Abstract
Cryptococcal disease most commonly occurs in patients with an underlying immune deficit, most commonly HIV infection, and is due to Cryptococcus neoformans var. grubii. Occasionally disease due to this variety occurs in apparently immunocompetent patients. The relationship between strains infecting immunosuppressed and immunocompetent patients is not clear. Amplified fragment length polymorphism (AFLP) analysis was used to characterize the relationship between strains infecting HIV-infected and uninfected patients. Isolates from 51 HIV-uninfected patients and 100 HIV-infected patients with cryptococcal meningitis were compared. C. neoformans var. grubii VNI was responsible for infections in 73% of HIV-uninfected and 100% of HIV-infected patients. AFLP analysis defined two distinct clusters, VNIγ and VNIδ. The majority (84%) of isolates from HIV-uninfected patients were VNIγ, compared with only 38% of isolates from HIV-infected patients (odds ratio, 8.30; 95% confidence interval [CI], 3.04 to 26.6; P < 0.0001). In HIV-uninfected patients, underlying disease was less frequent in those with VNIγ infections. Two clusters of C. neoformans var. grubii VN1 are responsible for the majority of cases of cryptococcal meningitis in Vietnam. The distribution of these clusters differs according to the immune status of the host.
There are an estimated 1 million cases of cryptococcal meningitis, resulting in 625,000 deaths, every year (42). Most disease occurs in patients with underlying immunosuppressive conditions, including HIV infection, malignancy, organ failure, immune-modulating therapy, or autoimmune disease. Two species of Cryptococcus cause almost all human infections: Cryptococcus neoformans and Cryptococcus gattii (25, 28). Host and geographical factors influence which species is responsible for disease in different settings (8, 26). C. neoformans most commonly affects immunocompromised patients, and two varietal forms are known: C. neoformans var. grubii and C. neoformans var. neoformans. C. neoformans var. grubii is distributed worldwide and is associated with pigeon guano (14). The vast majority of disease in HIV-infected patients is due to this variety, and while C. neoformans var. neoformans accounts for up to 25% of cases in Western Europe, it is rarely isolated outside this region (11, 12, 17–19, 22). C. gattii occurs mainly in the tropics and subtropics, is associated with eucalyptus and other tree species, and is most frequently described as causing disease in immunocompetent patients (9, 15, 16, 23, 27, 29–31, 38, 39, 49). An outbreak of disease due to C. gattii has been ongoing in British Columbia, Canada, since 1999, and there is evidence of autochthonous spread, suggesting that it is exploiting new environments and climatic zones (24, 51). Despite the high rates of HIV infection in the tropics, disease due to C. gattii is uncommon; in South Africa and Rwanda it is responsible for only 1 to 2% of cryptococcosis in HIV-infected patients, although the rate is higher (between 14 and 30%) in Botswana and Malawi (3, 34, 40, 50). Thus, due to the high prevalence of HIV infection, most cases of cryptococcal meningitis occur in the tropics and are caused by C. neoformans var. grubii (8, 22, 34, 37, 40, 42).
HIV-uninfected patients with cryptococcal meningitis due to C. neoformans usually have some other comorbidity present that is associated with immunosuppression (1, 8, 11, 13, 26, 39, 44, 47, 49). Occasionally, disease occurs in patients without any clear underlying immune deficit or predisposing condition. In Australia and New Zealand, up to 20% of all C. neoformans cryptococcosis has been reported to occur in immunocompetent individuals (8, 49). This contrasts with a recent report from China, where although disease is rare, most cases occur in patients without an obvious underlying immune deficit and are due to C. neoformans var. grubii. The strains found in China are closely related and of a single multilocus sequence type (MLST) (7).
The different species and varietal forms within the C. neoformans species complex can be distinguished by serological and molecular methods. PCR-restriction fragment length polymorphism (PCR-RFLP) analysis of the URA5 gene reliably divides C. neoformans and C. gattii into eight molecular genotypes (VNI to VNIV and VGI to VGIV) (37). These genotypes are supported by PCR fingerprinting, amplified fragment length polymorphism (AFLP) analysis, and multigene sequencing studies (2, 5, 36, 41). More recently, AFLP analysis and MLST have successfully identified a novel genotype of C. neoformans var. grubii from Botswana that is closely related to the VNI genotype (35). While not all environmental strains of C. neoformans are able to cause disease, the pathological significance of differences in genotypes of clinical isolates at the subvarietal level has not been determined (33).
It is not clear why immunocompetent patients are occasionally infected with C. neoformans. The pathogen may exploit an unrecognized immune deficit in the host or have some quality that increases its pathogenicity. In Vietnam, most HIV-uninfected patients with cryptococcal meningitis do not have obvious underlying conditions associated with immunosuppression. We investigated the molecular epidemiological relationship between strains of C. neoformans from HIV-infected and uninfected patients in Vietnam using PCR-RFLP analysis of the URA5 gene and AFLP analysis in order to test the hypothesis that strains from HIV-uninfected patients are a closely related population.
MATERIALS AND METHODS
Setting, participants, and strains.
