Abstract
Genetically diverse community-associated methicillin resistant Staphylococcus aureus (CA-MRSA) can harbor a bacteriophage encoding Panton-Valentine leukocidin (PVL) lysogenized into its chromosome (prophage). Six PVL phages (ΦPVL, Φ108PVL, ΦSLT, ΦSa2MW, ΦSa2USA, and ΦSa2958) are known, and single-nucleotide polymorphisms (SNPs) in the PVL genes have been reported. We sought to determine the distribution of lysogenized PVL phages among MRSA strains with PVL (PVL-MRSA strains), the PVL gene sequences, and the chromosomal phage insertion sites in 114 isolates comprising nine clones of PVL-MRSA that were selected for maximal underlying genetic diversity. The six PVL phages were identified by PCR; ΦSa2USA was present in the highest number of different lineages (multilocus sequence type clonal complex 1 [CC1], CC5, CC8, and sequence type 93 [ST93]) (n = 37 isolates). Analysis of 92 isolates confirmed that PVL phages inserted into the same chromosomal insertion locus in CC22, -30, and -80 but in a different locus in isolates of CC1, -5, -8, -59, and -88 and ST93 (and CC22 in two isolates). Within the two different loci, specific attachment motifs were found in all cases, although some limited inter- and intralineage sequence variation occurred. Overall, lineage-specific relationships between the PVL phage, the genes that encode the toxin, and the position at which the phage inserts into the host chromosome were identified. These analyses provide important insights into the microepidemiology of PVL-MRSA, will prove a valuable adjunct in outbreak investigation, and may help predict the emergence of new strains.
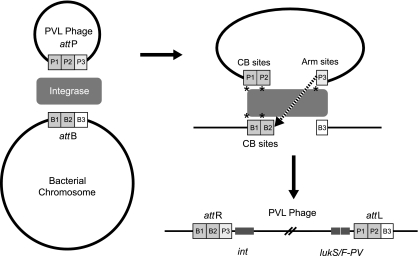
Panton-Valentine leukocidin (PVL) is a bacteriophage-encoded bicomponent leukotoxin found in some strains of Staphylococcus aureus, most notably community-associated methicillin-resistant S. aureus (CA-MRSA). The PVL-encoding genes (lukS-PV and lukF-PV) reside in the genomes of several icosahedral- or elongated-head-shape temperate bacteriophages, i.e., ΦSa2958, ΦSa2MW, ΦPVL, Φ108PVL, ΦSLT, and ΦSa2USA (11, 29). Transmission of the different PVL phages between lineages of S. aureus is likely to be limited by phage/bacterial specificity factors, including restriction-modification systems. After infection, phages lysogenize into the bacterial chromosome via the integrative pathway (14) (Fig. 1), but the chromosomal loci at which lysogeny of the PVL phages occurs have not been identified across multiple lineages of S. aureus, and the specific attachment motifs are unknown for the majority of the PVL-encoding phages. Despite the number of different PVL phages, the genes that encode PVL have been shown to be relatively conserved. Twelve single-nucleotide polymorphisms (SNPs) have been identified (31), including nonsynonymous changes that convert histidine to arginine at amino acid 176 (nucleotide 527) (described as H and R variants). The possible functional implications of this change have not been clearly defined (15, 31, 33), but this substitution is not thought to impair leukotoxicity (2).
Fig. 1.
Insertion of a PVL-encoding bacteriophage into the S. aureus host chromosome via the attP and attB sites forms two new attachment sites, attR and attL. PB sites, phage binding sites (P1, P2, and P3). CB sites, chromosome binding sites (B1, B2, and B3).
CA-MRSA causes infections predominantly among previously healthy individuals, often with few or no traditional health care-associated risk factors for MRSA (23). However, more than 10 years after its emergence, CA-MRSA is an increasing concern as an agent of health care-associated infection (34). CA-MRSA harbors small Staphylococcus cassette chromosome mec (SSCmec) (type IV, V, or VII) (4, 9, 24, 25, 30) and is typically susceptible to more antibiotics than health care-associated MRSA (HA-MRSA). While MRSA strains with PVL (PVL-MRSA strains) are typically associated with pyogenic skin and soft tissue infections (SSTI) (28), they can cause life-threatening disease, most notably necrotizing pneumonia (20). Although the role of PVL as a virulence determinant has been questioned (5, 6, 12, 39), some animal model-based studies have demonstrated its virulence (8, 10, 13, 27, 38). Further, a clear epidemiological association between the PVL genes and successful CA-MRSA lineages is apparent (1, 11, 20, 37). Worldwide, PVL-MRSA strains are mostly polyclonal within a particular region. Better knowledge of the correlation between different PVL-encoding phages and MRSA lineages could help predict the evolutionary pathway of PVL-positive strains and elucidate the potential for PVL phage acquisition by epidemic or endemic hospital-acquired MRSA strains. Here, for the first time, representatives of international lineages of PVL-MRSA circulating in England and Wales were analyzed in detail to determine whether polymorphisms in PVL gene sequence and/or PVL phage insertion locus correlates with PVL-encoding phages within and across lineages of PVL-MRSA.
