Abstract
Carbapenem resistance mediated by plasmid-borne Klebsiella pneumoniae carbapenemases (KPC) is an emerging problem of significant clinical importance in Gram-negative bacteria. Multiple KPC gene variants (blaKPC) have been reported, with KPC-2 (blaKPC-2) and KPC-3 (blaKPC-3) associated with epidemic outbreaks in New York City and various international settings. Here, we describe the development of a multiplex real-time PCR assay using molecular beacons (MB-PCR) for rapid and accurate identification of blaKPC variants. The assay consists of six molecular beacons and two oligonucleotide primer pairs, allowing for detection and classification of all currently described blaKPC variants (blaKPC-2 to blaKPC-11). The MB-PCR detection limit was 5 to 40 DNA copies per reaction and 4 CFU per reaction using laboratory-prepared samples. The MB-PCR probes were highly specific for each blaKPC variant, and cross-reactivity was not observed using DNA isolated from several bacterial species. A total of 457 clinical Gram-negative isolates were successfully characterized by our MB-PCR assay, with blaKPC-3 and blaKPC-2 identified as the most common types in the New York/New Jersey metropolitan region. The MB-PCR assay described herein is rapid, sensitive, and specific and should be useful for understanding the ongoing evolution of carbapenem resistance in Gram-negative bacteria. As novel blaKPC variants continue to emerge, the MB-PCR assay can be modified in response to epidemiologic developments.
Carbapenems are a class of β-lactam antibiotics with broad-spectrum antibacterial activity, often used to treat infections caused by extended-spectrum β-lactamase (ESBL)-producing Gram-negative bacteria (29). Resistance to carbapenems poses serious challenges in the treatment of such infections, with pan-resistant phenotypes described for an increasing number of Enterobacteriaceae. In particular, Klebsiella pneumoniae carbapenemases (KPCs) constitute a new variant of class A β-lactamase enzymes capable of hydrolyzing all known β-lactam antibiotics and displaying resistance to β-lactamase inhibitors. As with other class A enzymes, they are carried on a variety of plasmids, thereby facilitating horizontal transmission of blaKPC genes. Since their initial description in 2001, KPC-producing strains of K. pneumoniae and other Enterobacteriaceae have spread rapidly in the New York metropolitan region, with increasing numbers of cases reported across the United States (4, 36, 46). Similar reports have been described in other locations, including Argentina, Brazil, China, Colombia, Demark, Finland, France, Germany, Greece, Hungary, Italy, Israel, Norway, Sweden, Puerto Rico, and the United Kingdom (12, 14, 15, 23–26, 39, 43).
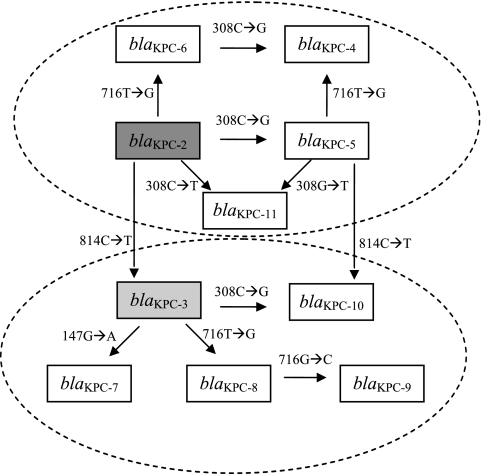
Ten KPC gene variants have been described to date, classified in sequential numeric order from blaKPC-2 to blaKPC-11 (Table 1 and Fig. 1). KPC-1 was initially reported in 2001 from a K. pneumoniae strain isolated in North Carolina (48), but subsequent revision of the blaKPC-1 sequence demonstrated that blaKPC-1 and blaKPC-2 are identical (47). As shown in Table 2, the KPC-2 to KPC-11 genes are characterized by nonsynonymous single nucleotide substitutions within four codons (nucleotides [nt] 147, 308, 716, and 814). A hypothetical scenario depicting the stepwise evolution of all subsequent variants from the KPC-2 gene is shown in Fig. 1. International reports of KPC-2 (25), KPC-3 (25), KPC-4 (20, 27, 31–32), KPC-5 (45), KPC-6 (33), KPC-7 (30), KPC-8 (15), and KPC-10 (31) have been described in numerous countries, with KPC-2 and KPC-3 accounting for most epidemic outbreaks (25). Sequence data for KPC-9 and KPC-11 have been deposited in GenBank (http://www.ncbi.nlm.nih.gov), but no clinical information is available as of this writing.
Table 1.
Discovery of blaKPC variants described to datea
| blaKPC gene | KPC enzyme | Species | Yr isolated | Location | GenBank accession no. | Reference(s) |
|---|---|---|---|---|---|---|
| blaKPC-1b | KPC-1b | Klebsiella pneumoniae | 1996 | North Carolina | AF297554 | 48 |
| blaKPC-2 | KPC-2 | K. pneumoniae | 1998–1999 | Maryland | AY034847 | 36 |
| blaKPC-3 | KPC-3 | K. pneumoniae | 2000–2001 | New York | AF395881 | 46 |
| blaKPC-4 | KPC-4 | Enterobacter cancerogenus | 2003 | Scotland | AY700571 | 27 |
| blaKPC-5 | KPC-5 | Pseudomonas aeruginosa | 2006 | Puerto Rico | EU400222 | 44 and 45 |
| blaKPC-6 | KPC-6 | K. pneumoniae | 2003 | Puerto Rico | EU555534 | 33 |
| blaKPC-7 | KPC-7 | K. pneumoniae | 2007–2008 | Ohio | EU729727 | 30 |
| blaKPC-8 | KPC-8 | K. pneumoniae | 2008 | Puerto Rico | FJ234412 | 15 |
| blaKPC-9 | KPC-9 | Escherichia coli | 2009 | Israel | FJ624872 | Unpublished |
| blaKPC-10 | KPC-10 | Acinetobacter baumannii | 2009 | Puerto Rico | GQ140348 | 31 |
| blaKPC-11 | KPC-11 | K. pneumoniae | 2010 | HM066995 | Unpublished |
Species, year, and location of initial report for each variant.
blaKPC-1 and KPC-1 are no longer considered valid designations, as their sequences are identical to those of blaKPC-2 and KPC-2, respectively (47).
