Abstract
Nosocomial norovirus (NoV) infection is common and may lead to complications in vulnerable hospitalized patients. Understanding sources and modes of transmission of noroviruses within health care settings will support the design of evidence-based strategies for reducing introduction and further spread. We sequenced a highly variable segment of the genome to identify possible clusters in patients with and without acute gastroenteritis who were hospitalized in the period 2002-2007. Admission and sampling dates were used to separate patients with nosocomial infection from those without nosocomial infection. Epidemiological clustering retrieved 22 clusters, defined as ≥2 patients with nosocomial infection on the same ward within 5 days. In total, 264 patients (of 2,458 tested) were diagnosed with NoV infection, and 61% of the patient strains could be genotyped. Of those, 51% (n = 82) belonged to GII.4, 34% (n = 54) belonged to GII.3, and 15% (n = 24) belonged to other genotypes (GI.6B, GII.17, GII.7, and GII.2). In children's wards, GII.3 strains were associated with nosocomial spread more often than other viruses were, whereas in adults this was the case for GII.4 strains. Sequence alignment recognized 11 new clusters based on identical P2 domains (4 GII.3 and 7 GII.4 clusters), involving patients in different wards. This increased the total number of recognized clusters by 50%. Five of these clusters involved at least one outpatient, providing a possible target for improvement of infection control. We concluded that the use of sequence-based typing should be considered for identifying hidden nosocomial clusters of NoV infections within health care settings.
Noroviruses (NoV) belong to the family Caliciviridae and are the most common cause of acute viral gastroenteritis worldwide (11). Noroviruses have a positive-sense RNA genome with an average length of 7.5 kb (4, 8). Noroviruses are genetically highly variable and are classified into 5 genogroups (GI, GII, GIII, GIV, and GV), 3 of which are found in humans (12, 24).
NoV are usually transmitted from person to person but may also spread via contaminated surfaces, food, and water (17). NoV outbreaks are common, particularly affecting health care institutions such as nursing homes and hospitals, but their impact and modes of transmission have not been assessed systematically (1, 6, 18). Previously, we described a high frequency of nosocomial infections by comparing the times of diagnosis and dates of hospitalization of newly diagnosed patients (1). There is evidence for increased health expenditures and possible complications in high-risk patients for nosocomial norovirus infections, showing that studies are required to develop effective methods for reducing nosocomial infections (23). A study examining the efficacy of control strategies found that implementation within 3 days after the first cases was the only factor that significantly reduced the size and duration of NoV outbreaks in nursing homes, regardless of the infection control protocol that was followed (5). Furthermore, another study monitoring gastroenteritis outbreaks in England demonstrated the potential effectiveness of ward closure in hospitals (15). This shows that timely detection of nosocomial spread is a key determinant of successful control activities (7, 9, 21). We therefore investigated the possible use of molecular typing in addition to routine monitoring for nosocomial infections to detect transmission pathways of norovirus in a hospital environment. Sequencing of the norovirus P2 domain, which is located in the ORF 2 capsid gene, was used to link patients with identical strains into clusters (24, 25). This approach identified possible clusters that would be missed by standard epidemiological cluster analysis.
MATERIALS AND METHODS
Data collection and fecal specimens.
Data on norovirus-positive cases diagnosed between 2002 and 2007 were retrieved from the database of the hospital laboratory and grouped as nosocomial cases, outpatient cases, and community-acquired cases (1). We used a conservative estimate to ensure high specificity by considering the possibility of nosocomial transmission only if a patient was diagnosed with NoV infection for the first time >4 days after admission. Patients who tested positive for NoV 0 to 1 day after admission were defined as having community-acquired cases. Patients with NoV-positive stools diagnosed 2 to 4 days after hospitalization were classified as indeterminate. On the basis of the >4-day cutoff, 22 nosocomial clusters had previously been obtained using epidemiological criteria (defined as ≥2 patients with nosocomial infection with NoV on the same ward within 5 days) (1). Background data listing the age of the patient, sex of the patient, ward where the patient was hospitalized, date of hospitalization, and date of onset of diarrhea were drawn from the hospital database. This extraction was done by an authorized person who also made the records anonymous prior to use by the research team, in compliance with regulations on use of patient data.
Outline.