The work was conducted at the Hospital for Tropical Diseases, Ho Chi Minh City, the tertiary referral center for infectious diseases in southern Vietnam. Strains were obtained from patients with cryptococcal meningitis enrolled into two clinical studies: a prospective descriptive study of cryptococcal meningitis in HIV-uninfected patients and a randomized controlled trial (RCT) of combination antifungal therapy in HIV-associated cryptococcal meningitis (http://www.controlled-trials.com/ISRCTN95123928/farrar). Cryptococcal meningitis was defined through the identification of C. neoformans or C. gattii from cerebrospinal fluid (CSF) by India ink staining, culture, or antigen testing or as a clinical syndrome consistent with cryptococcal meningitis and a positive blood cryptococcal antigen or culture. After giving informed consent, all patients underwent treatment and follow-up on the dedicated research ward. Ethical approval for the studies was obtained from the Hospital for Tropical Diseases, the Liverpool School of Tropical Medicine Research Ethics Committee, and the Oxford University Tropical Ethics Committee, United Kingdom. All patients were older than 15 years. Exclusion criteria for the randomized controlled trial were a previous episode of cryptococcal meningitis, liver or renal failure, and pregnancy.
The strains used were the initial isolates identified at diagnosis. Strains from HIV-uninfected patients were from patients sequentially enrolled into the study between April 1996 and June 2009. Strains from HIV-infected patients were from patients sequentially enrolled into the RCT between April 2004 and June 2009. All strains were confirmed as Cryptococcus species using classical mycological methods, including colony morphology, microscopy, growth on bird seed agar, urease production, sugar assimilation tests, and biotyping with canavanine-glycine bromothymol blue agar. Control strains representing the eight defined molecular genotypes were kindly provided by Wieland Meyer, Westmead Millennium Institute for Medical Research, Sydney, Australia. Isolates were stored at minus 30°C (Microbank beads; Pro-Lab Diagnostics, Neston, United Kingdom). HIV infection was confirmed or excluded through antibody and antigen testing (Determine HIV1/2, HIV AXSYM HIV1/2 gO; Abbott, Maidenhead, United Kingdom).
DNA Extraction.
Colonies were revived on Sabouraud's agar at 30°C for 72 h. Single colonies were spread for confluent growth and incubated at 30C for 24 h. Chromosomal DNA was extracted from approximately 0.5 g (wet weight) of yeast cells according to the method described by Wen et al. (54). The DNA pellet was resuspended in 100 μl of Tris-EDTA (TE) buffer containing 100 μg of RNase.
URA5 PCR-RFLP analysis.
RFLP analysis of the URA5 gene was carried out according to the methods of Meyer et al. (37). The final product was separated by electrophoresis on a 3% agarose gel at 100 V for 3 h. RFLP patterns were assigned visually by comparison with known standards.
AFLP analysis.
Amplified fragment length polymorphism (AFLP) analysis was based on the modification of the method of Vos et al. described by Halliday et al. (20, 53). Consistency was determined by performing multiple (between 8 and 16) experiments on single isolates, from DNA extraction through to fragment analysis. To determine the relationship between C. neoformans var. grubii strains from HIV-infected and uninfected patients, an experimental set was created containing all the isolates from HIV-uninfected patients (termed HIV-negative strains), two C. neoformans var. grubii control strains (molecular groups VNI and VNII), one Cryptococcus neoformans var. neoformans control strain (VNIV), one C. neoformans var. grubii-C. neoformans var. neoformans hybrid (VNIII), and 100 strains from HIV-infected patients (termed HIV-associated strains). The HIV-associated strains were selected randomly from those obtained from patients sequentially entered into the RCT using random numbers generated in a Microsoft Excel spreadsheet (Microsoft). In addition, the experimental set contained three C. gattii strains from HIV-uninfected patients as an outgroup. The complete set of 144 isolates was randomly divided into six batches using random numbers generated in a Microsoft Excel spreadsheet. AFLP analysis, from DNA extraction up to the final round of amplification, was performed by batch. Fragment analysis was performed simultaneously on all isolates from the experimental set in a single sequencer run, and all HIV-negative and control strains were analyzed in duplicate.
Restriction/ligation.
Each reaction mix contained 20 μl of genomic DNA (20 to 60 ng), 13 U of EcoRI, and 7 U of MseI (Helena Biosciences Europe, United Kingdom), 4 μl of One-Phor-All buffer (Amersham Biosciences, United Kingdom), and double-distilled water (ddH2O) to a final reaction mix volume of 40 μl. Adequate digestion was checked with a lambda DNA control (Promega UK Ltd.) (50 ng/μl). Samples and digestion checks were incubated at 37°C overnight. Ligation reactions with EcoRI and MseI double-stranded-adapter sequences were incubated at 22°C overnight (20).
PCR amplification.