MATERIALS AND METHODS
Bacterial isolates.
The England and Wales Staphylococcus Reference Unit (SRU) receives isolates from diverse infection types for epidemiological typing and outbreak investigation. In this work, 114 MRSA strains previously characterized by multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), SCCmec, spa, and arginine catabolic mobile element (ACME) PCR (19) and belonging to MLST clonal complex 1 (CC1), CC5, CC22, CC30, CC59, CC80, and CC88 and sequence type 93 (ST93) (3, 16–18, 32) were selected to maximize genetic and geographic diversity.
PVL gene and insertion locus sequencing.
Genomic DNA was extracted using the QIAxtractor (Qiagen, Crawley, United Kingdom) according to the manufacturer's instructions. Three primers pairs were designed to amplify the 1,918-bp lukSF-PV genes (Eurofins MWG, London, United Kingdom), and additional PCRs spanning the proximal and distal junctions of the phage/chromosome DNA targeted the insertion sites of the six known PVL phages (Eurofins MWG, London, United Kingdom). PCR mixtures contained 0.3 μM primers (Table 1) and AccuPrime Pfx DNA polymerase (Invitrogen Ltd., Paisley, United Kingdom) with 1× reaction buffer, according to the manufacturer's instructions. Reactions were thermocycled as follows: 94°C for 2 min; 35 cycles of 95°C for 15 s, 51°C for 30 s, and 68°C for 2 min; and 68°C for 10 min. PCR products were DNA sequenced on both strands. MEGA4.1 (36) was used for DNA sequence alignments, editing, and initial neighbor-joining analysis. These initial trees were used as the seed for an optimized maximum-likelihood (ML) tree generated in PhyML 2.4.4 (21), using 500 nonparametric bootstraps, the Hasegawa-Kishino-Yano (HKY) model (22) of nucleotide substitution, empirical estimation of base frequencies, a fixed transition transversion ratio, and empirically derived proportions of invariant sites.
Table 1.
Primers used in this study
| Use and primer name | Sequence (5′ to 3′) | PCR fragment |
|---|---|---|
| ΦSa2USA identification | ||
| Sa2USA_F | GGTATTACCCAACAACACAACAATTACG | ΦSa2USA phage |
| Sa2USA_R | CCTCAGGCGCCATCACCAATA | |
| Sequencing of PVL genes | ||
| PVL1F | ATGGTCAAAAAAAGACTATTAG | PVL1 |
| PVL1R | GTCTCTCGGATTTTGACTA | |
| PVL2F | GTAAAATGTCTGGACATGA | PVL2 |
| PVL2R | CTACAACGTTTACTGAGTC | |
| PVL3F | TTTATTGGGGTTCTAAGTAC | PVL3 |
| PVL3R | TTAGCTCATAGGATTTTTTTC | |
| Sequencing of PVL phage junctions | ||
| Start(phage) | TAAGAAAGTTAC CACGCACACA | Proximal fragment (CC1, -5, -8, -59, and -88 and ST93) |
| Start(chrom) | TACAATTCCCAAAATACGTAAG | |
| Start(phage) | TA AGAAAGTTAC CACGCACACA | Proximal fragment (CC22, -30, and -80) |
| Start(chrom2) | TAACATCTCAGAAAATGAAAATAAA | |
| End(phage) | GAAAAAAATCCTATGAGCTAA | Distal fragment (CC1, -5, -8, -59, and -88 and ST93) |
| End(chrom) | GTTTGACCGT ATTAGAGAGG | |
| End(phage) | GAAAAAAATCCTATGAGCTAA | Distal fragment (CC22, -30, and -80) |
| End(chrom2) | AGT ATACAGCTAG TTTTTCTAAT |
PVL phage identification.