Stored fecal specimens (stored at −80°C) were retrieved, viral RNA was extracted, and strains were typed using a two-step approach. First, viruses were assigned to a genotype by sequencing region A of the polymerase gene (22). Subsequently, the corresponding P2 domains in the capsid gene were sequenced, using a specific P2 primer set for each genotype (24). This approach was necessary because the genetic diversity of norovirus P2 regions is so high that a single set of primers has an inherently low sensitivity.
RNA extraction and RT.
Fecal samples were suspended (200 mg/200 μl) in Hanks' medium (800 μl) containing penicillin and were clarified for 30 min at 3,000 rpm and 4°C (8,000 relative centrifugal force [RCF]; Eppendorf 4515R centrifuge). Two hundred microliters of the supernatant was transferred to a Magna Pure LC plate for reverse transcription-PCR (RT-PCR) (total nucleic acid extraction program, performed according to the manufacturer's instructions) with an elution volume of 50 μl (Roche Diagnostics GmbH). For genotyping, 20 μl of RNA extract was reverse transcribed to cDNA with random hexamers by use of a MultiScribe reverse transcriptase kit (Applied Biosystems) according to the manufacturer's instructions. All of the obtained threshold cycle (CT) values in our study correspond with real-time PCR results, as previously mentioned (1).
Genotyping and P2 domain sequencing.
A seminested PCR was performed on the polymerase region (region A) to type the NoV strains. First-round PCRs were performed with the primer set FW-JV12 (ATACCACTATGATGCAGATTA) and COGREV (TCGACGCCATCTTCATTCACA) (10); 10 μl cDNA was added to a 40-μl reaction mix containing 5 μl PCR buffer, 1 μl (each) of forward and reverse primers (70 pmol), 2 μl MgCl2, 29.5 μl Adest, 1 μl deoxynucleoside triphosphates (dNTPs) (10 mM), and 0.5 μl Hotstart Taq polymerase (2 units). The second-round PCR (seminested) was performed with primer set JV12-FW and JV15-REV (CTCATCCAYCTRAACATNGNYTCYTG). Two microliters of the first PCR product was added to a 48-μl reaction mix containing 5 μl PCR buffer, 1 μl (each) of forward and reverse primers (70 pmol), 2 μl MgCl2, 37.5 μl Adest, 1 μl dNTPs (10 mM), and 0.5 μl Hotstart Taq polymerase (2 units). Both PCRs were performed using a Gene Amp 9700 thermal cycler (Applied Biosystems) under the following cycling conditions: 96°C for 15 min, 40 cycles of 96°C, 52°C, and 72°C for 1 min each, and 72°C for 10 min. PCR products were loaded on 2% agarose gels (stained with Syber Safe). When the target band was observed (approximately 650 bp), the PCR products were purified with ExoSAP-IT (USB Corporation, Cleveland, OH) (2 μl ExoSAP-IT for 5 μl PCR product), followed by sequencing with the same primers used for PCR, using the ABI Prism BigDye Terminator 3.1 approach (Applied Biosystems model 3730 DNA analyzer), with denaturation at 96°C for 10 s and 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. After assignment of the genotype, type-specific primers were used to sequence a 794- to 818-nucleotide (nt) target covering the P2 domain (24). This was done only for the two most common genotypes, GII.3 and GII.4, for which sufficient background data were available for sequence similarity comparisons. Primers and PCR conditions were as described previously (24).
Sequence analysis.
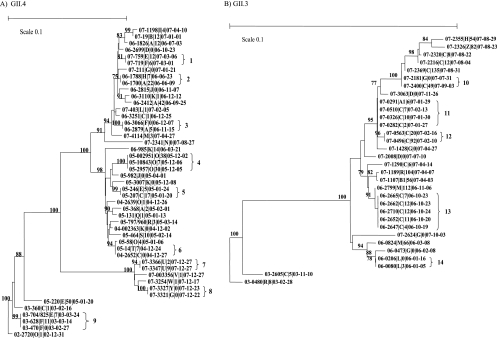
The obtained sequences were entered and aligned in Bionumerics (software package 5.1; Applied Maths) and were typed with a genotyping tool for noroviruses (http://www.rivm.nl/mpf/norovirus/typingtool). The sequences were connected with the available background data file listing age, sampling date, date of discharge from the hospital, and location (ward) where the patient stayed while hospitalized (19). The P2 domain sequences, with an average length of 550 nt, were compared using the neighbor-joining method (TREECON for Windows) to identify patients who had identical sequences in order to create molecular clusters. Sets of identical sequences were defined as clusters (Fig. 1). The community-acquired cases not only served as background sequence data in the comparison but also were used to link with nosocomial cases in order to detect the introduction of strains into the hospital.