The primer pairs used for preamplification were 5′-GACTGCGTACCAATTCA-3′/5′-GATGAGTCCTGAGTAAG-3′ and 5′-GACTGCGTACCAATTCG-3′/5′-GATGAGTCCTGAGTAAG-3′. Two selective primer combinations were used: EcoRI-GT–MseI-GT (5′-6FAM-GACTGCGTACCAATTCGT-3′ and 5′-GATGAGTCCTGAGTAAGT-3′) and EcoRI-AC–MseI-G (5′-6FAM-GACTGCGTACCAATTCAC-3′ and 5′-GATGAGTCCTGAGTAAG-3′). Amplification reactions were performed in a final volume of 25 μl. The preamplification reaction mixture contained 1× PCR buffer, a 5 nM concentration of each deoxyribonucleoside triphosphate, 2 μM concentrations of the appropriate EcoRI and MseI preamplification primer pairs or selective primers, 5 U of Taq DNA polymerase (Amersham Biosciences, United Kingdom), and 1 μl of digestion ligation product. The selective amplification mixture contained a 0.5 μM final concentration of each primer. The selective EcoRI primer was labeled at the 5′ end with 6-carboxyfluorescein (6FAM). Preamplification reaction conditions were 94°C for 2 min followed by 20 cycles of 94°C for 20 s, 56°C for 30 s, and 72°C for 2 min. Selective amplification conditions were 94°C for 2 min followed by 10 cycles of 94°C for 20 s, 66°C for 30 s (decreasing 1°C every cycle), and 72°C for 120 s, followed by 20 cycles consisting of 94°C for 20 s, 56°C for 30 s, and 72°C for 120 s. All amplification reactions were performed in a Bio-Rad C1000 thermal cycler (Bio-Rad Laboratories Ltd., Hemel Hempstead, United Kingdom).
AFLP fragment detection.
Fragment analysis was performed with an Applied Biosystems 3130X 16-capillary (50-cm) sequencer with Genemapper version 4.0 software (Applied Biosystems Inc., CA). The amplification product was diluted as necessary, mixed with LIZ 500 size standard and HiDi formamide (AB Biosystems Inc., CA), vortexed, heated to 95°C for 3 min, and snap-cooled on ice before loading and using the default AFLP electrophoresis settings as recommended by the manufacturer.
Analysis.
Raw file data were imported into Bionumerics version 5.1 (Applied Maths, Belgium). All 144 files were imported and normalized simultaneously. The similarities between the densitometric curves of the fingerprint data for each isolate were analyzed using the Pearson product moment correlation, and neighbor-joining trees (NJT) were constructed. The statistical significance of the clusters was tested by cophenetic correlation (48).
Statistical methods.
Statistical analysis was performed with R version 2.9.0 (R Foundation for Statistical Computing, Vienna, Austria) (43). Fisher's exact test was used to compare proportions of categorical or dichotomized parameters.
RESULTS
URA5 PCR-RFLP analysis of Cryptococcus isolates from HIV-infected and uninfected patients.
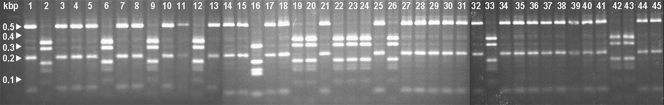
Seventy HIV-uninfected patients were enrolled into the descriptive study. Viable isolates were available from 51 patients. Patient characteristics are shown in Table 1. Thirty-seven isolates were Cryptococcus neoformans var. grubii molecular genotype VNI, and 14 were Cryptococcus gattii (13 C. gattii molecular genotype VGI and one C. gattii molecular genotype VGII). A total of 238 HIV-infected patients were enrolled into the randomized controlled trial between April 2004 and June 2009. The median age of the patients was 26 years (range, 15 to 62 years), and the median CD4 count was 15 cells/μl. A total of 211 baseline CSF isolates were available from these patients. All were C. neoformans var. grubii URA5 molecular genotype VNI. Representative strains and the characteristic banding patterns produced by URA5 PCR-RFLP analysis are shown in Fig. 1.
Table 1.
HIV-uninfected patient and isolate characteristics
| Patient | Age (yr) | Gendera | Underlying disease | RFLP groupb | AFLP VNI genotype | 6-mo outcome |
|---|---|---|---|---|---|---|
| 1 | 37 | M | VNI | γ | Died | |
| 2 | 29 | M | VGI | Alive | ||
| 3 | 16 | M | VNI | γ | Alive | |
| 4 | 52 | F | VNI | γ | Alive | |
| 5 | 24 | F | Idiopathic thrombocytopenic purpura | VNI | δ | Alive |
| 6 | 49 | F | VGI | Alive | ||
| 7 | 20 | M | VNI | γ | Alive | |
| 8 | 15 | M | VNI | γ | Alive | |
| 9 | 23 | M | VGI | Alive | ||
| 10 | 16 | F | VNI | γ | Alive | |
| 11 | 25 | M | VNI | γ | Alive | |
| 12 | 38 | F | VGI | Alive | ||
| 13 | 24 | F | VNI | γ | Alive | |
| 14 | 34 | M | VNI | δ | Alive | |
| 15 | 22 | M | Nephrotic syndrome | VNI | γ | Alive |
| 16 | 44 | M | VGII | Died | ||
| 17 | 63 | F | Lupus | VNI | γ | Died |
| 18 | 28 | F | VNI | γ | Alive | |
| 19 | 53 | M | VGI | Alive | ||
| 20 | 51 | M | VGI | Alive | ||
| 21 | 20 | M | VNI | γ | Alive | |
| 22 | 53 | F | Polyarthritis | VGI | Died | |
| 23 | 16 | M | VGI | Alive | ||
| 24 | 31 | F | VGI | Died | ||
| 25 | 28 | M | VNI | γ | Alive | |
| 26 | 74 | F | VNI | γ | Alive | |
| 27 | 40 | M | VNI | γ | Alive | |
| 28 | 51 | M | VNI | γ | Alive | |
| 29 | 64 | F | Cushing's syndrome | VNI | δ | Died |
| 30 | 22 | F | VNI | γ | Alive | |
| 31 | 18 | M | VGI | Alive | ||
| 32 | 47 | M | VNI | γ | Alive | |
| 33 | 61 | F | Cirrhosis | VNI | γ | Died |
| 34 | 67 | M | Diabetes | VNI | γ | Died |
| 35 | 35 | F | VNI | γ | Alive | |
| 36 | 53 | M | VNI | γ | Died | |
| 37 | 21 | M | VNI | γ | Alive | |
| 38 | 30 | M | Gastric cancer | VNI | δ | Died |
| 39 | 37 | M | VGI | Alive | ||
| 40 | 43 | M | VGI | Alive | ||
| 41 | 66 | M | VNI | δ | Alive | |
| 42 | 46 | F | Lupus | VNI | δ | Alive |
| 43 | 61 | F | Mitral valve disease | VNI | γ | Alive |
| 44 | 42 | M | VNI | γ | Not known | |
| 45 | 19 | M | VGI | Alive | ||
| 46 | 32 | M | VNI | γ | Alive | |
| 47 | 19 | F | Hemolytic anemia | VNI | γ | Alive |
| 48 | 32 | M | VNI | γ | Alive | |
| 49 | 28 | F | Lupus | VNI | γ | Died |
| 50 | 53 | M | VNI | γ | Alive | |
| 51 | 31 | M | VNI | γ | Alive |
F, female; M, male.
