing on these screening media can be verified as NDM-1 producers with molecular methods as described herein.
NDM-1 is a broad-spectrum β-lactamase (carbapenemase) that is able to inactivate all β-lactams except aztreonam (18). However, most NDM-1-positive strains also express the CMY-4 and CTX-M-15 β-lactamases, which confer resistance to all β-lactams (8). Although discovered in 2008, NDM-1-positive Enterobacteriaceae have attracted worldwide media attention due to their high incidence in India and because many United Kingdom isolates can be directly tracked to India and Pakistan (5, 8). Several reports indicate further spread of this resistance determinant worldwide, including in the United States (three isolates [3]) and Australia (11), with relationships in most of the cases with the Indian subcontinent. In contrast to many other resistance mechanisms, NDM-1 is not associated with a single strain but has spread very rapidly to non-clonally related isolates (8). It has been identified mostly in Escherichia coli and Klebsiella pneumoniae and to a lesser degree in other enterobacterial species (5, 8). Plasmids carrying the blaNDM-1 gene also carry a number of other genes conferring resistance to all aminoglycosides, macrolides, and sulfamethoxazole, thus making these isolates multidrug resistant or, because of other non-plasmid-mediated resistances, resistant in some cases to all antibiotics, including tigecycline and colistin (8). Although carbapenemase-producing Enterobacteriaceae isolates have been increasingly identified worldwide, such as K. pneumoniae carbapenemase (KPC) producers (9, 10), NDM-1 producers bring several additional factors which are deeply disconcerting for public health worldwide. First, the blaNDM-1 gene has been identified in not a single species but in unrelated species (including Acinetobacter spp. [7]), indicating that this gene can spread at an unprecedented rate. Second, it is present not only in K. pneumoniae, a typical nosocomial pathogen, but also in E. coli, which is also a major community-acquired pathogen. Third, E. coli is also the number one cause of diarrhea in children in India and Pakistan, increasing the risk of resistant strains being released into the environment, and the blaNDM-1 gene is primarily from the Indian subcontinent, which has the second largest population in the world (1.3 billion people). Overpopulation, lack of basic sanitation and access to clean water, a tropical climate (the southern part of the peninsula), and poor control of antibiotic use are all compounding factors that may promote the transfer of NDM-1-positive bacteria among the populations of India and Pakistan and then internationally. Therefore, there is an urgent international need to detect NDM-1 producers in any health care facility to prevent their further spread.
We report here several techniques for the detection of NDM-1 producers—whether for managing clinical infections or screening for colonizers. We have gathered a collection of international nonclonal strains, including several enterobacterial species demonstrating various susceptibilities to carbapenems, which were used to assess these methods (Table 1).
Table 1.
Sensitivity of detection of 27 NDM-1 producers by ChromID ESBL and CHROMagar KPC media, results of the combined disk testa and the Etest MBL, and MICs of several β-lactams
| Isolate | Country of isolation | MIC (μg/ml)b |
Etest MBL | IMP/IMP + EDTA | Lower limit of detection (CFU/ml) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CTX | CAZ | IMP | ETP | MER | DOR | ChromID ESBL | CHROMagar KPC | ||||