Fig. 1.
Hypothetical stepwise evolution of blaKPC variants, based on single nucleotide changes and order of discovery. In this scenario, blaKPC-2 is the prototype from which all subsequent variants have arisen (44) and gives rise by stepwise nonsynonymous substitution to blaKPC-3 (46), blaKPC-5 (44), blaKPC-6 (33), and blaKPC-11 (GenBank accession no. HM066995). Similarly, blaKPC-3 gives rise to blaKPC-7 (30), blaKPC-8 (15), and blaKPC-10 (31), while blaKPC-4 (27) and blaKPC-9 (FJ624872) each differ by two nucleotides from blaKPC-2 and blaKPC-3, respectively. Alternatively, blaKPC-10 and blaKPC-11 may be derived from blaKPC-5, blaKPC-4 may be derived from either blaKPC-5 or blaKPC-6, and blaKPC-9 may be derived from blaKPC-8. Nucleotide sequences for each blaKPC variant are deposited in GenBank (NCBI) under the following accession numbers: AY034847 (blaKPC-2), AF395881 (blaKPC-3), AY700571 (blaKPC-4), EU400222 (blaKPC-5), EU555534 (blaKPC-6), EU729727 (blaKPC-7), FJ234412 (blaKPC-8), FJ624872 (blaKPC-9), GQ140348 (blaKPC-10), and HM066995 (blaKPC-11).
Table 2.
Nucleotide and amino acid differences between KPC enzymes (blaKPC variants)
| KPC variant | Nonsynonymous substitutiona |
Molecular beacon probeb |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| nt 147 (aa 49) | nt 308 (aa 103) | nt 716 (aa 239) | nt 814 (aa 272) | 147G | 308C | 716T | 716Cc/308T | 814C | |
| KPC-2 | ATG (Met) | CCG (Pro) | GTG (Val) | CAC (His) | + | + | + | − | + |
| KPC-3 | ATG (Met) | CCG (Pro) | GTG (Val) | TAC (Tyr) | + | + | + | − | − |
| KPC-4 | ATG (Met) | CGC (Arg) | GGG (Gly) | CAC (His) | + | − | − | − | + |
| KPC-5 | ATG (Met) | CGC (Arg) | GTG (Val) | CAC (His) | + | − | + | − | + |
| KPC-6 | ATG (Met) | CCG (Pro) | GGG (Gly) | CAC (His) | + | + | − | − | + |
| KPC-7 | ATA (Ile) | CCG (Pro) | GTG (Val) | TAC (Tyr) | − | + | + | − | − |
| KPC-8 | ATG (Met) | CCG (Pro) | GGG (Gly) | TAC (Tyr) | + | + | − | − | − |
| KPC-9 | ATG (Met) | CCG (Pro) | GCG (Ala) | TAC (Tyr) | + | + | − | + | − |
| KPC-10 | ATG (Met) | CGC (Arg) | GTG (Val) | TAC (Tyr) | + | − | + | − | − |
| KPC-11 | ATG (Met) | CTG (Leu) | GTG (Val) | CAC (His) | + | − | + | + | + |
Nucleotide and amino acid (aa) positions of nonsynonymous substitutions in blaKPC variants. Single nucleotide polymorphisms used to design molecular beacon probes are underlined, and amino acids specified by each codon are displayed.
Molecular beacon probe dyes and quenchers are listed in Table 3. Plus and minus signs denote the presence and absence of the target, respectively.
Mixture of MB308T and MB716C, each labeled with the same fluorescent dye (Quasar 670). Positive amplification will be observed if either blaKPC-9 (716C) or blaKPC-11 (308T) is present.
Worrisomely, KPC gene variants have been described in numerous Gram-negative species, including Klebsiella spp. (3, 36, 49), Enterobacter spp. (20, 27), Escherichia coli (4), Pseudomonas aeruginosa (44), Acinetobacter spp. (31), Salmonella enterica (22), Serratia marcescens (7), Citrobacter freundii (50), Proteus mirabilis (38), and most recently Raoultella spp. (8). The successive appearance of distinct blaKPC variants in different species is suggestive of adaptation in response to antibiotic pressure and highlights the ease with which plasmid-borne variants can spread among Enterobacteriaceae, thereby accelerating the global dissemination of carbapenem-resistant strains. Consequently, timely identification of KPC variants is critical for understanding the ongoing evolution of carbapenem resistance in Gram-negative bacteria and can inform infection control measures directed against this emerging public health threat.
In this study, a multiplex real-time PCR scheme was developed which can identify and classify all 10 current blaKPC gene variants (blaKPC-2 to blaKPC-11) in a single reaction using molecular beacon probes. Molecular beacons are short oligonucleotide probes with a hairpin stem-loop structure which exhibit fluorescence only when the loop region anneals to a complementary target (41). The presence of the hairpin significantly enhances specificity (40), enabling detection of single nucleotide polymorphisms (SNPs) in an unambiguous and reproducible manner (21). The assay is rapid and simple to perform, with a high degree of sensitivity and specificity, and may be expanded to accommodate the discovery of novel variants.