Eight PCRs were performed to detect five of the PVL-encoding phages (ΦSa2958, ΦSa2MW, ΦPVL, Φ108PVL, and ΦSLT), as described previously (3, 29). Within this scheme two PCRs detect two morphologically distinct phage types by targeting genes encoding icosahedral or elongated head shape. Two further PCRs linked these morphologies to the PVL genes. The remaining PCRs classified individual PVL phages (29). Phages that were not identifiable by the characterization PCRs but were positive for the icosahedral head type were described as “icosahedral”; similarly, those positive for the elongated head type were described as “elongated.” PCR-negative samples were labeled “unknown.” A PCR (fragment size, 680 bp) was designed to detect a sixth PVL phage, ΦSa2USA (Table 1); PCR conditions were as follows: 94°C for 2 min; 36 cycles of 94°C for 30 s, 62°C for 30 s, and 72°C for 1 min; and 72°C for 10 min.
RESULTS
The 114 PVL-MRSA strains studied clustered by lineage based on MLST and PFGE profile; they had 23 different spa types, harbored SCCmec IV, V (5C2), or V (5C2 and 5), and were resistant to oxacillin (MIC of ≥4 mg/liter) (Table 2). The patient demographics and disease presentations for the 114 affected individuals were typical for PVL-MRSA (23). Patients were between 11 days and 99 years old (median, 20 years); 56.1% were male (n = 64), and 39.5% were female (n = 45, sex unknown = 5). There was a wide geographic distribution throughout England and Wales. Four patients presented with upper respiratory tract infection, two with bacteremia, and three presented with community-onset pneumonia; three patients subsequently died. Overall, each isolate had one of the known PVL-encoding phages detected by PCR, i.e., ΦSa2USA (n = 37), ΦPVL (n = 33), ΦSa2MW (n = 10), Φ108PVL (n = 8), ΦSa2958-like phage (n = 2) (Table 2), or had an unidentifiable phage that had an icosahedral head morphology (n = 12) or an elongated head morphology (n = 8). Four isolates encoded PVL but had no detectable phage (described as unknown), highlighting the limitations of the current PCR scheme (29). Using the proposed progenitor PVL gene as a reference sequence in ΦSLT/ST30 (35, 42), six SNPs in lukS-PV and two in lukF-PV were identified (Table 2). Of the eight SNPs, two were nonsynonymous; these have been previously described as an arginine-to-histidine change at amino acid 176 (nucleotide 527) (n = 37) and a lysine-to-glutamic acid change at amino acid 577 (nucleotide 1729) (n = 40).
Table 2.
Characteristics of 114 Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus isolates, including PVL phage identification and PVL gene SNPs
| Lineage (n) | Haplotype groupa | Total no. of isolates | PVL phage | Nucleotide (reference sequence, ΦSLT) |
spa type (n) | SCCmec type (n) | GenBank accession no. | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
lukS-PV |
lukF-PV |
|||||||||||||
| 33 (G) | 105 (T) | 345 (C) | 470 (T) | 527 (A)b | 663 (G) | 1396 (A) | 1729 (A)b | |||||||
| CC1 (7), CC5 (7), CC8 (12), ST93 (11) | R | 37 | ΦSA2USA | G | T | C | T | G | T | A | G | t002 (7), t008 (11), t024 (1), t128 (7), t202 (10), t3949 (1) | IVa (29),IVb (1), IVc (7) | HM584700 |
| CC22 (47), CC5 (1) | H1 | 48 | ΦPVL (33), Φ108PVL (8), icosahedral (6), unknown (1) | G | T | C | T | A | G | G | A | t005c (42), t1790 (2), t849 (1), t2816 (1), t1516 (1), t002 (1) | IVc (42), IVd (3), IV-NT (1), V (5C2) (1), IV-NT (1) | HM584701 |
| CC80 (7) | H2 | 7 | ΦSa2MW | A | T | T | T | A | G | A | A | t044c (7) | IVc (7) | HM584703 |
| CC88 (5) | H2 | 5 | Elongated phi88 | G | C | C | T | A | G | A | A | t690 (2), t448 (2), t186 (1) | IVa (5) | HM584704 |
| CC1 (ST772; 1), CC30 (8),CC5 (1) | H2 | 10 | Unknown (2), icosahedral (5), elongated (3) | G | T | C | T | A | G | A | A | t657 (1), t012 (1), t318 (1), t019 (6), t002 (1) | V (5C2 and 5) (1), IVa (2), IVc (5), IVd (1), IVh (1) | HM584705 |
| CC30 (1) | H2 | 1 | Icosahedral (1) | G | T | C | A | A | G | A | A | t318 (1) | IVa (1) | HM584706 |
| CC59 (3) | H2 | 3 | ΦSa2958 like (2), unknown (1) | G | T | C | T | A | G | A | G | t437 (3) | V (5C2 and 5) (3) | HM584707 |
| CC1 (3) | H3 | 5 | ΦSa2MW (3) | G | T | C | T | A | T | A | A | t127 (3) | IVc (3) | HM584702 |
From reference 31.