Fig. 1.
Phylogenetic trees representing the clusters (1 to 14) of GII.4 (A) and GII.3 (B) strains from both community-acquired and nosocomial cases detected in hospitalized patients. Each strain is labeled as follows: SS-TTTT|U|V|WW-XX-YY, where SS is the year, TTTT is the unique case code, U identifies the ward, V is the number of days after admission at the time of diagnosis, and WW (year)-XX (month)-YY (day) is the sampling date. Strains from patients who were not hospitalized but were sampled while visiting the outpatient clinic or emergency department are indicated with a V of 0.
Statistical analysis.
To test for differences in proportions of nosocomial infection, the following steps were taken. First, the proportion of nosocomial cases within all cases (based on the cutoff of onset of norovirus illness >4 days after admission) was calculated for the genotype categories GII.4, GII.7, GII.3, remaining genotypes, and unknown. These were calculated separately for young children (0 to 5 years) and the remainder of patients (>5 years), as young children are potentially at increased risk for norovirus infection and therefore virus introduction into a hospital is more common for this age group. In these calculations, we excluded patients who had been diagnosed between 2 and 4 days after hospitalization, as the distinction between hospital-acquired and community-acquired infection was not always possible (n = 44). Second, using the chi-square test of independence, we tested whether the proportion of nosocomial infection was independent of (i) genotype, within each age group; (ii) age, within each genotype; (iii) genotype, within all ages; and (iv) age, within all genotypes. Because we were testing multiple hypotheses (nine in total), we needed to adjust the chi-square P values to control for false discoveries. We used the Benjamini-Hochberg method, as this method has more power than other Bonferroni-type procedures (2). A relationship was considered significant if the adjusted P value did not exceed 0.05.
RESULTS
In total, 264 patients (of 2,458 that were evaluated) tested positive for noroviruses during the 5-year period. Among these, 61% of the infecting strains (n = 160) could be genotyped. Viruses belonging to GII.3 (34%; n = 54) and GII.4 (51%; n = 82) were identified most commonly, followed by viruses of GII.7 (9%; n = 15), GII.2 (4%; n = 6), and GI.6B/II.17 (2%; n = 3). The samples that could not be genotyped were retested using different diagnostic PCRs. Mean CT values did not differ between samples that could or could not be genotyped.
Overall, 48% (n = 128) of NoV-positive patients most likely had hospital-acquired infection, according to the cutoff. Patients with newly diagnosed cases (17%; n = 44) had an onset of illness within 2 to 4 days after admission, but the exact source of infection could not be established. Finally, 35% (n = 92) of patients tested NoV positive 0 to 1 day after admission and were classified as having community-acquired cases. In Table 1, the proportions of nosocomial cases for several groups are shown, based on age and genotype. These proportions varied from 0 (for GII.7 strains in adults) to 0.737 (for GII.3 strains in children younger than 5 years of age). As shown in Table 1, genotype GII.3 in children represented a large proportion of nosocomial infections, whereas for the group of patients of >5 years of age, this was the case for GII.4 strains (Table 2). Testing the relationship between the proportion of nosocomial transmission and genotypes, on one hand, and age, on the other hand, showed that the proportion of nosocomial transmission was significantly different in the older age group but not in children. Overall, nosocomial NoV was observed more commonly in young children. Viruses of genotype II.3 were found more often in young children.
Table 1.
Proportions of nosocomial infection by age and genotypea
| Group | Genotype | Age | Proportion of nosocomial infection | No. of patients |
|---|---|---|---|---|
| 1 | II.7 | 0–5 | 0.286 | 7 |
| 2 | II.7 | Rest | 0.000 | 6 |
| 3 | II.4 | 0–5 | 0.478 | 23 |
| 4 | II.4 | Rest | 0.553 | 47 |
| 5 | II.3 | 0–5 | 0.737 | 38 |
| 6 | II.3 | Rest | 0.143 | 7 |
| 7 | Unknown | 0–5 | 0.688 | 48 |
| 8 | Unknown | Rest | 0.694 | 36 |
| 9 | Rest | 0–5 | 0.500 | 6 |
| 10 | Rest | Rest | 0.000 | 2 |
| A | All genotypes | 0–5 | 0.631 | 122 |
| B | All genotypes | Rest | 0.531 | 98 |
| C | II.7 | All ages | 0.154 | 13 |
| D | II.4 | All ages | 0.529 | 70 |
| E | II.3 | All ages | 0.644 | 45 |
| F | Unknown | All ages | 0.690 | 84 |
| G | Rest | All ages | 0.375 | 8 |
Groups are defined as any combination of age group and genotype. Patients who tested positive for NoV after 2 to 4 days of admission were classified as indeterminate and were omitted from the analysis (n = 44).