MATERIALS AND METHODS
Bacterial strains.
K. pneumoniae strains BK26633 and BK26625 were selected as positive controls for blaKPC from our collection at the Public Health Research Institute Tuberculosis (PHRI TB) Center. Sequence analysis determined that the blaKPC genes of BK26633 and BK26625 are identical to those of Klebsiella oxytoca strain 3127 (blaKPC-2, GenBank accession no. AY210886) and K. pneumoniae strain CL 5761 (blaKPC-3, GenBank accession no. AF395881) (46), respectively. Positive-control strains containing blaKPC-4 to blaKPC-8 were created using a site-saturation mutagenesis method as described in reference 28 at the Louis Stokes Cleveland Department of Veterans Affairs Medical Center in Cleveland, OH (see below for details). Control strains E. coli ATCC 25922 and P. aeruginosa ATCC 27853 were acquired from the American Type Culture Collection (ATCC). All other clinical isolates used in this study were selected from the PHRI TB Center strain repository, obtained from 17 hospitals in New York and New Jersey between 1998 and 2010 (five hospitals from New York City, six from northern New Jersey, and six from southern New Jersey).
Molecular beacon design and synthesis.
Molecular beacon probes were designed according to guidelines described at http://www.molecular-beacons.org and obtained from Biosearch Technologies (Novato, CA). Oligonucleotide primers were designed using Primer3 software (34) and obtained from Integrated DNA Technologies (Coralville, IA). BLAST (http://blast.ncbi.nlm.nih.gov) similarity searches were performed to rule out potential cross-reactivity between primers/probes and nonspecific targets. Six molecular beacons were synthesized with the following 5′ modifications: FAM (fluorescein), HEX (hexachlorofluorescein), CAL Fluor Red 610, Quasar 670, and amino (Table 3); molecular beacon probes MB716C and MB308T utilize the same dye (Quasar 670) (Table 2). For probe MB716T, the 5′ amino modification was coupled with fluorophore Atto 425 as described on the ATTO-TEC GmbH website. Briefly, a molecular beacon probe with a 3′ DABCYL [4-(4′-dimethylaminophenylazo)benzoic acid] quencher and a 5′ amino modification was obtained from Biosearch Technologies and then labeled with Atto 425–N-hydroxysuccinimide (NHS) ester (Sigma-Aldrich, St. Louis, MO) in 0.1 M sodium carbonate buffer (pH 9). All molecular beacons were purified by high-performance liquid chromatography (HPLC) as described elsewhere (42).
Table 3.
Oligonucleotide primers and molecular beacon probes used in this studya
| Primer/probeb | Sequence (5′ to 3′)c | Size (bp) | 5′ fluorescent dye | 3′ quencher |
|---|---|---|---|---|
| KPC-F107 | TCGAACAGGACTTTGGCGGCT | 260 | ||
| KPC-R366 | GGACAGCTCCGCCACCGTCATG | |||
| MB147G | gcgct CGATGGATACCGGCTCA agcgc | FAM | DABCYL | |
| MB308C | cgcga CTGGTTCCGTGGTCAC tcgcg | HEX | DABCYL | |
| MB308T | cgcga CTGGTTCTGTGGTC tcgcg | Quasar 670d | BHQ-2 | |
| KPC-F684 | GGCAGTCGGAGACAAAACC | 177 | ||
| KPC-R860 | CCCTCGAGCGCGAGTCTA | |||
| MB716T | cgcga AACCTGCGGAGTGTATGG tcgcg | Atto 425 | DABCYL | |
| MB716C | cgcga AACCTGCGGAGCGTATGG tcgcg | Quasar 670d | BHQ-2 | |
| MB814C | cgacg GACAAGCACAGCGAGG cgtcg | CAL Fluor Red 610 | BHQ-2 |
FAM, fluorescein; HEX, hexachlorofluorescein; DABCYL, 4-(4′-dimethylaminophenylazo)benzoic acid; BHQ, black hole quencher.
Oligonucleotide primer sets spanning nucleotide positions 107 to 366 and 684 to 860 were used to amplify two segments, each containing two molecular beacon target positions. Positions 308 and 716 are targeted by more than one molecular beacon probe.
Molecular beacon hairpin sequences are shown in lowercase letters. Single nucleotide polymorphism target positions are underlined, as in Table 2.
Quasar 670 dye is used for both MB308T and MB716C, as explained in Table 2.
DNA isolation.
DNA was isolated from bacterial colonies using the boiling lysis method described elsewhere (9). Bacterial strains were grown on Luria-Bertani (LB) agar and incubated overnight at 37°C. A loopful of bacterial growth was suspended in 100 μl of sterile double-distilled water (ddH2O) and boiled for 10 min in a water bath. Following centrifugation at 16,000 × g for 5 min, the supernatant was diluted 10-fold into ddH2O, and 1 μl was used as the template in a 10-μl PCR. For analytical sensitivity based on bacterial CFU, DNA isolation was performed using the NucliSENS easyMAG system (bioMérieux, Durham, NC). In brief, overnight bacterial cultures were serially diluted 10-fold in LB broth, and 200 μl of each dilution was used to extract total nucleic acid (TNA) according to the manufacturer's instructions. Bacterial TNA was eluted in 50 μl elution buffer and stored at −20°C.
Real-time PCR.