| E. coli | |||||||||||
| A | Australia | >32 | >256 | 6 | 4 | 4 | >32 | + | + | 2 × 101 | 3 × 101 |
| B | France | >32 | >256 | 3 | 3 | 2 | 2 | + | + | 2 × 101 | 3 × 105 |
| C | India | >32 | >256 | 2 | >32 | 24 | 6 | + | + | 3 × 101 | 3 × 101 |
| D | India | >32 | >256 | 16 | >32 | 16 | 3 | + | + | 1 × 101 | 1 × 101 |
| E | India | >32 | >256 | 32 | >32 | >32 | 24 | + | + | 2 × 102 | 1 × 101 |
| F | India | >32 | >256 | >32 | >32 | 16 | 16 | + | + | 1 × 101 | 3 × 101 |
| G | India | >32 | >256 | 4 | >32 | 8 | 3 | + | + | 3 × 101 | 4 × 101 |
| H | India | >32 | >256 | 1.5 | 8 | 2 | 1 | + | + | 1 × 101 | 2 × 105 |
| I | India | >32 | >256 | 3 | >32 | 6 | 2 | + | + | 1 × 101 | 1 × 101 |
| J | India | >32 | >256 | 2 | 4 | 2 | 1.5 | + | + | 8 × 100 | 1 × 104 |
| E. cloacae | |||||||||||
| A | India | >32 | >256 | 8 | 6 | 6 | 4 | + | + | 1 × 101 | 5 × 101 |
| B | India | >32 | >256 | >32 | 12 | 8 | 12 | + | + | 1 × 101 | 1 × 101 |
| C | India | >32 | >256 | 2 | 16 | 2 | 1 | + | + | 3 × 102 | 4 × 104 |
| D | India | >32 | >256 | 0.75 | 3 | 1.5 | 1 | —c | + | 1 × 101 | 1 × 102 |
| K. pneumoniae | |||||||||||
| A | Kenya | >32 | >256 | >32 | >32 | >32 | >32 | + | + | 2 × 101 | 1 × 101 |
| B | Sultanate of Oman | >32 | >256 | >32 | >32 | >32 | >32 | + | + | 1 × 101 | 2 × 101 |
| C | Sultanate of Oman | >32 | >256 | 1 | 6 | 2 | 2 | + | + | 1 × 101 | 3 × 102 |
| D | India | >32 | >256 | >32 | >32 | >32 | >32 | + | + | 2 × 101 | 3 × 101 |
| E | India | >32 | >256 | 0.75 | 8 | 2 | 1.5 | — | + | 1 × 101 | 3 × 102 |
| F | India | >32 | >256 | >32 | >32 | >32 | >32 | + | + | 1 × 101 | 8 × 101 |
| G | India | >32 | >256 | 2 | 6 | 2 | 1.5 | + | + | 1 × 101 | 1 × 102 |
| H | India | >32 | >256 | >32 | >32 | >32 | >32 | + | + | 1 × 101 | 5 × 102 |
| I | India | >32 | >256 | >32 | >32 | >32 | >32 | + | + | 1 × 101 | 5 × 102 |
| J | India | >32 | >256 | 6 | >32 | 16 | 12 | + | + | 1 × 101 | 1 × 101 |
| K. oxytoca A | India | >32 | >256 | 2 | 4 | 3 | 3 | + | + | 1 × 101 | 1 × 104 |
| C. freundii A | France | >32 | >256 | >32 | >32 | >32 | >32 | + | + | 1 × 101 | 1 × 101 |
| P. rettgeri A | India | >32 | >256 | 3 | 0.5 | 1.5 | 0.75 | + | + | 5 × 102 | 5 × 105 |
IMP, IMP plus EDTA.
CTX, cefotaxime; CAZ; ceftazidime; IMP, imipenem; ETP, ertapenem; MER, meropenem; DOR, doripenem.
—, not interpretable.
Identification of NDM-1 producers.
Susceptibility testing was performed by determining MICs by Etest (AB bioMérieux, Solna, Sweden) on Mueller-Hinton agar plates at 37°C, and results of susceptibility testing were recorded according to the CLSI guidelines as modified in June 2010 (4). The breakpoints for imipenem are as follows: susceptible (S), ≤1 μg/ml; resistant (R), ≥4 μg/ml. Those for meropenem are as follows: S, ≤1 μg/ml; R, ≥4 μg/ml. Those for ertapenem are as follows: S, ≤0.25 μg/ml; R, ≥1 μg/ml. Those for doripenem are as follows: S, ≤1 μg/ml; R, ≥4 μg/ml. Twenty-six of the 27 NDM-1 strains were resistant to at least one carbapenem, and 8 were resistant only to ertapenem (Table 1). A single isolate (Providencia rettgeri A) was only of intermediate susceptibility to imipenem and ertapenem (Table 1). Various susceptibilities to carbapenems have been reported for producers of other types of carbapenemases, such as KPC and VIM producers (9, 10, 12, 15), particularly E. coli. Ertapenem has also been proposed to be a most appropriate carbapenem for detecting KPC producers with low-level resistance to carbapenems (2, 15) and also seems to be the most appropriate carbapenem for detecting NDM-1 producers.