VGI or -2, C. gattii; VNI, C. neoformans var. grubii molecular group VNI.
Fig. 1.
Representative PCR-RFLP profiles of the URA5 genes of 45 Cryptococcus isolates. Typical molecular types: lane 1, C. neoformans var. grubii VNI; lane 2, C. gattii VG1; lane 16, C. gattii VG2.
AFLP analysis of 144 representative Cryptococcus isolates from HIV-infected and uninfected patients.
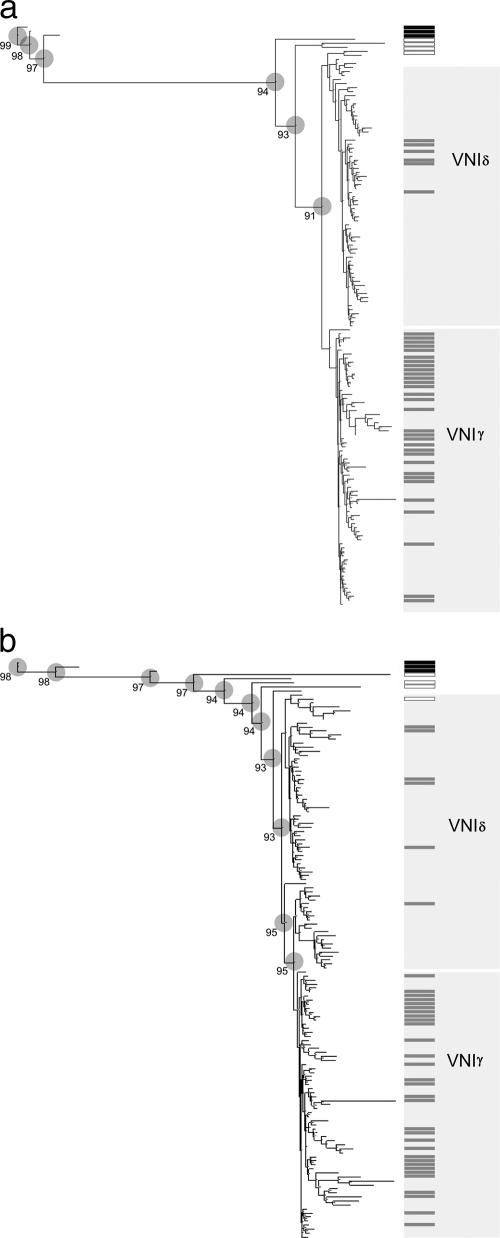
AFLP analysis was performed with 144 of the isolates described above. These included all 37 C. neoformans var. grubii isolates from HIV-uninfected patients, 100 randomly selected C. neoformans var. grubii isolates from HIV-infected patients, the four C. neoformans control strains (C. neoformans var. grubii VNI and VNII, C. neoformans var. neoformans VNIV, and an AD hybrid VNIII), and three C. gattii strains. The median similarities of fingerprint patterns from duplicate experiments on identical isolates were 95.1% (interquartile range [IQR], 94.2 to 98.1%) for the GT/GT primer set and 92.5% (IQR, 88.85 to 99.4%) for the AC/G primer set. Both primer sets reliably distinguished strains by species and RFLP molecular group. The neighbor-joining trees generated with each primer pair for the experimental set are shown in Fig. 2a and b.
Fig. 2.
(a) AFLP-derived neighbor-joining tree obtained using the GT/GT primer set for Vietnamese C. neoformans var. grubii clinical isolates. Gray bars, isolates from HIV-uninfected patients; white bars, C. neoformans controls; black bars, C. gattii controls. Circles represent well-supported clusters; the numbers are the cophenetic correlation values (%). (b) AFLP-derived neighbor-joining tree obtained using the AC/G primer set for Vietnamese C. neoformans var. grubii clinical isolates Gray bars, isolates from HIV-uninfected patients; white bars, C. neoformans controls; black bars, C. gattii controls. Circles represent well-supported clusters, the numbers are the cophenetic correlation values (%).