A multiplex real-time PCR scheme using molecular beacons (MB-PCR) was designed for rapid detection of variants blaKPC-2 to blaKPC-11 (Table 2). The combination of two sets of primers and six molecular beacons (Table 3) results in a minimum of two specific targets for each blaKPC gene variant. Real-time PCR amplification was performed using the Stratagene Mx3005P multiplex quantitative PCR system (Agilent Technologies, Santa Clara, CA). PCR was performed in 10-μl reactions consisting of 0.5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA), 4 mM MgCl2, 250 μM each deoxynucleoside triphosphate (dNTP), and 0.5 μM each primer (Table 3) in 1× PCR buffer (Applied Biosystems). Optimized molecular beacon concentrations were as follows: 0.4 μM for MB716T and MB716C; 0.2 μM for MB308C, MB308T, and MB814C; and 0.1 μM for MB147G. Optimal cycling conditions included an initial denaturation step of 95°C for 10 min, followed by 40 cycles of 95°C for 20 s (denaturation) and 58°C for 45 s (annealing and extension). The entire MB-PCR can be performed in less than 1.5 h on the Mx3005P platform. K. pneumoniae strains BK26633 and BK26625 were used as positive controls in each PCR assay, while E. coli ATCC 25922 and P. aeruginosa ATCC 27853 were used as negative controls.
Analytical sensitivity.
The analytical sensitivity of the MB-PCR assay was estimated by serial dilution experiments using purified synthesized DNA templates. At the time the study was performed, only bacterial strains harboring blaKPC-2 and blaKPC-3 were available. Overlap extension PCR was therefore used to generate the complete coding sequences of blaKPC-4 to blaKPC-11 (17). Briefly, site-directed mutagenesis was performed using oligonucleotide primers containing single nucleotide polymorphisms (SNPs) specific to each blaKPC variant. Overlapping PCR fragments were then fused together in successive PCR extension reactions. For consistency, the complete coding sequences of blaKPC-2 and blaKPC-3 were similarly generated using the same primer set. The resulting synthesized targets (blaKPC-2 to blaKPC-11) were further purified using the QIAquick gel extraction kit (Qiagen, Hilden, Germany), and DNA concentrations were measured using the Invitrogen Quant-iT PicoGreen dsDNA assay kit (Molecular Probes, Eugene, OR). DNA templates were serially diluted in ddH2O, and l μl of each dilution was used for MB-PCR as described above. Limits of detection for each MB-PCR target were then calculated based on the molecular weight and concentration of each target.
In order to estimate analytical sensitivity using bacterial CFU, transformants carrying blaKPC-4 through blaKPC-8 were created by site-saturation mutagenesis using the Stratagene QuikChange XL site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA), as described elsewhere (28). Plasmids pET24a(+)-blaKPC-4 through pET24a(+)-blaKPC-8 were electroporated into E. coli DH10B competent cells, following which their respective blaKPC regions were sequenced in order to verify the presence of the correct variants. Overnight cultures of the resulting transformants, along with K. pneumoniae strains BK26633 (blaKPC-2) and BK26625 (blaKPC-3), were harvested by centrifugation and serially diluted 10-fold into sterile LB medium. DNA was isolated from 200 μl of each dilution using the NucliSENS easyMAG instrument as described above, and 1 μl of each sample was used for MB-PCR amplification. The numbers of CFU per suspension (CFU/ml) were estimated using standard plating procedures and were used to calculate MB-PCR limits of detection for molecular beacon probes MB147G, MB308C, MB716T, and MB814C. The analytical sensitivities of MB716C (blaKPC-9) and MB308T (blaKPC-11) were not determined by this method, since these targets are present only in blaKPC-9 and blaKPC-11, and transformants containing these variants were not created for this study.
Analytical specificity.
Analytical specificities for each molecular beacon probe were initially tested using the synthesized blaKPC-2 to blaKPC-11 targets described above. DNA was isolated by boiling lysis from the aforementioned blaKPC-4 to blaKPC-8 E. coli DH10B transformants, as well as BK26633 (blaKPC-2) and BK26625 (blaKPC-3), and then used as a template to rule out cross-reactivity between different blaKPC gene variants. In addition, the analytical specificities of all primer/probe combinations were tested using several other species, including E. coli ATCC 25922, P. aeruginosa ATCC 27853, Enterococcus spp. (n = 2 isolates), Staphylococcus spp. (n = 8 isolates), Streptococcus spp. (n = 2 isolates), and Candida albicans (n = 4 isolates). All strains except ATCC 25922 and ATCC 27853 were selected from a previous study (9).
Detection of blaKPC variants in clinical isolates.
Fifty-three blaKPC-positive K. pneumoniae isolates were obtained from three hospitals in the New York/New Jersey metropolitan area and tested using the multiplex real-time MB-PCR assay described here. The isolates were collected from multiple infection sites, including blood, wound, sputum, urine, bone, bile, and bronchial fluid (Table 4). Carbapenemase production was determined for all isolates using the modified Hodge test (19), and the presence of blaKPC was confirmed by conventional PCR (35). Additional Gram-negative bacterial strains (n = 404 isolates) were randomly chosen from the PHRI TB Center collection and also subjected to blaKPC analysis by the MB-PCR assay. The latter were collected between 1998 and 2010 from 17 hospitals in New Jersey and New York and included K. pneumoniae, K. oxytoca, E. coli, Enterobacter spp., Acinetobacter spp., S. marcescens, P. aeruginosa, and C. freundii (Table 5).
Table 4.