Table 2.
Results of testing the relationship between the proportion of nosocomial transmission and genotype or age
| Subgroup | Null hypothesis | Adjusted P value | Significant relationship |
|---|---|---|---|
| Age | |||
| 0 to 5 yr | Independent of genotype | 0.14 | No |
| >5 yr | Independent of genotype | <0.01 | Yes |
| Genotype | |||
| GII.7 | Independent of age | 0.69 | No |
| GII.4 | Independent of age | 0.69 | No |
| GII.3 | Independent of age | 0.02 | Yes |
| Unknown | Independent of age | >0.99 | No |
| Rest | Independent of age | 0.60 | No |
| All ages | Independent of genotype | <0.01 | Yes |
| All genotypes | Independent of age | 0.30 | No |
Molecular clustering.
Based on clustering of cases in time and place (two or more cases on the same ward within a 5-day interval), 22 clusters were previously identified in the original data set (1). Viruses from the two major genotypes of NoV (GII.4 and GII.3) were analyzed further. Sequence comparison of amplified P2 domains showed nine clusters of GII.4 strains, involving 17 different patients, and five clusters of GII.3 strains, involving 8 different patients (Fig. 1 and Table 3). Of the molecular clusters, three (two GII.3 and one GII.4) had previously been identified through epidemiological observation, as shown in Table 3. The other 11 identified clusters of patients had not previously been identified as such. This was explained by the fact that all clusters included patients from different wards and ages. Remarkably, for 5 patients, this included a link with a patient who had visited the hospital outpatient care department but was not admitted (clusters 3, 6, 9, 10, and 14).
Table 3.
Overview of molecular clustering versus epidemiological clustering
| Molecular cluster | Presence of epidemiological cluster | Ward(s) | Presence within 5 days | No. of nosocomial infections | No. of indeterminate cases | No. of community-acquired cases |
|---|---|---|---|---|---|---|
| 1 | No | Different | Yes | 2 | 0 | 0 |
| 2 | No | Different | No | 2 | 0 | 0 |
| 3 | No | Different | No | 1 | 0 | 1 |
| 4 | Yes | Same | Yes | 3 | 0 | 0 |
| 5 | No | Different | Yes | 2 | 0 | 0 |
| 6 | No | Different | Yes | 1 | 0 | 1 |
| 7 | No | Same | Yes | 1 | 1 | 0 |
| 8 | No | Different | Yes | 0 | 0 | 2 |
| 9 | No | Different | No | 2 | 0 | 1 |
| 10 | No | Different | No | 1 | 0 | 1 |
| 11 | No | Different | No | 3 | 1 | 0 |
| 12 | Yesa | Same | Yes | 2 | 0 | 0 |
| 13 | Yesa | Same | Yes | 4 | 1 | 0 |
| 14 | No | Same | No | 0 | 1 | 1 |
Cluster was identified by both methods, but the size (number of patients) differed.
DISCUSSION
We describe the results of a systematic evaluation of patients diagnosed with NoV in a large hospital between 2002 and 2007 to look for evidence of nosocomial outbreaks through sequence-based clustering of cases. This approach was done as part of a study aimed at mapping the sources of virus introduction that may be amenable to intervention strategies, as NoV outbreaks in hospitals may have significant health impacts. The use of sequence analysis in this study identified 11 clusters that had not been recognized through earlier defined epidemiological clustering analysis (1), increasing the number of probable nosocomial clusters by 50%. Almost half of these involved links with a patient who had visited the hospital but was not admitted, suggesting the introduction of virus into wards through staff movement or contaminated surfaces. Since we used a rather conservative cutoff for the definition of nosocomial infection, we may have underestimated the prevalence.