The Etest MBL strip is one of the methods advocated for detecting metallo-β-lactamase (MBL) producers based on inhibition of MBL activity by EDTA (16). The Etest MBL using imipenem and imipenem-EDTA was reliable for their detection, except in two cases (Enterobacter cloacae D and K. pneumoniae E), in which interpretation of the results was not possible because the imipenem MICs were too low (Table 1). Therefore, we have evaluated another technique, the combined disk test, which is also based on inhibition of MBL activity by EDTA (6). Two imipenem disks (10 μg), one containing 10 μl of 0.1 M anhydrous EDTA (292 μg), were placed 25 mm apart on a Mueller-Hinton plate (6). A strain producing a diameter of >4 mm around the disk with IMP-EDTA and not around the disk with IMP alone was considered positive for MBL. All 27 NDM-1 producers were positively detected by this technique (Table 1), which is reliable for detecting MBL producers on a daily basis. The Etest MBL may be used in labs which do not screen for MBL producers on a daily basis or in those which perform susceptibility testing using liquid medium techniques and rarely use disk diffusion susceptibility techniques.
The collection of NDM-1 producers was then examined by a commercial automated system designed for antibiotic susceptibility testing, Vitek 2 (expert system version 04.02; bioMérieux, La-Balme-Les-Grottes, France). The tested carbapenem in card AST-N128 is imipenem, with breakpoints of ≤2 μg/ml for susceptibility and >8 μg/ml for resistance. All NDM-1 producers had MICs of imipenem by Vitek2 that were higher than those obtained by the Etest method. They were of intermediate susceptibility or resistant to carbapenems and were flagged as producers of carbapenemases. Our data mirror and extend the results obtained in another study that evaluated three automated systems, BD Phoenix (BD Diagnostics), Microscan (Siemens), and Vitek 2 (bioMérieux), for the detection and inference of mechanisms responsible for carbapenem resistance in carbapenem-resistant Enterobacteriaceae (17). That study included six NDM-1 producers likely of United Kingdom origin (17).
Although carbapenemase detection by spectrophotometric assay has been suggested for detecting carbapenemase producers (10, 17) and the results were positive for all of the NDM-1 producers we tested here (data not shown), we do not recommend this method on a routine basis as it is time-consuming and does not discriminate between NDM-1 and other MBLs. Similarly, we do not recommend MBL activity detection by the Hodge test, which is not only time-consuming but may also lack specificity (15). Finally, the detection of the blaNDM-1 gene based on a PCR molecular technique was established. The primers assessed in this study were NDM-Fm (5′-GGTTTGGCGATCTGGTTTTC-3′, positions 133 to 153) and NDM-Rm (5′-CGGAATGGCTCATCACGATC-3′, positions 734 to 754), which amplified an internal fragment of 621 bp of the blaNDM-1 gene. Primers were designed according to the sequence of the blaNDM-1 gene in the GenBank database under accession no. FN392876. Total DNA was extracted from bacterial isolates by alkaline lysis. Negative controls included reference strains producing the MBL blaIMP, blaVIM, blaSPM, blaGIM, and blaSIM genes. Two microliters of extracted total DNA was subjected to PCR in a 50-μl reaction mixture. The PCR mixture for the detection of MBL genes contained 1× PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl), 1.5 mM MgCl2, 0.125 mM each deoxynucleotide triphosphate, 0.1 μM each primer, and 2 U of AmpliTaq Gold polymerase (Roche, Meylan, France). Amplification was carried out under the following thermal cycling conditions: 10 min at 94°C; 36 cycles of amplification consisting of 30 s at 94°C, 40 s at 52°C, and 50 s at 72°C; and 5 min at 72°C for the final extension. DNA fragments were visualized by electrophoresis in a 2% agarose gel at 100 V for 1 h in 1× TAE (40 mM Tris-HCl [pH 8.3], 2 mM acetate, 1 mM EDTA) containing 0.05 mg/liter ethidium bromide. Using this technique, the blaNDM-1-positive isolates were detected within less than 3 h with 100% sensitivity and excellent specificity, as deduced by testing a few reference strains expressing other types of carbapenemase genes (data not shown).
Screening for carriers of NDM-1 producers.
