Abstract
We assessed the usefulness of mycobacterial interspersed repetitive-unit–variable-number tandem-repeat loci 26, 31, ETR-A, Mtub30, and Mtub02 and a deletion-targeted multiplex PCR in identifying pediatric Mycobacterium tuberculosis Beijing lineage strain infection. We found that the Beijing lineage isolates accounted for ∼62% (130/210) of the study isolates.
The wide spread of the Mycobacterium tuberculosis Beijing lineage strain infection worldwide and its associations with multidrug resistance, treatment failure, and HIV coinfection have been well documented (2, 6, 12). The observation that Beijing family isolates can multiply rapidly in human macrophages further suggests the likelihood of hypervirulence of this genetic family of M. tuberculosis (12). Given the importance of Beijing family strains in the global tuberculosis (TB) epidemic, a rapid and accurate identification of Beijing lineage isolates can significantly impact the global TB control.
However, spoligotyping, known as the gold standard for identifying Beijing lineage isolates, is more technically demanding and time-consuming than PCR-based assays (5). Therefore, several PCR-based methods for the rapid identification of Beijing family isolates have been developed (3, 9, 10). In this study, we assessed the usefulness of PCR-based assays targeting several previously described mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) loci (4, 9, 10) and the deletion-targeted multiplex PCR (DTM-PCR) assay developed by Chen et al. (3) in the identification of Beijing family isolates from pediatric TB cases.
The study sample comprised 210 isolates from 210 pediatric TB cases diagnosed in the Children's Hospital of Chongqing Medical University from January 2002 to December 2009. All the isolates were spoligotyped, as described previously by Kamerbeek et al. (8). They were then subjected to five PCR-based assays to evaluate five MIRU-VNTR loci (MIRU 26, MIRU 31, ETR-A, Mtub02, and Mtub30) as well as to the DTM-PCR assay (3, 4, 9, 10), using primers and programs described previously (1). The PCR products were analyzed by 2% agarose gel electrophoresis in 0.5× Tris-borate-EDTA (TBE) buffer (Fig. 1).
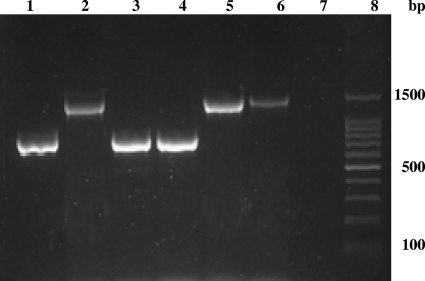
Fig. 1.
Examples of the Mtub02 locus PCR results. Lanes 1 and 8, 100-bp DNA ladder; lane 2, laboratory reference strain H37Rv; lanes 3, 4, and 5, non-Beijing family isolates; lane 6, Beijing family isolate; lane 7, negative control (double-distilled water).
Forty-four spoligotypes were observed among the 210 isolates, of which 30 involved only a single isolate, while the remaining 12 were shared by 180 of the 210 isolates. Based on their spoligotypes, 130 (61.9%) of the 210 study isolates were determined to belong to the Beijing family, while the remaining 80 (38.09%) were classified as non-Beijing family isolates.
Despite the well-recognized global spread of the Beijing family strain infection, its contribution to the burden of pediatric TB is largely unknown. Previous studies conducted in East Asia countries, including China, reported much higher rates (approximately 80% to 90%) of Beijing family strain infection than those found in our study (7, 9, 11). Our observation raised the question of whether the rates of Beijing family strain infection differ between the adult and the pediatric TB patient populations. Future population-based studies will allow a more thorough investigation to answer this question.
The five MIRU-VNTR PCR assays were performed on the same set of isolates double blindly. A seven-repeat allele at the MIRU 26 locus was found in 84.62% (110/130) of the Beijing family isolates and 12.50% (10/80) of the non-Beijing family isolates, while 15.38% (20/130) of the Beijing family isolates and 87.50% (70/80) of the non-Beijing family isolates had an allele with less or more than seven repeats. A four-repeat allele at the ETR-A locus was found in 87.69% (114/130) of the Beijing family isolates and 32.50% (26/80) of the non-Beijing family isolates. At the Mtub30 locus, a four-repeat allele was observed in 97.69% (127/130) of the Beijing family isolates and 10.00% (8/80) of the non-Beijing family isolates. For the MIRU 31 locus, 81.54% (106/130) of the Beijing family isolates and 12.50% (10/80) of the non-Beijing genotype isolates had an allele with five repeats, while 18.46% (24/130) of the Beijing family isolates and 87.50% (70/80) of the non-Beijing family isolates had an allele with less than five repeats. For the Mtub02 locus, 98.46% (128/130) of the Beijing family isolates and 1.25% (1/80) of the non-Beijing family isolates showed a negative result, and the rest of the isolates had a positive result (Fig. 1).
Using spoligotyping as the gold standard for defining Beijing family isolates, we evaluated the sensitivity and specificity of the PCR assays targeting the five MIRU-VNTR loci in the detection of Beijing family isolates. We also evaluated the concordance between spoligotyping and each of the five MIRU-VNTR PCR typings (Table 1).
Table 1.