MB-PCR results for 53 blaKPC-positive K. pneumoniae isolates
| Isolate source | No. of isolates with: |
|
|---|---|---|
| blaKPC-2 | blaKPC-3 | |
| Blood | 2 | 3 |
| Wound | 1 | 4 |
| Sputum | 1 | 7 |
| Urine | 5 | 25 |
| Othera | 0 | 5 |
Isolated from bone (n = 1 isolate), bile (n = 1 isolate), or bronchial fluid culture (n = 3 isolates).
Table 5.
MB-PCR results for 404 randomly selected clinical Gram-negative isolates
| Species | No. of isolates with: |
Total no. of isolates | |
|---|---|---|---|
| blaKPC-2 | blaKPC-3 | ||
| Acinetobacter spp. | 0 | 0 | 67 |
| Citrobacter freundii | 0 | 1 | 1 |
| Escherichia coli | 3 | 3 | 35 |
| Enterobacter aerogenes | 1 | 0 | 1 |
| Enterobacter cloacae | 0 | 3 | 13 |
| Enterobacter sakazakii | 0 | 0 | 3 |
| Klebsiella pneumoniae | 72 | 97 | 230 |
| Klebsiella oxytoca | 0 | 0 | 3 |
| Pseudomonas aeruginosa | 0 | 0 | 25 |
| Serratia marcescens | 0 | 0 | 26 |
RESULTS
Classification of blaKPC variants by MB-PCR assay.
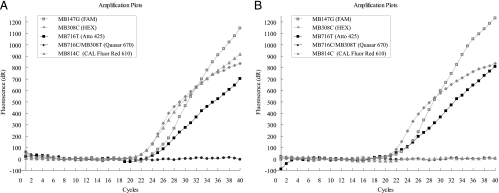
All 10 currently described blaKPC variants were successfully detected using a combination of two oligonucleotide primer sets and six molecular beacon probes (Table 3). Each variant displayed at least two positive amplification signals (Table 2), with four signals displayed by blaKPC-2 and blaKPC-11; three signals by blaKPC-3, blaKPC-5, blaKPC-6, and blaKPC-9; and two signals by blaKPC-4, blaKPC-7, blaKPC-8, and blaKPC-10. All blaKPC variants were positive for target 147G, with the exception of blaKPC-7. Most of the variants could be classified using only four probes/dyes, except for two closely related pairs which required the use of a fifth target. A mixture of MB308T and MB716C was therefore included to distinguish between blaKPC-5/blaKPC-11 and blaKPC-8/blaKPC-9, respectively (Table 3). Characteristic MB-PCR amplification plots for blaKPC-2 and blaKPC-3 are depicted in Fig. 2.
Fig. 2.
Real-time PCR amplification results for blaKPC-2 (A) and blaKPC-3 (B), obtained after 40 cycles on the Stratagene Mx3005P multiplex quantitative PCR system. The multiplex real-time MB-PCR assay consists of six molecular beacon probes labeled with five different fluorescent dyes, as indicated in the legend.
Analytical sensitivity.
The sensitivity of the real-time PCR platform was initially evaluated using synthesized blaKPC-2 to blaKPC-11 targets. Limits of detection based on DNA copy number were obtained for each molecular beacon using purified synthesized DNA, as described in Materials and Methods. Lower limits were reliably detected within 45 cycles as follows: 5 to 10 copies per PCR for targets MB147G, MB308C, MB308T, and MB814C and 20 to 40 copies for targets MB716T and MB716C. Limits of detection based on bacterial CFU were also estimated using DNA isolated from serial dilutions of bacterial cultures. The multiplex assay was capable of reproducibly detecting 2 CFU per PCR for MB147G, MB308C, and MB814C and 4 CFU for MB716T. The overall detection limit of the MB-PCR assay was therefore approximately 4 CFU per reaction.
Analytical specificity.
All molecular beacon primer/probe sets used in this study displayed good specificity for their respective blaKPC targets, with no cross-reactivity observed between individual beacons and synthesized targets. Comparable results were obtained using K. pneumoniae strains harboring blaKPC-2 and blaKPC-3 and E. coli DH10B transformants carrying blaKPC-4 through blaKPC-8. No amplification was observed among blaKPC-negative Enterobacteriaceae isolates (Table 5), while analysis of other organisms, including E. coli ATCC 25922, P. aeruginosa ATCC 27853, Enterococcus spp., Staphylococcus spp., Streptococcus spp., and Candida albicans yielded negative results, in agreement with BLAST similarity searches performed during the assay design phase.
Detection of blaKPC variants in clinical isolates.
Fifty-three blaKPC-positive clinical K. pneumoniae isolates were selected in order to validate the multiplex real-time MB-PCR assay. All isolates were initially shown to produce carbapenemase using the modified Hodge test, with blaKPC carriage confirmed by conventional PCR. Isolates were obtained from diverse specimen sources, including urine (n = 30), sputum (n = 8), blood (n = 5), wound (n = 5), bronchial fluid (n = 3), bone (n = 1), and bile (n = 1). All 53 isolates were positive by MB-PCR, in agreement with conventional PCR results (Table 4). Among the 53 isolates, 44 (83.0%) harbored the blaKPC-3 variant, while only 9 (17.0%) harbored blaKPC-2.