The GT/GT primer set divided the VNI population into two clusters designated VNIγ and VNIδ (Fig. 2a). The cophenetic correlation value at the separating node was 0.91, demonstrating robust clustering. Cluster VNIγ consisted of 69 isolates, and cluster VNIδ consisted of 68 isolates. The HIV serostatus of the source patient was related to isolate cluster (Table 2). Thirty-one isolates (84%) from HIV-uninfected patients were VNIγ, compared with 38 (38%) of isolates from HIV-infected patients (odds ratio, 8.30; 95% confidence interval [CI], 3.04 to 26.6; P < 0.0001). Additionally, in HIV-uninfected patients, other underlying diseases were more common in those with VNIδ infections than in those with VNIγ infections (4 of 6 [67%] versus 7 of 31 [23%]; odds ratio, 6.43; 95% CI, 0.75 to 85.1; P = 0.05). In HIV patients the clusters did not segregate by HIV risk group; 36% of intravenous drug users had VNIγ infections, compared with 41% of those believed to have had a sexual route of acquisition of HIV infection (P = 0.67). While a larger proportion of HIV-infected patients were male (87%), there was no difference in cluster distribution by patient gender (38% versus 45% VNIγ for males and females, respectively; P = 0.75). HIV-infected patients were more likely to be from Ho Chi Minh City than the provinces (64% versus 29%; P = 0.007). However, within groups (HIV infected or uninfected), the patient address (Ho Chi Minh City versus province) had no impact on the distribution of isolate cluster; for HIV-associated isolates, 25 of 63 (40%) of isolates from patients with Ho Chi Minh City addresses were VNIγ, compared with 13 of 36 (36%) of isolates from province dwellers (P = 0.83); for HIV-uninfected patients, 9 of 10 (90%) of Ho Chi Minh City residents had VNIγ infections versus 20 of 25 (80%) of those from the provinces (P = 0.65). Isolates from HIV-negative patients were collected over a period of 13 years. There was no change in the relative proportion of VNI clusters over the study period (P = 1.0).
Table 2.
Distribution of C. neoformans var. grubii isolates between clusters by HIV status of host
| Genotype | No. of patientsa |
||
|---|---|---|---|
| HIV infected | HIV uninfected | Total | |
| VNIγ | 38 | 31 | 69 |
| VNIδ | 62 | 6 | 68 |
| Total | 100 | 37 | 137 |
Odds ratio, 8.30; 95% CI, 3.04 to 26.6; P < 0.0001 (Fisher's exact test).
The AC/G primer set defined a monophyletic cluster that included the same 31 HIV-negative isolates assigned to the VNIγ genotype cluster by the GT/GT primer set (Fig. 2b). The cophenetic correlation value of the node defining this cluster was 0.96, again supporting robust clustering.
The results of AFLP typing were not perfectly consistent between primer sets: five strains from HIV-infected patients identified in cluster VNIγ by the GT/GT primer set were not preserved in the monophyletic cluster using the AC/G primer set, and three HIV-associated strains that the AC/G primer set identified within the monophyletic cluster were assigned to cluster VNIδ by the GT/GT primer set. The AC/G-derived dendrogram suggested more variability within this population than that derived from the GT/GT primer set. All the major nodes along the tree were well supported, with cophenetic correlation values of greater than 0.9.
DISCUSSION
Our findings demonstrate that the vast majority of cases of cryptococcal meningitis in Vietnamese patients, irrespective of HIV infection status, are due to C. neoformans var. grubii genotype VNI. Most HIV-uninfected patients in Vietnam (76% of all patients and 68% of patients with C. neoformans var. grubii infection) have no obvious underlying immunosuppressive disease. In this tropical location, it might have been predicted that C. gattii would account for more infections in HIV-negative patients, yet we found it to account for only 25% of cases. This is higher than is reported in locations in the Far East, albeit with more temperate climates (7, 10).
AFLP analysis with the GT/GT primer set defined two clusters of VNI in the C. neoformans var. grubii population, which we have termed VNIγ and VNIδ. The majority of strains from HIV-uninfected patients clustered within the VNIγ cluster. The rate of underlying disease was lower in HIV-uninfected patients infected with strains from this cluster. Typing using the AC/G primer set confirmed this observation, with the strains from HIV-uninfected patients being closely related to each other and forming a monophyletic cluster. In contrast, the VNIγ cluster caused only 38% of infections in HIV-infected patients. A small number of strains were allocated to different clusters according to the primer set used. Whether these are genuine differences or the result of experimental variability is not clear. A sequence-based typing method, such as MLST, might resolve these differences.
The difference in cluster distributions by HIV serostatus could be explained by a number of factors. Differences in cluster presumably relate to some difference in environmental niche adaptation. Different behaviors of different patient groups could lead to different risks of exposures to each cluster. For example, a large proportion of HIV-infected patients in Vietnam are intravenous drug users (59% in this cohort). However, we found no difference in the prevalence of cluster by HIV risk factor. Moreover, the HIV-uninfected patients are demographically diverse, implying that shared risk factors are unlikely, yet they are predominantly infected with a single cluster.
The geographical location of patients might explain different exposures. A sophisticated geospatial analysis is not possible with our data, and uncertainty over the incubation period with cryptococcal disease in any case makes this difficult, but a simple analysis suggests that there is no difference in cluster spread according to residency in Ho Chi Minh City or the provinces. The HIV-infected and HIV-uninfected groups contained patients from all provinces in southern Vietnam, as would be expected in our hospital, which is the tertiary referral center for all infectious diseases in the region.