Nonsynonymous SNPs.
Fewer than five repeat differences between this and other spa types (the evolutionary significance of repeat differences in spa is currently unknown).
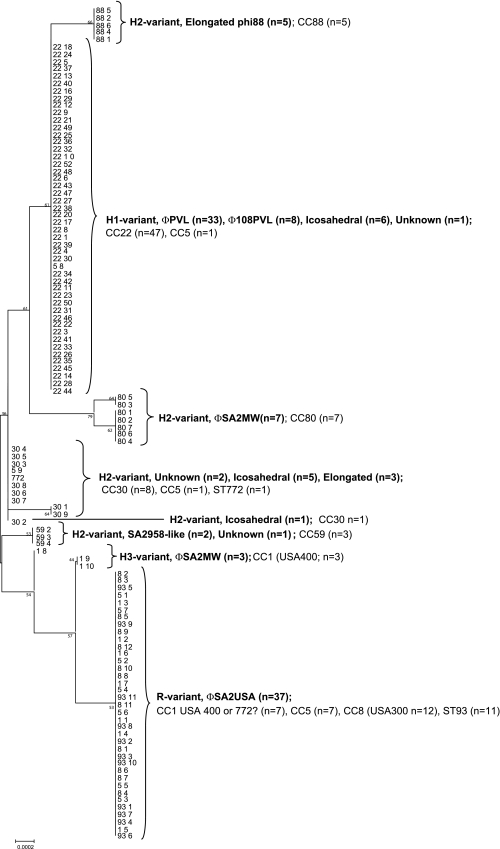
The ML analysis tree was derived as described in Materials and Methods and showed relationships between the PVL-encoding phage and genetic lineage (Fig. 2). Assignation of the PVL gene sequences to one of the four previously described PVL gene haplotype groups, the R, H1, H2, and H3 variants (31), indicated that the PVL phage type often had the same haplotype sequence, and in most cases this relationship was direct (Table 2). For example, 37 isolates had the same R variant SNP profile with three substitutions compared to ΦSLT, and this profile was associated with the ΦSa2USA phage in all cases. Exceptions occurred where ΦSa2MW had lukSF-PV with H2 or H3 SNP profiles in different MRSA lineages. Heterogeneity within SNP profiles with respect to PVL-encoding phage occurred where it was not possible to fully characterize the PVL phage due to known limitations of the PCR scheme (3, 33); for example, 47 isolates with one of three different PVL phages (ΦPVL, Φ108PVL, and an icosahedral phage of unknown type) had the same H1 SNP profile (Table 2).
Fig. 2.
ML analysis tree with bootstrap values based on comparative sequence analysis; this suggests a possible evolutionary relationship to explain the variation in PVL sequence data for 114 PVL-MRSA strains. SNP profile groups are shown to cluster for PVL-encoding phages. For each of the 114 PVL-MRSA isolates, the first number corresponds to the MLST clonal complex (CC) and the second to the isolate number. Branching numbers represent bootstrap values.
Relationships between the SNP profile, PVL-encoding phage, and genetic lineage were, as expected, less exclusive. In PVL-MRSA strains belonging to CC1, there were a number of different SNP profiles correlating with different PVL phages: ΦSa2USA in a typical United Kingdom-derived strain of ST1 isolates, ΦSa2MW in ST1 isolates similar to USA400, and an unknown phage associated with isolates of the double-locus variant of ST1 (ST772), also known as the Bengal Bay clone (Table 2). This highlights the potential for a single lineage to include isolates which have different PVL gene SNP profiles in combination with different PVL phages, suggesting that PVL phages circulating in a population of S. aureus may have different lysogenic potentials.