Following validation of the MB-PCR assay, an additional 404 clinical isolates from 17 hospitals in New Jersey and New York were also screened for blaKPC variants. These isolates, collected from 1998 to 2010, were comprised of several Gram-negative species, including K. pneumoniae, K. oxytoca, E. coli, Enterobacter spp., Acinetobacter spp., S. marcescens, P. aeruginosa, and C. freundii (Table 5). Within this collection, 104 (25.7%) isolates were positive for blaKPC-3, while 76 (18.8%) harbored blaKPC-2; no other blaKPC variants were identified in this study. Among the 230 K. pneumoniae isolates tested, a total of 169 (73.5%) harbored blaKPC genes, of which 97 (57.4%) were blaKPC-3 and 72 (42.6%) were blaKPC-2. In contrast, only six (17.1%) blaKPC-positive isolates were identified among 35 E. coli strains tested, with blaKPC-2 and blaKPC-3 represented equally. Three Enterobacter cloacae isolates also harbored blaKPC-3 genes, while one E. aerogenes isolate was positive for blaKPC-2. Similarly, one C. freundii isolate was positive for blaKPC-3, whereas no blaKPC variants were found among the rest of the species included in this study. In order to further validate these findings, we sequenced the entire blaKPC coding regions from 40 randomly selected blaKPC-positive strains and observed 100% concordance with our MB-PCR results (data not shown).
DISCUSSION
Carbapenem resistance among Gram-negative bacteria is an emerging concern of significant clinical and public health importance worldwide (25). In light of the potential for rapid horizontal and vertical transmission of these genes, prompt recognition of KPC-producing organisms is critical for controlling their spread in nosocomial and long-term-care settings. Unfortunately, detection of KPC carbapenemases by susceptibility testing is challenging, due to the heterogeneous expression of β-lactam resistance by multiple determinants (18, 25). Various studies have reported that carbapenem-resistant bacteria may be incorrectly identified as carbapenem susceptible, resulting in inappropriate selection of therapy (2, 5, 37). In order to overcome such shortcomings when treating infections caused by Enterobacteriaceae, the Clinical and Laboratory Standards Institute (CLSI) has recently lowered the susceptibility breakpoints for meropenem, imipenem, and doripenem to ≤1 μg/ml and to ≤0.25 μg/ml for ertapenem (10). Nevertheless, despite the new breakpoints, susceptibility testing results still vary among different methods (6).
Given these limitations, molecular detection of blaKPC genes by PCR has been proposed as the gold standard (25) for detection of KPC-bearing organisms. To date, several PCR-based detection methods have been described (35), including two real-time PCR assays (11, 16), as well as a method that uses PCR in conjunction with electrospray ionization mass spectrometry (PCR/ESI-MS) (13). However, most of these methods do not distinguish between different blaKPC variants, thereby failing to convey information of epidemiologic as well as evolutionary significance. In addition, evidence suggests that some nonsynonymous mutations within blaKPC variants may be of functional significance as well (44). KPC-3, for instance, exhibits a higher rate of ceftazidime hydrolysis than KPC-2 but has lower affinity for cefoxitin (1). Similarly, the activity spectra of KPC-4 and KPC-5 for certain β-lactam antibiotics may differ from that of KPC-2 (44). KPC enzymes may therefore be evolving through stepwise mutation (Fig. 1) in a manner that alters their functional activity (44). Consequently, identification of specific blaKPC variants may be relevant to selection of appropriate therapies for infections caused by multidrug-resistant Gram-negative organisms.
Currently, DNA sequencing is the definitive method for identification of blaKPC variants. However, sequencing is impractical for studies involving large sample sizes, as well as for rapid identification in clinical settings. In contrast, methods such as real-time PCR offer rapid, robust, and cost-efficient alternatives to DNA sequencing for known blaKPC variants. Previously, Cole et al. (11) developed an assay for blaKPC detection using TaqMan real-time PCR, with subsequent differentiation of blaKPC-2 and blaKPC-3 by restriction enzyme analysis. However, the assay is based on one SNP (nt 944, equivalent to nt 814 in the MB-PCR assay described here), corresponding to an RsaI restriction site in blaKPC-3. Consequently, as those authors point out, their assay can potentially misclassify blaKPC-4 to blaKPC-6 as either blaKPC-2 or blaKPC-11, while blaKPC-7 to blaKPC-10 may be misclassified as blaKPC-3. Moreover, this method requires postamplification restriction analysis and electrophoresis (11), which is time-consuming and unsuitable for large-scale studies.
In this study, we describe a multiplex real-time PCR assay for rapid detection and classification of all currently known blaKPC variants. This platform includes two sets of oligonucleotide primers and six molecular beacon probes, enabling detection and differentiation of 10 blaKPC gene variants in a single reaction. Each variant exhibits at least two positive amplification curves, increasing the reliability of blaKPC detection. Using synthesized blaKPC targets, our MB-PCR assay was successfully able to detect all blaKPC gene variants, with no cross-reactivity observed between mutant and wild-type targets. Further analysis using E. coli DH10B transformants harboring blaKPC-4 to blaKPC-8, as well as blaKPC-2- and blaKPC-3-positive K. pneumoniae strains, confirmed the assay's ability to specifically identify these variants. The MB-PCR assay also demonstrated very good analytical sensitivity, detecting as few as 5 to 40 DNA copies and 4 CFU per reaction, similar to the sensitivity reported (1 CFU) by Hindiyeh et al. (16). Lastly, the assay successfully classified blaKPC variants in 53 K. pneumoniae isolates from diverse clinical specimens, while screening of 404 Gram-negative strains uncovered blaKPC genes in four additional species. As shown in Table 5, while blaKPC-2 and blaKPC-3 may be found in the same species, no other blaKPC gene variants were detected in this study.
Aside from greater sensitivity and specificity, real-time PCR does not require postamplification analysis, thereby facilitating results with significantly less time and labor. Similarly, the relatively high price of molecular beacons is offset by savings in sample processing costs and labor, as described previously (9). Moreover, based on the regional frequencies of different blaKPC variants, the MB-PCR scheme may be modified to include a smaller number of molecular beacons. In our region, for example, blaKPC-2 and blaKPC-3 appear to be predominant, requiring only four molecular beacons (MB147G, MB308C, MB716T, and MB814C) for definitive identification (Table 2). Such modifications may be of practical significance given the limited availability of real-time PCR platforms capable of recognizing more than four dyes.