Another possibility is that the clusters we identified are artifactual and a consequence of error somewhere in the experimental process. AFLP analysis is a multistage procedure, and error could enter through different efficiencies of digestion, ligation, PCR amplification, or fragment detection (45). We performed multiple experiments on single strains to determine the rate of experimental variability, and we randomized the selection of strains for experimentation from DNA extraction through to final amplification. Fragment analysis was performed simultaneously in all experiments and in duplicate on all isolates from HIV-uninfected patients to minimize the effect of sequencer-induced noise. While the dendrograms were slightly different for each primer set, the positions of strains within the trees are similar, with the same HIV-negative isolates found in a monophyletic cluster in both, suggesting that the grouping of a majority of HIV-negative isolates in cluster VNIγ is genuine.
A further possibility is that the VNIγ cluster may represent strains with increased pathogenicity. If this is the case, an explanation is needed as to why it is not the dominant genotype causing disease in HIV-infected patients. This is explainable if VNIγ occurs less frequently than VNIδ in the environment. Differences in environmental prevalence correlating with differences in pathogenicity are biologically plausible. C. neoformans is a facultative pathogen, and human-to-human transmission is not important in the disease cycle. Differences in pathogenicity are likely bystander effects of adaptation to particular environmental niches. It is already known that not all environmental strains are pathogenic (32, 33, 55). The prevalence and site of a given niche in the environment, along with dispersal from the niche as an infectious propagule, will determine the risk of a potentially infectious contact between the pathogen and susceptible hosts. Thus, a strain with increased pathogenicity may be infrequent in the environment because its niche is rare, explaining a low overall disease prevalence. Alternatively, the low prevalence may reflect an additional need for an as-yet-unidentified host immune defect for successful infection in HIV-uninfected patients.
Host and geographical factors are known to influence the species of Cryptococcus that cause meningitis in particular groups (8, 29, 46, 52). However, a subgroup of C. neoformans var. grubii that is associated with HIV-uninfected individuals has been described only once before, despite extensive study using numerous typing methods (4, 6, 21, 32, 37, 52). This may be explained by the relatively small number of isolates from HIV-uninfected patients, particularly from those without underlying disease. Only Varma et al. found a segregation of genotype by HIV status (52). In a study of 45 clinical isolates (23 from HIV-uninfected patients and 25 from HIV-infected patients), one of nine genotypes was found to be more frequent in HIV-uninfected patients, but no association was found for isolates from HIV-infected patients. It is not clear whether all isolates in this study were of the same variety.
The fact that we were able to demonstrate an association between a subgroup of C. neoformans var. grubii and the HIV infection status of the isolate source, in contrast to the case in most of the studies described above, may reflect the fact that all of our isolates were from a small, well-defined geographical area, reducing the “noise” that might be found in a more diverse population. More recently, it has been reported that in 91 immunocompetent patients with cryptococcal meningitis in China, disease was due to infection with a highly homogeneous group of C. neoformans var. grubii VNI strains, a finding consistent with our work (7). However, no comparison was made with strains from HIV-infected patients. This same genotype is also reported from Korea, although most affected patients have serious underlying disease (10). The strains described had variable virulence in a mouse infection model despite the genetic similarity, suggesting that they are not a single hypervirulent clone. Assessing the virulence of the Vietnamese isolates from cluster VNIγ, and determining their relationship to the Chinese isolates from HIV-uninfected patients will be interesting areas for further study.
Conclusions.
We have identified a cluster of C. neoformans var. grubii strains that is responsible for the majority of disease in HIV-uninfected patients in Vietnam, most of whom have no comorbidities. This genotype accounts for only one-third of cases in HIV-infected patients. Whether this difference is due to host or pathogen factors is not yet clear.
ACKNOWLEDGMENTS
Wieland Meyer (Molecular Mycology Research Laboratory, Centre for Infectious Disease and Microbiology, University of Sydney at the Westmead Millennium Institute, Westmead, NSW, Australia) kindly provided Cryptococcus neoformans and Cryptococcus gattii control strains.
This work was funded by the Wellcome Trust UK. J. N. Day was in part supported by a British Infection Society Fellowship.
The authors have no conflicts of interest.
Footnotes
Published ahead of print on 15 December 2010.