Efficient prevention of the spread of NDM-1 producers requires a rapid screening method that can detect NDM-1 producers as colonizers (mostly in the gastrointestinal tract) when the carrier patient is admitted to any health care facility. We have shown recently that the screening culture medium ChromID ESBL culture medium (bioMérieux) containing cefpodoxime as a selector, which is routinely used to screen for extended-spectrum β-lactamase (ESBL) producers (13), may also be used to detect carbapenemase-producing Enterobacteriaceae (1). This culture medium may also provide a preliminary bacterial identification (E. coli; K. pneumoniae, etc.) at the species level based on the differential chromogenic properties of the enterobacterial species (13). Producers of IMP-, VIM-, and KPC-type carbapenemases with high-level resistance to cephalosporins and carbapenems were easily detected by ChromID ESBL (1, 13). In this same study, we had evaluated another screening medium, CHROMagar KPC (CHROMagar Company, Paris, France), which contains a carbapenem as the selector for resistance and was first specifically designed for screening for KPC producers (1, 14). Using the CHROMagar medium, carbapenemase-producing isolates with MICs of <4 μg/ml were detected with much higher detection limits (1).
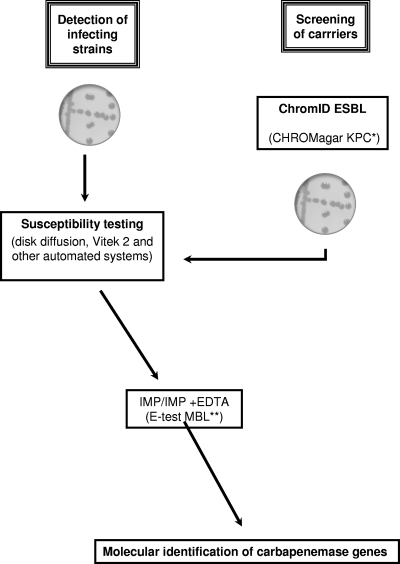
Since NDM-1 producers were not included in that study, we have now evaluated both screening media using our collection of NDM-1 producers. An inoculum of ∼2 × 106 (range, 1.5 × 105 to 3.5 × 107) CFU/ml was used, serial 10-fold dilutions were made in normal saline buffer, and then 100 μl was plated onto sheep blood-containing tryptic soy agar, ChromID ESBL medium, and CHROMagar KPC medium. Viable bacteria were counted after 24 and 48 h of culture at 37°C, and growth on selective medium was compared to growth on standard culture medium. The lower limit of detection of NDM-1 producers ranged from 8 × 100 to 5 × 102 CFU/ml for ChromID ESBL (Table 1). The ability of that medium to detect NDM-1 producers is based on the fact that those producers are also resistant to expanded-spectrum cephalosporins, in part due to the broad-spectrum hydrolytic properties of the β-lactamase NDM-1 (Table 1). Therefore, this detection was possible using ChromID ESBL even though the strain may not express an ESBL (E. coli B, for example [data not shown]). For CHROMagar KPC, the lower limit of detection of NDM-1 producers ranged from 1 × 101 to 4 × 105 CFU/ml (Table 1). Those detection limits were higher than those of the ChromID ESBL medium, especially for several strains with low MICs of carbapenems (E. coli B, E. coli H, E. coli J, E. cloacae C, Klebsiella oxytoca A, Providencia rettgeri A). After the screening of putative NDM-1 producers, definitive identification of the NDM-1 producers is needed using the techniques developed above (susceptibility testing, Etest, and PCR) (Fig. 1). This detection strategy may help to prevent the development of clinically significant outbreaks of infections with NDM-1-positive clinical isolates, in particular NDM-1+ K. pneumoniae.
Fig. 1.
Strategy for identification of NDM-1 producers as a source of clinical infections and for detecting carriers of NDM-1 producers. *, this culture medium can be used for surveillance of outbreaks of infections with NDM-1 producers after validation of its detection sensitivity for the specific strain responsible for an outbreak. **, Etest MBL is reliable when the MIC of imipenem is not too low.
Conclusion.