In conclusion, we have described a multiplex real-time PCR assay for the identification and classification of blaKPC gene variants from Gram-negative bacterial isolates. As many as 96 strains may be classified easily within 3 h, including DNA isolation, PCR cycling, and data analysis. The MB-PCR assay is robust, sensitive, and specific, allowing for high-throughput detection and classification of all presently described blaKPC variants. Nevertheless, as novel blaKPC variants continue to emerge, the typing system described herein can be modified in response to epidemiologic developments.
ACKNOWLEDGMENTS
We thank Salvatore Marras for assistance with molecular beacon design and labeling.
This study was supported by grants (to B.N.K.) from the New York Community Trust and Merck & Co., Inc. R.A.B. was supported by the Veterans Affairs Merit Review Program, the Geriatric Research Education and Clinical Center (VISN 10), and the National Institutes of Health (R01 A1063517-01).
Footnotes
Published ahead of print on 1 December 2010.
REFERENCES
- 1. Alba J., Ishii Y., Thomson K., Moland E. S., Yamaguchi K. 2005. Kinetics study of KPC-3, a plasmid-encoded class A carbapenem-hydrolyzing beta-lactamase. Antimicrob. Agents Chemother. 49:4760–4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson K. F., et al. 2007. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J. Clin. Microbiol. 45:2723–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bradford P., et al. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin. Infect. Dis. 39:55–60 [DOI] [PubMed] [Google Scholar]
- 4. Bratu S., et al. 2007. Detection and spread of Escherichia coli possessing the plasmid-borne carbapenemase KPC-2 in Brooklyn, New York. Clin. Infect. Dis. 44:972–975 [DOI] [PubMed] [Google Scholar]
- 5. Bratu S., et al. 2005. Carbapenemase-producing Klebsiella pneumoniae in Brooklyn, NY: molecular epidemiology and in vitro activity of polymyxin B and other agents. J. Antimicrob. Chemother. 56:128–132 [DOI] [PubMed] [Google Scholar]
- 6. Bulik C. C., et al. 2010. Comparison of meropenem MICs and susceptibilities for carbapenemase-producing Klebsiella pneumoniae isolates by various testing methods. J. Clin. Microbiol. 48:2402–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cai J. C., Zhou H. W., Zhang R., Chen G. X. 2008. Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli isolates possessing the plasmid-mediated carbapenem-hydrolyzing beta-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob. Agents Chemother. 52:2014–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castanheira M., et al. 2009. First descriptions of blaKPC in Raoultella spp. (R. planticola and R. ornithinolytica): report from the SENTRY Antimicrobial Surveillance Program. J. Clin. Microbiol. 47:4129–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen L., et al. 2009. Multiplex real-time PCR for rapid staphylococcal cassette chromosome mec (SCCmec) typing. J. Clin. Microbiol. 47:3692–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. CLSI 2010. Performance standards for antimicrobial susceptibility testing; twentieth informational supplement (June 2010 update), M100-S20-U. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 11. Cole J. M., Schuetz A. N., Hill C. E., Nolte F. S. 2009. Development and evaluation of a real-time PCR assay for detection of Klebsiella pneumoniae carbapenemase genes. J. Clin. Microbiol. 47:322–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cuzon G., Naas T., Demachy M. C., Nordmann P. 2008. Plasmid-mediated carbapenem-hydrolyzing beta-lactamase KPC-2 in Klebsiella pneumoniae isolate from Greece. Antimicrob. Agents Chemother. 52:796–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Endimiani A., et al. 2010. Rapid identification of blaKPC-possessing Enterobacteriaceae by PCR/electrospray ionization-mass spectrometry. J. Antimicrob. Chemother. 65:1833–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fontana C., et al. 2010. Emergence of KPC-producing Klebsiella pneumoniae in Italy. BMC Res. Notes 3:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gregory C. J., et al. 2010. Outbreak of carbapenem-resistant Klebsiella pneumoniae in Puerto Rico associated with a novel carbapenemase variant. Infect. Control Hosp. Epidemiol. 31:476–484 [DOI] [PubMed] [Google Scholar]
- 16. Hindiyeh M., et al. 2008. Rapid detection of blaKPC carbapenemase genes by real-time PCR. J. Clin. Microbiol. 46:2879–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59 [DOI] [PubMed] [Google Scholar]
- 18. Kitchel B., et al. 2010. Genetic factors associated with elevated carbapenem resistance in KPC-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 54:4201–4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee K., et al. 2001. Modified Hodge and EDTA-disk synergy tests to screen metallo-beta-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 7:88–91 [DOI] [PubMed] [Google Scholar]
- 20. Livermore D. M., et al. 2008. Non-susceptibility trends among Enterobacteriaceae from bacteraemias in the United Kingdom and Ireland, 2001-06. J. Antimicrob. Chemother. 62(Suppl. 2):ii41–ii54 [DOI] [PubMed] [Google Scholar]
- 21. Mhlanga M. M., Malmberg L. 2001. Using molecular beacons to detect single-nucleotide polymorphisms with real-time PCR. Methods 25:463–471 [DOI] [PubMed] [Google Scholar]