REFERENCES
- 1. Bennett J. E., Kwon-Chung K. J., Howard D. H. 1977. Epidemiologic differences among serotypes of Cryptococcus neoformans. Am. J. Epidemiol. 105:582–586 [DOI] [PubMed] [Google Scholar]
- 2. Boekhout T., et al. 2001. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology 147:891–907 [DOI] [PubMed] [Google Scholar]
- 3. Bogaerts J., et al. 1999. AIDS-associated cryptococcal meningitis in Rwanda (1983–1992): epidemiologic and diagnostic features. J. Infect. 39:32–37 [DOI] [PubMed] [Google Scholar]
- 4. Bovers M., Hagen F., Boekhout T. 2008. Diversity of the Cryptococcus neoformans-Cryptococcus gattii species complex. Rev. Iberoam. Micol. 25:S4–12 [DOI] [PubMed] [Google Scholar]
- 5. Bovers M., Hagen F., Kuramae E. E., Boekhout T. 2008. Six monophyletic lineages identified within Cryptococcus neoformans and Cryptococcus gattii by multi-locus sequence typing. Fungal Genet. Biol. 45:400–421 [DOI] [PubMed] [Google Scholar]
- 6. Brandt M. E., et al. 1996. Molecular subtype distribution of Cryptococcus neoformans in four areas of the United States. Cryptococcal Disease Active Surveillance Group. J. Clin. Microbiol. 34:912–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen J., et al. 2008. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg. Infect. Dis. 14:755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen S., et al. 2000. Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Australasian Cryptococcal Study Group. Clin. Infect. Dis. 31:499–508 [DOI] [PubMed] [Google Scholar]
- 9. Chen S. C., et al. 1997. Cryptococcus neoformans var. gattii infection in northern Australia: existence of an environmental source other than known host eucalypts. Trans. R. Soc. Trop. Med. Hyg. 91:547–550 [DOI] [PubMed] [Google Scholar]
- 10. Choi Y. H., et al. 2010. Prevalence of the VNIc genotype of Cryptococcus neoformans in non-HIV-associated cryptococcosis in the Republic of Korea. FEMS Yeast Res. 10:769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dromer F., Mathoulin-Pelissier S., Launay O., Lortholary O. 2007. Determinants of disease presentation and outcome during cryptococcosis: the CryptoA/D study. PLoS Med. 4:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dromer F., Mathoulin S., Dupont B., Laporte A. 1996. Epidemiology of cryptococcosis in France: a 9-year survey (1985–1993). French Cryptococcosis Study Group. Clin. Infect. Dis. 23:82–90 [DOI] [PubMed] [Google Scholar]
- 13. Dromer F., Mathoulin S., Dupont B., Letenneur L., Ronin O. 1996. Individual and environmental factors associated with infection due to Cryptococcus neoformans serotype D. French Cryptococcosis Study Group. Clin. Infect. Dis. 23:91–96 [DOI] [PubMed] [Google Scholar]
- 14. Ellis D. H., Pfeiffer T. J. 1990. Ecology, life cycle, and infectious propagule of Cryptococcus neoformans. Lancet 336:923–925 [DOI] [PubMed] [Google Scholar]
- 15. Ellis D. H., Pfeiffer T. J. 1990. Natural habitat of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 28:1642–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fisher D., Burrow J., Lo D., Currie B. 1993. Cryptococcus neoformans in tropical northern Australia: predominantly variant gattii with good outcomes. Aust. N. Z. J. Med. 23:678–682 [DOI] [PubMed] [Google Scholar]
- 17. Franzot S. P., Fries B. C., Cleare W., Casadevall A. 1998. Genetic relationship between Cryptococcus neoformans var. neoformans strains of serotypes A and D. J. Clin. Microbiol. 36:2200–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Franzot S. P., Hamdan J. S., Currie B. P., Casadevall A. 1997. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for both local genetic differences and a global clonal population structure. J. Clin. Microbiol. 35:2243–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Franzot S. P., Salkin I. F., Casadevall A. 1999. Cryptococcus neoformans var. grubii: separate varietal status for Cryptococcus neoformans serotype A isolates. J. Clin. Microbiol. 37:838–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Halliday C. L., Carter D. A. 2003. Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var. gattii isolates from Australia. J. Clin. Microbiol. 41:703–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Illnait-Zaragozi M. T., Martinez-Machin G. F., Fernandez-Andreu C. M., Boekhout T., Meis J. F., Klaassen C. H. 2010. Microsatellite typing of clinical and environmental Cryptococcus neoformans var. grubii isolates from Cuba shows multiple genetic lineages. PLoS One 5:e9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jain N., et al. 2005. Molecular epidemiology of clinical Cryptococcus neoformans strains from India. J. Clin. Microbiol. 43:5733–5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kidd S. E., et al. 2007. Characterization of environmental sources of the human and animal pathogen Cryptococcus gattii in British Columbia, Canada, and the Pacific Northwest of the United States. Appl. Environ. Microbiol. 73:1433–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kidd S. E., et al. 2004. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc. Natl. Acad. Sci. U. S. A. 101:17258–17263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kwon-Chung J., Boekhout T., Fell J. W., Diaz M. 2002. Proposal to conserve the name Cryptococcus gattii against C. hondurianus and C. bacillisporus (Basidiomycota, Hymenomycetes, Tremellomycetidae). Taxon 51:804–806 [Google Scholar]
- 26. Kwon-Chung K. J., Bennett J. E. 1984. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am. J. Epidemiol. 120:123–130 [DOI] [PubMed] [Google Scholar]