We analyzed the virus diversity in relation to date of hospitalization. This provided the opportunity to compare the diversity of the viruses in nosocomial patients with that of viruses causing illness in the community. This comparison is essential, as widespread community outbreaks may occur, in which case finding an identical sequence in the hospital may not signify a hospital-acquired event. An example where this occurred is cluster 8 in Fig. 1A, showing 2 apparently connected patients with community-acquired illness. However, since all other community-acquired cases were distinct, this strengthened the support for the observed approach and thus the clusters that were identified.
Our findings clearly show the limitations of commonly used epidemiological clustering, where these clusters would not be noticed. In this study, patients were identified as possible linked cases when they were hospitalized within the same wards and within the same time frame (5 days).
A limitation of our study is the number of samples for which the genotype could not be determined. Since mean CT values did not differ between stool samples with and without genotyping results, the quantity of virus in the original specimens is not an explanation. A reasonable explanation could be the different PCRs used for diagnosis and genotyping: the former uses a smaller amplicon size, and fragmentation of RNA during preparation and freeze-thawing could preferentially influence the genotyping PCR, with its longer target fragment. Alternatively, it is possible that the nontypeable samples contained different norovirus genotypes, but we could not find any evidence of this possibility.
In the current approach, we used stringent selection based on 100% similarity among strains defining a link (24, 25). This may be too stringent, as NoV is rapidly evolving and mutations are accumulated rapidly (20). Therefore, allowing one or even two nucleotide differences between sequences could potentially increase the sensitivity of outbreak detection. However, this remains to be proven, as few studies have addressed the evolution of NoV over different chains of transmission (3, 20).
Interestingly, we found that the proportion of nosocomial infections seemed to depend on the particular strain involved. In particular, the GII.3 strains showed a significantly larger proportion of nosocomial infection, regardless of age, than the other genotypes. This illustrates the complexity of NoV epidemiology, showing that NoV should not be viewed as “a” virus but rather as a group of related viruses with different properties. This comes as no surprise given the huge diversity of NoV: viruses belonging to GII.7 and GII.4 are quite distinct. Taking these differences into account, one could possibly speculate that each specific genotype could be associated with a particular disease burden. As shown within the family Picornaviridae, another family of positive-strand RNA viruses, genetically related viruses can cause quite distinct spectra of diseases (13). In addition to the virus genotype, age group needs to be factored in: GII.3 strains were found more often in children and in nosocomial cases, in contrast to GII.4 strains, for which this age difference was not found.
Our findings suggest differences in either susceptibility or severity of GII.3 infection for different age groups, as has been described for group A rotaviruses (16). A plausible explanation would be the development of herd immunity, given the widespread circulation of these viruses. For GII.4 strains, rapid evolution of viruses into new antigenic variants has been shown to be an explanation for repeated epidemics involving all age groups (14).
The age-related probability of transmission in a health care setting is something to be aware of. The generally higher rate of nosocomial infection in the young is easily explained by hygienic conditions: young children may wear diapers, and the handling thereof is associated with higher exposure to stools. Without proper hand-washing hygiene, this may constitute a greater risk of transmission. A second factor could be that viral loads are higher in young children, as has been observed for other viruses against which immunity develops. Be that as it may, nonviral factors such as behavior (e.g., hygiene) seem to be important in contributing to transmission.
In conclusion, we show the usefulness of molecular information as a basis for detecting transmission events in the hospital setting. We show that the use of molecular typing may increase the early detection of clusters by 50%, and we were able to identify introductions from the outpatient department. This indicates that a careful review of movements of people between outpatient clinics and wards could potentially identify areas for improvement. The significantly increased proportions of nosocomial transmission of GII.4 and GII.3 strains compared with those of NoV belonging to other genotypes show that an early warning system that rapidly identifies an increasing prevalence of new variants of these genotypes could be used to guide enhanced infection control policies.
ACKNOWLEDGMENTS
We thank Georgina Aron for her assistance during sample collection and Erwin Duizer, Annelies Kroneman, and Linda Verhoef for their useful comments and feedback.
This study was financed by ZonMw, Netherlands.
Footnotes
Published ahead of print on 15 December 2010.