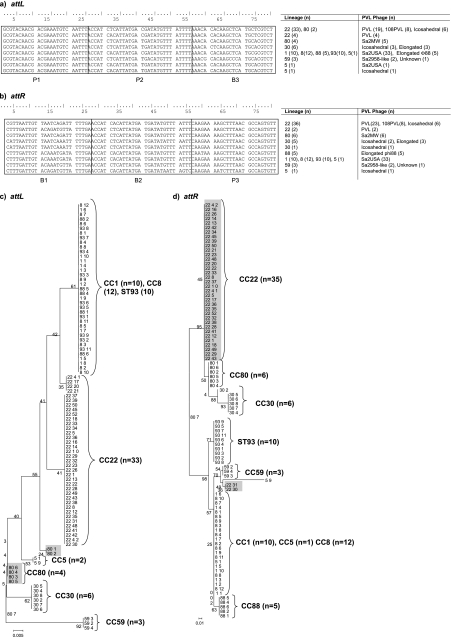
DNA sequencing of the junctions between the S. aureus chromosome and the ends of the lysogenized PVL phage chromosome showed a high degree of conservation at the 5′ and 3′ ends of the PVL phage chromosomes (all PVL phage types) in all isolates studied. In contrast, significant variations occurred in the chromosomal region proximal to the PVL phage, and, to a lesser extent, there was some variation in the distal region. The concatenated S. aureus chromosomal sequences indicated the existence of two different PVL insertion sites (Ins1 and Ins2; GenBank accession numbers HQ020531 to HQ020554), and these occurred in specific MRSA lineages: Ins1 in CC22, -30, and -80 (n = 48) and Ins2 in CC1, -5, -8, -59, and -88 and ST93 (n = 44) (Fig. 3). It was not possible to identify the insertion site sequence in 22 of the isolates. BLAST searches showed the presence of only one insertion site in each lineage; i.e., where Ins1 was present, Ins2 was not and vice versa. Moreover, insertion site sequences could be identified in BLAST searches of sequences from PVL-negative isolates in GenBank. From previous work on sequencing the ΦPVL phage, we were able to identify the specific attachment sites for lysogeny, sites attR and attL (Fig. 4) (26), and these conserved sites can occur in different genes on the host chromosome (depending on lineage). ML analysis tree clustering of the attR and attL attachment sites corresponded to the different lineages of PVL-MRSA, and nine SNP profiles for attR and eight for attL were apparent. Lineage-specific SNPs were identified in both attachment sites (for example, in CC22 and CC80 [highlighted in Fig. 4c and d]), as was some intralineage variation, although overall there was broad conservation of Ins site sequences across the different PVL-MRSA lineages tested (Fig. 4).
Fig. 3.
ML analysis tree with bootstrap values analyzing sequence data spanning proximal and distal phage/chromosomal regions of the PVL phage insertion sites of 92 PVL-MRSA strains. It was not possible to sequence across the junction in 22 of the PVL-MRSA isolates. For each of the 114 PVL-MRSA isolates, the first number corresponds to the MLST clonal complex (CC) and the second to the isolate number. Branching numbers represent bootstrap values.
Fig. 4.
ClustalW sequence alignment of attachment motifs from 92 PVL-MRSA strains. (a) attL attachment site; (b) attR attachment site; (c and d) ML analysis tree with bootstrap values for attL (c) and attR (d). Regions of intralineage variation are shaded (CC80 and CC22). For each of the 114 PVL-MRSA isolates, the first number corresponds to the MLST clonal complex (CC) and the second to the isolate number. Branching numbers represent bootstrap values.
DISCUSSION
CA-MRSA strains which produce PVL first emerged in the 1990s (20, 23), have subsequently disseminated globally, and are recognized as being polyclonal in nature. The genes encoding PVL are borne on bacteriophages, the population dynamics of which are poorly understood. Collectively, the data presented here show that PVL-MRSA strains encode a number of different PVL phages which can incorporate into the host chromosome at one of two sites (depending on lineage). Sequence variation in the PVL genes is generally lineage specific but suggests that most derive from a recent common ancestor. This work demonstrates hitherto-unrecognized diversity in PVL phage genotype and allows us to gain a better understanding of the dynamics of the spread of PVL phages within clonal strains of PVL-MRSA.