- 27. Kwon-Chung K. J., Bennett J. E. 1984. High prevalence of Cryptococcus neoformans var. gattii in tropical and subtropical regions. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 257:213–218 [PubMed] [Google Scholar]
- 28. Kwon-Chung K. J., Varma A. 2006. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 6:574–587 [DOI] [PubMed] [Google Scholar]
- 29. Lalloo D., et al. 1994. Cryptococcal meningitis (C. neoformans var. gattii) leading to blindness in previously healthy Melanesian adults in Papua New Guinea. Q. J. Med. 87:343–349 [PubMed] [Google Scholar]
- 30. Laurenson I. F., et al. 1996. Meningitis caused by Cryptococcus neoformans var. gattii and var. neoformans in Papua New Guinea. Trans. R. Soc. Trop. Med. Hyg. 90:57–60 [DOI] [PubMed] [Google Scholar]
- 31. Lehmann P. F., Morgan R. J., Freimer E. H. 1984. Infection with Cryptococcus neoformans var. gattii leading to a pulmonary cryptococcoma and meningitis. J. Infect. 9:301–306 [DOI] [PubMed] [Google Scholar]
- 32. Litvintseva A. P., Kestenbaum L., Vilgalys R., Mitchell T. G. 2005. Comparative analysis of environmental and clinical populations of Cryptococcus neoformans. J. Clin. Microbiol. 43:556–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Litvintseva A. P., Mitchell T. G. 2009. Most environmental isolates of Cryptococcus neoformans var. grubii (serotype A) are not lethal for mice. Infect. Immun. 77:3188–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Litvintseva A. P., Thakur R., Reller L. B., Mitchell T. G. 2005. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in Sub-Saharan Africa. J. Infect. Dis. 192:888–892 [DOI] [PubMed] [Google Scholar]
- 35. Litvintseva A. P., Thakur R., Vilgalys R., Mitchell T. G. 2006. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics 172:2223–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meyer W., et al. 2009. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med. Mycol. 47:561–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meyer W., Castaneda A., Jackson S., Huynh M., Castaneda E. 2003. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg. Infect. Dis. 9:189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mitchell D. H., Sorrell T. C. 1992. Pancoast's syndrome due to pulmonary infection with Cryptococcus neoformans variety gattii. Clin. Infect. Dis. 14:1142–1144 [DOI] [PubMed] [Google Scholar]
- 39. Mitchell D. H., et al. 1995. Cryptococcal disease of the CNS in immunocompetent hosts: influence of cryptococcal variety on clinical manifestations and outcome. Clin. Infect. Dis. 20:611–616 [DOI] [PubMed] [Google Scholar]
- 40. Morgan J., et al. 2006. Cryptococcus gattii infection: characteristics and epidemiology of cases identified in a South African province with high HIV seroprevalence, 2002–2004. Clin. Infect. Dis. 43:1077–1080 [DOI] [PubMed] [Google Scholar]
- 41. Ngamskulrungroj P., et al. 2009. Genetic diversity of the Cryptococcus species complex suggests that Cryptococcus gattii deserves to have varieties. PLoS One 4:e5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park B. J., et al. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. Aids 23:525–530 [DOI] [PubMed] [Google Scholar]
- 43. R Foundation for Statistical Computing 2009, posting date Accessed 10 October 2009 R: a language and environment for statistical computing (version 2.9.0). http://www.R-project.org/ [Google Scholar]
- 44. Rozenbaum R., Goncalves A. J. 1994. Clinical epidemiological study of 171 cases of cryptococcosis. Clin. Infect. Dis. 18:369–380 [DOI] [PubMed] [Google Scholar]
- 45. Savelkoul P. H., et al. 1999. Amplified-fragment length polymorphism analysis: the state of an art. J. Clin. Microbiol. 37:3083–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Seaton R. A., Verma N., Naraqi S., Wembri J. P., Warrell D. A. 1997. Visual loss in immunocompetent patients with Cryptococcus neoformans var. gattii meningitis. Trans. R. Soc. Trop. Med. Hyg. 91:44–49 [DOI] [PubMed] [Google Scholar]
- 47. Shih C. C., Chen Y. C., Chang S. C., Luh K. T., Hsieh W. C. 2000. Cryptococcal meningitis in non-HIV-infected patients. QJM. 93:245–251 [DOI] [PubMed] [Google Scholar]
- 48. Sokal R., Rohlf J. 1962. The comparison of dendrograms by objective methods. Taxon 11:33–40 [Google Scholar]
- 49. Speed B., Dunt D. 1995. Clinical and host differences between infections with the two varieties of Cryptococcus neoformans. Clin. Infect. Dis. 21:28–36 [DOI] [PubMed] [Google Scholar]
- 50. Steele K. T., Thakur R., Nthobatsang R., Steenhoff A. P., Bisson G. P. In-hospital mortality of HIV-infected cryptococcal meningitis patients with C. gattii and C. neoformans infection in Gaborone, Botswana. Med. Mycol. 48:1112–1115 [DOI] [PubMed] [Google Scholar]
- 51. Stephen C., Lester S., Black W., Fyfe M., Raverty S. 2002. Multispecies outbreak of cryptococcosis on southern Vancouver Island, British Columbia. Can. Vet. J. 43:792–794 [PMC free article] [PubMed] [Google Scholar]
- 52. Varma A., Swinne D., Staib F., Bennett J. E., Kwon-Chung K. J. 1995. Diversity of DNA fingerprints in Cryptococcus neoformans. J. Clin. Microbiol. 33:1807–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vos P., et al. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wen H., Caldarelli-Stefano R., Tortorano A. M., Ferrante P., Viviani M. A. 1996. A simplified method to extract high-quality DNA from Cryptococcus neoformans. J. Mycol. Med. 6:136–138 [Google Scholar]
- 55. Zhu J., et al. 2010. Comparison of genotypes between environmental and clinical isolates of Cryptococcus neoformans var. grubii based on microsatellite patterns. Mycopathologia. 169:47–55 [DOI] [PubMed] [Google Scholar]