Early identification of NDM-1 producers in bacteria causing clinical infections and/or colonizers is mandatory to prevent their spread. Identification of NDM-1 producers in clinical infections will be suspected on any decreased susceptibility to carbapenems in Enterobacteriaceae, especially in E. coli, where resistance to carbapenems may not always be apparent. The ChromID ESBL is a reliable culture medium for screening for carriers of NDM-1 producers. Since ChromID ESBL may also detect ESBL producers, its use for detecting carbapenemase producers may be the source of an extra work load. However, the higher sensitivity of ChromID ESBL than CHROMagar argues for its use. In addition, ChromID ESBL offers the economical advantage of using a single plate to detect ESBL and carbapenemase producers.
CHROMagar culture medium may be proposed for the follow-up of an outbreak of NDM-1 producers after identification of the first cases using ChromID ESBL and after checking that this medium is sensitive enough to detect the specific NDM-1 producer strain responsible for the outbreak. We believe that a screening strategy based on early detection of NDM-1 producers for any international transfer of hospitalized patients on a worldwide scale and for at-risk patients in areas of endemicity may prevent outbreaks of infections with NDM-1 producers.
Acknowledgments
This work was funded mostly by a grant from the INSERM (U914); by a grant-in-aid from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, Paris, France; and by a grant from the European Community (TEMPOtest-QC, HEALTH-2009-241742).
We thank A. Ros for the gift of clinical isolates.
Footnotes
Published ahead of print on 1 December 2010.
REFERENCES
- 1. Carrër A., Fortineau N., Nordmann P. 2010. Use of ChromID ESBL medium for detecting carbapenemase-producing Enterobacteriaceae. J. Clin. Microbiol. 48:1913–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Center for Disease Control and Prevention 2009. Guidance for control of infections with carbapenem-resistant or carbapenemase-producing Enterobacteriaceae in acute care facilities. Morb. Mortal. Wkly. Rep. 58:256–266 [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention 2010. Detection of Enterobacteriaceae isolates carrying metallo-beta-lactamase—United States. Morb. Mortal. Wkly. Rep. 25:750. [PubMed] [Google Scholar]
- 4. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing. CLSI M100-S20U. Update June 2010 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5. Deshpande P., et al. 2010. New Delhi Metallo-beta lactamase (NDM-1) in Enterobacteriaceae: treatment options with carbapenems compromised. J. Assoc. Physicians India 58:147–149 [PubMed] [Google Scholar]
- 6. Franklin C., Liolios L., Peleg Y. 2006. Phenotypic detection of carbapenem-susceptible metallo-β-lactamase-producing Gram-negative bacilli in the clinical laboratory. J. Clin. Microbiol. 44:3139–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Karthikeyan K., Thirunarayan M. A., Krishnan P. 2010. Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii in India. J. Antimicrob. Chemother. 65:2253–2254 [DOI] [PubMed] [Google Scholar]
- 8. Kumarasamy K. K., et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miriagou V., et al. 2010. Acquired carbapenemases in Gram-negative bacterial pathogens: detection and surveillance issues. Clin. Microbiol. Infect. 16:112–122 [DOI] [PubMed] [Google Scholar]
- 10. Nordmann P., Cuzon G., Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228–236 [DOI] [PubMed] [Google Scholar]
- 11. Poirel L., Lagrutta E., Taylor P., Pham J., Nordmann P. 2010. Emergence of metallo β-lactamase NDM-1 producing multidrug resistant Escherichia coli in Australia. Antimicrob. Agents Chemother. 54:4914–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Queenan A. M., Bush K. 2007. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20:440–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Réglier-Poupet H., et al. 2008. Performance of the ChromID ESBL, a chromogenic medium for detection of Enterobacteriaceae producing extended-spectrum β-lactamases. J. Med. Microbiol. 57(Pt. 3):310–315 [DOI] [PubMed] [Google Scholar]
- 14. Samra Z., et al. 2008. Evaluation of CHROMagar KPC for rapid detection of carbapenem-resistant Enterobacteriaceae. J. Clin. Microbiol. 46:3110–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thomson K. S. 2010. Extended-spectrum β-lactamase, AmpC and carbapenemase issues. J. Clin. Microbiol. 48:1019–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walsh T. R., Toleman M. A., Poirel L., Nordmann P. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Woodford N., et al. 2010. Comparison of BD Phoenix, Vitek2, and Microscan automated systems for detection and inference of mechanisms responsible for carbapenem resistance in Enterobacteriaceae. J. Clin. Microbiol. 48:2999–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yong D., et al. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]