Of the 114 CA-MRSA strains studied, 89 carried one of five PVL-encoding bacteriophages. The PVL phages in the remainder could not be fully characterized by PCR as described by Ma et al. (29), highlighting the limited specificity range of the scheme (3, 33) in addition to the potential for misleading data in detecting phage DNA, as S. aureus can carry multiple lysogenized phages (7). A seventh PVL-encoding phage, Φtp310-1, has recently been identified, and this may be harbored by some of the isolates where phage type was not identified (41). Nevertheless, the specific PCR developed in this work detected ΦSa2USA in a large number of diverse isolates (n = 37, including CC1, -5, and -8 and ST93). This assay is therefore an important adjunct, where USA300 and other PVL-MRSA strains harboring the ΦSa2USA phage are found in significant numbers. Further, the data suggest that the ΦSa2USA phage is to be the most competent at infiltrating different lineages of MRSA, in particular those which subsequently cause significant public health concerns (e.g., USA300. This phenomenon warrants further study.
The sequence variation in the PVL genes, and hence the PVL gene SNP profile designations, were related to PVL bacteriophages. Nonsynonymous SNPs were identified in the PVL genes, in accordance with the findings of others (15, 31, 33). The His176Arg change (nucleotide 527) in 37/114 isolates is known not to affect the leukotoxicity of PVL (2), while the functional implication of the Lys577Glu substitution (nucleotide 1729) found in CC1, -5, -8, and -59 (n = 40) merits further investigation. All other synonymous SNPs appear to be lineage and/or phage associated. The PVL gene SNPs correlate with those of another United Kingdom-based study by Otter et al. (33); differences from the SNPs described by O'Hara et al. (31) are likely to be attributable to variations in strain demographics. The conservation of DNA sequence among the PVL genes suggests that these have derived from a recent common ancestor, emphasizing the importance of horizontal transfer of PVL via different bacteriophages. While the totality of the factors underlying the susceptibility of S. aureus to the various PVL bacteriophages is not fully understood, host factors (e.g., the SauI restriction-modification system [40], bacterial host receptors, and the availability of insertion hot spots or phage lysogeny sites) are likely to contribute to the ability of the phages to insert into the genomes of specific lineages of S. aureus.
Analysis of the sequence diversity at the insertion site for the different PVL phages identified in this study showed that PVL phages inserted into one of two lineage-specific insertion sites within the S. aureus chromosome: Ins1 in CC22, -30, and -80 and Ins2 in CC1, -5, -8, -59, and -88 and ST93 (plus occasional CC22 [n = 2]). It was not possible to identify the insertion site sequence in 22 of the isolates; this may be due to sequence variation in the primer binding sites or may suggest there are more than two insertion sites for the PVL phages (yet to be indentified); this warrants further work. Notably, within these different insertion sites, specific attachment motifs were conserved across the lineages of PVL-MRSA tested, although some limited inter- and intralineage variation was present. Taken together, these data lend support to the notion of there being considerable lineage-phage specificity within PVL-MRSA. Waldron and Lindsay concluded that the SauI hsdS profile of an isolate is a key characteristic for phage infectivity and lysogeny and that this is likely to be lineage specific. This is likely to, at least in part, influence the type of PVL phage that a particular genetic lineage of PVL-MRSA harbors. However, while diversity in the HsdRSau1 system in MLST strain types which carry different PVL phages (e.g., ΦSa2USA or ΦSa2MW in ST1 [40]) remains, the relative contribution of the Sau restriction system or other factors remains unclear.
In a recent paper by Wirtz et al., treatment of an ST80 strain with mitomycin C showed unusual prophage excision, in the course of which bacterial DNA was incorporated in the phage genome (41). This phenomenon was not apparent in an MW2 strain. Thus, lineage-specific variation in upstream insertion site sequences identified in this study could be due to incorporation of host chromosomal DNA as part of the lysogenizing prophage in certain clonal complexes. This would correlate with the different upstream sequences seen in CC80 isolates in this study. This phenomenon may also extend to phages infecting CC22 and CC30, where a similar pattern exists; further work is warranted.