Concordance between spoligotyping and five different MIRU-VNTR PCR assays and the DTM-PCR in genotyping of 210 pediatric isolates of M. tuberculosis and sensitivity and specificity of the studied MIRU-VNTR PCR assays and the DTM-PCR for the identification of Beijing family isolates, using spoligotyping as the gold standard
| PCR assay(s) | Concordance (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|
| Mtub02 | 98.75 | 98.46 | 98.75 |
| DTM-PCR | 96.19 | 99.23 | 91.25 |
| Mtub30 | 94.76 | 97.69 | 90.00 |
| MIRU 26 | 85.71 | 84.62 | 87.50 |
| MIRU 31 | 83.33 | 83.08 | 87.50 |
| ETR-A | 81.43 | 87.69 | 67.50 |
| Mtub02 and DTM-PCR | 94.76 | 100 | 92.50 |
| Mtub02 and Mtub30 | 93.81 | 100 | 91.25 |
| DTM-PCR and Mtub30 | 91.90 | 100 | 86.25 |
| Mtub02, Mutb30, and DTM-PCR | 91.90 | 100 | 86.25 |
The Mtub02 locus PCR assay showed the highest concordance (98.57%), sensitivity (98.46%), and specificity (98.75%) among the five loci. The other MIRU-VNTR PCR that showed a high sensitivity, specificity, and concordance was the Mtub30 locus PCR (Table 1). Although the Mtub30 locus PCR has been used for M. tuberculosis genotyping, this is the first time it has been found to be a useful method for the detection of Beijing isolates. A good concordance with spoligotyping was found for PCR assays targeting the Mtub02 and Mtub30 loci.
The DTM-PCR resulted in a product of 1,466 bp in 85 of the 210 study isolates and a 761-bp product in the remaining 125 isolates (Fig. 2). Of the 125 isolates with the 761-bp PCR product, 123 (98.4%) were Beijing family isolates, accounting for 94.62% of the 130 Beijing family isolates. In contrast, the majority (78/85 [91.76%]) of the isolates with the 1,466-bp PCR product were non-Beijing family isolates. The DTM-PCR assay showed the highest sensitivity (99.23%) for the detection of Beijing family isolates compared with the sensitivities of all the other five MIRU-VNTR PCR assays. However, its specificity was lower than that of the Mtub02 PCR assay (98.75% versus 91.25%).
Fig. 2.
Examples of the deletion-targeted multiplex PCR results. Lanes 1, 3, and 4, Beijing family isolates; lanes 2 and 5, non-Beijing family isolates; lane 6, strain H37Rv; lane 7, negative control (double-distilled water); lane 8, 100-bp DNA ladder.
We also assessed the sensitivity, specificity, and concordance of the various combinations of Mtub02 PCR, Mtub30 PCR, and DTM-PCR (Table 1). A combination of any two or all of the three methods generated a sensitivity of 100%. However, the combined specificity and concordance were lower than those resulting from the Mtub02 PCR assay alone.
Taken together, the Mtub02 and Mtub30 locus PCR assays and the DTM-PCR assay can serve as some good alternatives to spoligotyping in resource-limited countries where the infection by Beijing family strains is prevalent. If resources allow, a combination of the Mtub02 locus PCR with DTM-PCR assays is recommended to achieve higher sensitivity and specificity. Nevertheless, because the Beijing family comprises a diverse population of clinical isolates and the population structure of M. tuberculosis can vary among geographic regions, the recommendation needs to be validated using isolates collected from a broader geographic region.
Acknowledgments
We thank Aihua Zhang in the Department of Infectious and Gastroenterology, Children's Hospital of Chongqing Medical University, for her assistance in collecting the study isolates.
Zhenhua Yang is grateful for the support of the Elizabeth Crosby Award of the University of Michigan that allowed her to travel to China from the United States for sabbatical, during which this collaborative study was developed.
Footnotes
Published ahead of print on 1 December 2010.
REFERENCES
- 1. Allix C., et al. 2006. Evaluation of the epidemiological relevance of variable-number–tandem-repeat genotyping of Mycobacterium bovis and comparison of the method with IS6110 restriction fragment length polymorphism analysis and spoligotyping. J. Clin. Microbiol. 44:1951–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bifani P. J., Mathema B., Kurepina N. E., Kreiswirth B. N. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45–52 [DOI] [PubMed] [Google Scholar]
- 3. Chen J., et al. 2007. Deletion-targeted multiplex PCR (DTM-PCR) for identification of Beijing/W genotypes of Mycobacterium tuberculosis. Tuberculosis (Edinb.) 87:446–449 [DOI] [PubMed] [Google Scholar]
- 4. Chin P. J., Chiu C. C., Jou R. 2007. Identification of Beijing lineage Mycobacterium tuberculosis with combined mycobacterial interspersed repetitive unit loci 26, 31, and ETR-A. J. Clin. Microbiol. 45:1022–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cousins D., et al. 1998. Evaluation of four DNA typing techniques in epidemiological investigation of bovine tuberculosis. J. Clin. Microbiol. 36:168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glynn J. R., Whiteley J., Bifani P. J., Kremer K., van Soolingen D. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiao W. W., et al. 2007. Molecular characteristics of rifampin and isoniazid resistant Mycobacterium tuberculosis strains from Beijing, China. Chin. Med. J. (Engl.) 120:814–819 [PubMed] [Google Scholar]
- 8. Kamerbeek J., et al. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li S., et al. 2007. Genotyping study of 216 Mycobacterium tuberculosis strains isolated from the patients in Tibet with MLVA and spoligotyping. Chin. J. Microbiol. Immunol. 27:711–718 [Google Scholar]
- 10. Rao K. R., Ahmed N., Srinivas S., Sechi L. A., Hasnain S. E. 2006. Rapid identification of Mycobacterium tuberculosis Beijing genotypes on the basis of the mycobacterial interspersed repetitive unit locus 26 signature. J. Clin. Microbiol. 44:274–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Soolingen D., et al. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 33:3234–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang M., et al. 1999. Enhanced capacity of a widespread strain of Mycobacterium tuberculosis to grow in human macrophages. J. Infect. Dis. 179:1213–1217 [DOI] [PubMed] [Google Scholar]