REFERENCES
- 1. Beersma M. F., et al. 2009. Norovirus in a Dutch tertiary care hospital (2002–2007): frequent nosocomial transmission and dominance of GIIb strains in young children. J. Hosp. Infect. 71:199–205 [DOI] [PubMed] [Google Scholar]
- 2. Benjamini Y. H., et al. 1995. Controling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289–300 [Google Scholar]
- 3. Bull R. A., Eden J. S., Rawlinson W. D., White P. A. 2010. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 6:e1000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dingle K. E. 2004. Mutation in a Lordsdale norovirus epidemic strain as a potential indicator of transmission routes. J. Clin. Microbiol. 42:3950–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Friesema I. H., et al. 2009. Norovirus outbreaks in nursing homes: the evaluation of infection control measures. Epidemiol. Infect. 137:1722–1733 [DOI] [PubMed] [Google Scholar]
- 6. Godoy P., et al. 2009. Norovirus outbreaks in hospitals and nursing homes in Catalonia, Spain. Rev. Esp. Salud Publica 83:745–750 [DOI] [PubMed] [Google Scholar]
- 7. Hansen S., et al. 2007. Closure of medical departments during nosocomial outbreaks: data from a systematic analysis of the literature. J. Hosp. Infect. 65:348–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iritani N., et al. 2008. Genetic analysis of the capsid gene of genotype GII.2 noroviruses. J. Virol. 82:7336–7345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Johnston C. P., et al. 2007. Outbreak management and implications of a nosocomial norovirus outbreak. Clin. Infect. Dis. 45:534–540 [DOI] [PubMed] [Google Scholar]
- 10. Kageyama T., et al. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaplan J. E., et al. 1982. Epidemiology of Norwalk gastroenteritis and the role of Norwalk virus in outbreaks of acute nonbacterial gastroenteritis. Ann. Intern. Med. 96:756–761 [DOI] [PubMed] [Google Scholar]
- 12. La Rosa G., et al. 2007. Molecular identification and genetic analysis of norovirus genogroups I and II in water environments: comparative analysis of different reverse transcription-PCR assays. Appl. Environ. Microbiol. 73:4152–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee T. C., et al. 2009. Diseases caused by enterovirus 71 infection. Pediatr. Infect. Dis. J. 28:904–910 [DOI] [PubMed] [Google Scholar]
- 14. Lopman B., et al. 2004. Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet 363:682–688 [DOI] [PubMed] [Google Scholar]
- 15. Lopman B. A., et al. 2004. Epidemiology and cost of nosocomial gastroenteritis, Avon, England, 2002–2003. Emerg. Infect. Dis. 10:1827–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mast T. C., et al. 2010. Burden of childhood rotavirus disease on health systems in the United States. Pediatr. Infect. Dis. J. 29:e19–e25 [DOI] [PubMed] [Google Scholar]
- 17. Onishi N., et al. 2008. Molecular epidemiology of norovirus gastroenteritis in Soma, Japan, 2001–2003. Pediatr. Int. 50:65–69 [DOI] [PubMed] [Google Scholar]
- 18. Pang X. L., Preiksaitis J. K., Wong S., Li V., Lee B. E. 2010. Influence of novel norovirus GII.4 variants on gastroenteritis outbreak dynamics in Alberta and the Northern Territories, Canada between 2000 and 2008. PLoS One 5:e11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Siebenga J. J., Vennema H., Duizer E., Koopmans M. P. 2007. Gastroenteritis caused by norovirus GGII.4, The Netherlands, 1994–2005. Emerg. Infect. Dis. 13:144–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Siebenga J. J., et al. 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J. Virol. 81:9932–9941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Siebenga J. J., et al. 2009. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001–2007. J. Infect. Dis. 200:802–812 [DOI] [PubMed] [Google Scholar]
- 22. Svraka S., et al. 2007. Etiological role of viruses in outbreaks of acute gastroenteritis in The Netherlands from 1994 through 2005. J. Clin. Microbiol. 45:1389–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van den Brandhof W. E., De Wit G. A., de Wit M. A., van Duynhoven Y. T. 2004. Costs of gastroenteritis in The Netherlands. Epidemiol. Infect. 132:211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xerry J., Gallimore C. I., Iturriza-Gomara M., Allen D. J., Gray J. J. 2008. Transmission events within outbreaks of gastroenteritis determined through analysis of nucleotide sequences of the P2 domain of genogroup II noroviruses. J. Clin. Microbiol. 46:947–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xerry J., Gallimore C. I., Iturriza-Gomara M., Gray J. J. 2009. Tracking the transmission routes of genogroup II noroviruses in suspected food-borne or environmental outbreaks of gastroenteritis through sequence analysis of the P2 domain. J. Med. Virol. 81:1298–1304 [DOI] [PubMed] [Google Scholar]