Although these data do not provide information on the rate of sequence change or degree of geographic variation, they provide an important insight into the microepidemiology of PVL-MRSA and highlight the role that bacteriophages play in the evolution of MRSA clones (40). In particular, this kind of analysis has already proven to be a valuable adjunct in elucidating the microepidemiology of PVL-MRSA during the course of an outbreak (32). It is probable that the variation highlighted here will also have implications for PVL expression and production, which may ultimately affect disease severity. The structural and functional implications of this variation and how this relates to disease severity warrant further study. PVL is yet to be identified in epidemic or endemic HA-MRSA strains, but this work suggests that the insertion sites for PVL phages may be present in PVL-negative strains. If an epidemic HA-MRSA strain were to acquire PVL, it could raise the morbidity and mortality among cases of nosocomial MRSA infection, with attendant public health impact. Our ability to monitor the spread of existing and potential new PVL phages provides an insight into the evolution of new lineages of PVL-MRSA as they emerge and will allow us to detect the acquisition of PVL phages by other MRSA strains, such as HA-MRSA or livestock-associated MRSA (3). Such knowledge will help in detection of the emergence of such strains and allow for the instigation of appropriate management strategies.
ACKNOWLEDGEMENT
The authors have no conflicts of interest.
Footnotes
Published ahead of print on 24 November 2010.
REFERENCES
- 1. Baba T., et al. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827 [DOI] [PubMed] [Google Scholar]
- 2. Besseyre des Horts T., et al. 2010. A histidine-to-arginine substitution in Panton-Valentine leukocidin from USA300 community-acquired methicillin-resistant Staphylococcus aureus does not impair its leukotoxicity. Infect. Immun. 78:260–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boakes E., et al. 2010. Molecular diversity within clonal complex 22 methicillin-resistant Staphylococcus aureus encoding Panton-Valentine leukocidin in England and Wales. Clin. Microbiol. Infect. doi:10.1111/j.1469-0691.2010.03199.x [DOI] [PubMed] [Google Scholar]
- 4. Boyle-Vavra S., Ereshefsky B., Wang C. C., Daum R. S. 2005. Successful multiresistant community-associated methicillin-resistant Staphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel Staphylococcal chromosome cassette mec (SCCmec) type VT or SCCmec type IV. J. Clin. Microbiol. 43:4719–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bubeck Wardenburg J., Bae T., Otto M., DeLeo F. R., Schneewind O. 2007. Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat. Med. 13:1405–1406 [DOI] [PubMed] [Google Scholar]
- 6. Bubeck Wardenburg J., Palazzolo-Ballance A. M., Otto M., Schneewind O., DeLeo F. R. 2008. Panton-Valentine leukocidin is not a virulence determinant in murine models of community-associated methicillin-resistant Staphylococcus aureus disease. J. Infect. Dis. 198:1166–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Canchaya C., Proux C., Fournous G., Bruttin A., Brussow H. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cremieux A. C., et al. 2009. Panton-Valentine leukocidin enhances the severity of community-associated methicillin-resistant Staphylococcus aureus rabbit osteomyelitis. PLoS One 4:e7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daum R. S., et al. 2002. A novel methicillin-resistance cassette in community-acquired methicillin-resistant Staphylococcus aureus isolates of diverse genetic backgrounds. J. Infect. Dis. 186:1344–1347 [DOI] [PubMed] [Google Scholar]
- 10. Diep B. A., et al. 2010. Polymorphonuclear leukocytes mediate Staphylococcus aureus Panton-Valentine leukocidin-induced lung inflammation and injury. Proc. Natl. Acad. Sci. U. S. A. 107:5587–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diep B. A., et al. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739 [DOI] [PubMed] [Google Scholar]
- 12. Diep B. A., Otto M. 2008. The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 16:361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Diep B. A., et al. 2008. Contribution of Panton-Valentine leukocidin in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. PLoS One 3:e3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dorgai L., Sloan S., Weisberg R. A. 1998. Recognition of core binding sites by bacteriophage integrases. J. Mol. Biol. 277:1059–1070 [DOI] [PubMed] [Google Scholar]
- 15. Dumitrescu O., et al. 2008. Polymorphism of the Staphylococcus aureus Panton-Valentine leukocidin genes and its possible link with the fitness of community-associated methicillin-resistant S. aureus. J. Infect. Dis. 198:792–794 [DOI] [PubMed] [Google Scholar]