- 22. Miriagou V., et al. 2003. Imipenem resistance in a Salmonella clinical strain due to plasmid-mediated class A carbapenemase KPC-2. Antimicrob. Agents Chemother. 47:1297–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Monteiro J., Santos A. F., Asensi M. D., Peirano G., Gales A. C. 2009. First report of KPC-2-producing Klebsiella pneumoniae strains in Brazil. Antimicrob. Agents Chemother. 53:333–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Naas T., Nordmann P., Vedel G., Poyart C. 2005. Plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC in a Klebsiella pneumoniae isolate from France. Antimicrob. Agents Chemother. 49:4423–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nordmann P., Cuzon G., Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228–236 [DOI] [PubMed] [Google Scholar]
- 26. Osterblad M., et al. 2009. First isolations of KPC-2-carrying ST258 Klebsiella pneumoniae strains in Finland, June and August 2009. Euro Surveill. 14:pii19349. [PubMed] [Google Scholar]
- 27. Palepou M. F., et al. 2005. Novel class A carbapenemase, KPC-4, in an Enterobacter isolate from Scotland, abstr. 1134_01_20. Prog. Abstr. 15th Eur. Cong. Clin. Microbiol. Infect. Dis., Copenhagen, Denmark [Google Scholar]
- 28. Papp-Wallace K. M., et al. 2010. Substrate selectivity and a novel role in inhibitor discrimination by position 237 in the KPC-2 β-lactamase. Antimicrob. Agents Chemother. 54:2867–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Paterson D. L., Bonomo R. A. 2005. Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perez F., et al. 2010. Carbapenem-resistant Acinetobacter baumannii and Klebsiella pneumoniae across a hospital system: impact of post-acute care facilities on dissemination. J. Antimicrob. Chemother. 65:1807–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robledo I. E., et al. 2010. Detection of KPC in Acinetobacter spp. in Puerto Rico. Antimicrob. Agents Chemother. 54:1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robledo I. E., et al. 2007. First report of a KPC-4 and CTX-M producing K. pneumoniae isolated from Puerto Rico (PR), abstr. C2-1933. Abstr. 47th Annu. Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC [Google Scholar]
- 33. Robledo I. E., et al. 2008. A novel KPC variant, KPC-6, in a Klebsiella pneumoniae (Kp) isolated in Puerto Rico (PR), abstr. C2-3738. Abstr. 48th Annu. Intersci. Conf. Antimicrob. Agents Chemother. (ICAAC)-Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet. American Society for Microbiology, Washington, DC [Google Scholar]
- 34. Rozen S., Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365–386 In Krawetz S., Misener S. (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 35. Schechner V., et al. 2009. Evaluation of PCR-based testing for surveillance of KPC-producing carbapenem-resistant members of the Enterobacteriaceae family. J. Clin. Microbiol. 47:3261–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith Moland E., et al. 2003. Plasmid-mediated, carbapenem-hydrolysing beta-lactamase, KPC-2, in Klebsiella pneumoniae isolates. J. Antimicrob. Chemother. 51:711–714 [DOI] [PubMed] [Google Scholar]
- 37. Tenover F. C., et al. 2006. Carbapenem resistance in Klebsiella pneumoniae not detected by automated susceptibility testing. Emerg. Infect. Dis. 12:1209–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tibbetts R., Frye J. G., Marschall J., Warren D., Dunne W. 2008. Detection of KPC-2 in a clinical isolate of Proteus mirabilis and first reported description of carbapenemase resistance caused by a KPC beta-lactamase in P. mirabilis. J. Clin. Microbiol. 46:3080–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Toth A., et al. 2010. Emergence of a colistin-resistant KPC-2-producing Klebsiella pneumoniae ST258 clone in Hungary. Eur. J. Clin. Microbiol. Infect. Dis. 29:765–769 [DOI] [PubMed] [Google Scholar]
- 40. Tyagi S., Bratu D. P., Kramer F. R. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49–53 [DOI] [PubMed] [Google Scholar]
- 41. Tyagi S., Kramer F. R. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303–308 [DOI] [PubMed] [Google Scholar]
- 42. Vet J. A., Marras S. A. 2005. Design and optimization of molecular beacon real-time polymerase chain reaction assays. Methods Mol. Biol. 288:273–290 [DOI] [PubMed] [Google Scholar]
- 43. Wendt C., et al. 2010. First outbreak of Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae in Germany. Eur. J. Clin. Microbiol. Infect. Dis. 29:563–570 [DOI] [PubMed] [Google Scholar]
- 44. Wolter D. J., et al. 2009. Phenotypic and enzymatic comparative analysis of the novel KPC variant KPC-5 and its evolutionary variants, KPC-2 and KPC-4. Antimicrob. Agents Chemother. 53:557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wolter D. J., et al. 2007. Detection of KPC carbapenem-hydrolyzing beta-lactamase in Pseudomonas aeruginosa from the Puerto Rico Medical Center Hospitals (PRMCHs), abstr. C2-1928. Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC [Google Scholar]
- 46. Woodford N., et al. 2004. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A beta-lactamase, KPC-3, in a New York Medical Center. Antimicrob. Agents Chemother. 48:4793–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yigit H., et al. 2008. Author's correction. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae Antimicrob. Agents Chemother. 52:809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yigit H., et al. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yigit H., et al. 2003. Carbapenem-resistant strain of Klebsiella oxytoca harboring carbapenem-hydrolyzing beta-lactamase KPC-2. Antimicrob. Agents Chemother. 47:3881–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang R., Yang L., Cai J. C., Zhou H. W., Chen G. X. 2008. High-level carbapenem resistance in a Citrobacter freundii clinical isolate is due to a combination of KPC-2 production and decreased porin expression. J. Med. Microbiol. 57:332–337 [DOI] [PubMed] [Google Scholar]