- 16. Ellington M. J., et al. 2010. First international spread and dissemination of the virulent Queensland community-associated methicillin-resistant Staphylococcus aureus strain. Clin. Microbiol. Infect. 16:1009–10012 [DOI] [PubMed] [Google Scholar]
- 17. Ellington M. J., Ganner M., Warner M., Cookson B. D., Kearns A. M. 2010. Polyclonal multiply antibiotic-resistant methicillin-resistant Staphylococcus aureus with Panton-Valentine leucocidin in England. J. Antimicrob. Chemother. 65:46–50 [DOI] [PubMed] [Google Scholar]
- 18. Ellington M. J., et al. 2009. Clinical and molecular epidemiology of ciprofloxacin-susceptible MRSA encoding PVL in England and Wales. Eur. J. Clin. Microbiol. Infect. Dis. 28:1113–1121 [DOI] [PubMed] [Google Scholar]
- 19. Ellington M. J., Yearwood L., Ganner M., East C., Kearns A. M. 2008. Distribution of the ACME-arcA gene among methicillin-resistant Staphylococcus aureus from England and Wales. J. Antimicrob. Chemother. 61:73–77 [DOI] [PubMed] [Google Scholar]
- 20. Gillet Y., et al. 2002. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 359:753–759 [DOI] [PubMed] [Google Scholar]
- 21. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 22. Hasegawa M., Kishino H., Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160–174 [DOI] [PubMed] [Google Scholar]
- 23. Herold B. C., et al. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593–598 [DOI] [PubMed] [Google Scholar]
- 24. Higuchi W., Takano T., Teng L. J., Yamamoto T. 2008. Structure and specific detection of staphylococcal cassette chromosome mec type VII. Biochem. Biophys. Res. Commun. 377:752–756 [DOI] [PubMed] [Google Scholar]
- 25. Ito T., et al. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaneko J., Kimura T., Narita S., Tomita T., Kamio Y. 1998. Complete nucleotide sequence and molecular characterization of the temperate staphylococcal bacteriophage phiPVL carrying Panton-Valentine leukocidin genes. Gene 215:57–67 [DOI] [PubMed] [Google Scholar]
- 27. Labandeira-Rey M., et al. 2007. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science 315:1130–1133 [DOI] [PubMed] [Google Scholar]
- 28. Lina G., et al. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128–1132 [DOI] [PubMed] [Google Scholar]
- 29. Ma X. X., et al. 2008. Two different Panton-Valentine leukocidin phage lineages predominate in Japan. J. Clin. Microbiol. 46:3246–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ma X. X., et al. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O'Hara F. P., et al. 2008. A geographic variant of the Staphylococcus aureus Panton-Valentine leukocidin toxin and the origin of community-associated methicillin-resistant S. aureus USA300. J. Infect. Dis. 197:187–194 [DOI] [PubMed] [Google Scholar]
- 32. Orendi J. M., et al. 2010. Community and nosocomial transmission of Panton-Valentine leucocidin-positive community-associated meticillin-resistant Staphylococcus aureus: implications for healthcare. J. Hosp. Infect. 75:258–264 [DOI] [PubMed] [Google Scholar]
- 33. Otter J. A., Kearns A. M., French G. L., Ellington M. J. 2010. Panton-Valentine leukocidin-encoding bacteriophage and gene sequence variation in community-associated methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 16:68–73 [DOI] [PubMed] [Google Scholar]
- 34. Popovich K. J., Weinstein R. A., Hota B. 2008. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin. Infect. Dis. 46:787–794 [DOI] [PubMed] [Google Scholar]
- 35. Takano T., et al. 2008. Novel characteristics of community-acquired methicillin-resistant Staphylococcus aureus strains belonging to multilocus sequence type 59 in Taiwan. Antimicrob. Agents Chemother. 52:837–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 37. Vandenesch F., et al. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Varshney A. K., et al. 2010. Augmented production of Panton-Valentine leukocidin toxin in methicillin-resistant and methicillin-susceptible Staphylococcus aureus is associated with worse outcome in a murine skin infection model. J. Infect. Dis. 201:92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Voyich J. M., et al. 2006. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J. Infect. Dis. 194:1761–1770 [DOI] [PubMed] [Google Scholar]
- 40. Waldron D. E., Lindsay J. A. 2006. SauI: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J. Bacteriol. 188:5578–5585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wirtz C., Witte W., Wolz C., Goerke C. 2010. Insertion of host DNA into PVL-encoding phages of the Staphylococcus aureus lineage ST80 by intra-chromosomal recombination. Virology 406:322–327 [DOI] [PubMed] [Google Scholar]
- 42. Wolter D. J., Tenover F. C., Goering R. V. 2007. Allelic variation in genes encoding Panton-Valentine leukocidin from community-associated Staphylococcus aureus. Clin. Microbiol. Infect. 13:827–830 [DOI] [PubMed] [Google Scholar]