Abstract
We genotyped 102 Borrelia burgdorferi sensu lato strains isolated from ticks, animals, and patients in 11 provinces in China by PCR–restriction fragment length polymorphism (PCR-RFLP) amplification of 5S (rrf)-23S (rrl) rRNA gene spacer amplicons and multilocus sequence analysis (MLSA). The results showed that Borrelia garinii was the main genotype in China (65/102) and that it was distributed mainly in northern China. Borrelia afzelii was the second most frequently found species (22/102), and it was distributed in both northern and southern China. All Borrelia valaisiana strains were isolated from Guizhou Province. Additionally, one B. burgdorferi strain was isolated from Hunan Province. Our results show the diversity and wide distribution of B. burgdorferi sensu lato in China.
Lyme disease is a multisystemic enzootic disease that is common in all temperate regions of the Northern Hemisphere. Its etiologic agent, Borrelia burgdorferi sensu lato, was originally thought to be homogeneous. However, many studies have demonstrated that B. burgdorferi sensu lato is phenotypically and genotypically heterogeneous (13, 14, 21). To date, at least 14 species of B. burgdorferi sensu lato have been described: B. burgdorferi sensu stricto, B. garinii, B. afzelii, B. japonica, B. valaisiana, B. lusitaniae, B. andersonii, B. tanukii, B. turdi, B. bissettii, B. sinica, B. spielmanii, B. californiensis, and B. carolinensis sp. nov. (2, 5, 8, 10, 11, 20).
In China, several B. burgdorferi sensu lato genotyping studies have been conducted (1, 16, 19, 22) (see Table S2 in the supplemental material), but there has been no analysis of Chinese strains in different provinces. We have been conducting an epidemiological investigation of Lyme disease in China since 1986; we have collected rodents and ticks as well as patient samples from 30 provinces in China and have isolated more than 100 Borrelia strains (18, 22). To understand the diversity and distribution of Borrelia burgdorferi species in China, 102 strains from 11 provinces were genotyped by means of PCR–restriction fragment length polymorphism (PCR-RFLP) of 5S-23S ribosomal rRNA gene spacer amplicons and multilocus sequence analysis (MLSA).
MATERIALS AND METHODS
Identification of ticks.
Ticks were observed under a microscope, and identification of ticks was based mainly on physical characteristics, especially the mouthpart and the shield. This work was done by Guilan Dou, who was in the Department of Vector Biology and Control, National Institute for Communicable Disease Control and Prevention, China CDC.
Collection of strains.
The origins of the isolates used in this study are detailed in Table 1. In total, 102 strains were obtained from 11 provinces: 33 strains from Jilin, 10 strains from Heilongjiang, 2 strains from Liaoning, 11 strains from Inner Mongolia (Neimeng), 15 strains from the Xinjiang Uygur Autonomous Region, 3 strains from Beijing, 1 strain from Hebei, 1 strain from Hunan, 6 strains from Chongqing, 3 strains from Guangdong, and 17 strains from Guizhou. Reference strains B31 (B. burgdorferi sensu stricto), 20047 (B. garinii), and VS461 (B. afzelii) were provided by R. C. Johnson and T. J. Quan. All isolates were cultured in Barbour-Stoenner-Kelly II (BSKII) medium at 33°C for 4 to 7 days, after which spirochetes were harvested by centrifugation at 12,000 × g for 30 min. The pellet was washed twice in 0.01 M phosphate-buffered saline (PBS; pH 7.4) and was finally resuspended in 1 ml of sterile PBS. The preparations were stored at −20°C until use.
Table 1.
PCR-RFLP analysis of the 5S-23S rRNA intergenic spacers of 102 Chinese strains
| Species | No. of isolates |
RFLP result for 5S-23S rRNA intergenic spacera |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients | Ticks |
Rodents |
Total | ||||||||||
| Ixodes persulcatus | Haemaphysalis longinus | Haemaphysalis bispinosa | Ixodes granulatus | Caprolagus sinensis | Apodemus agrarius | Rattus fulvescens | Rattus edwardsi | Rattus norvegicus | MseI | DraI | |||
| B. burgdorferi | 1 | 1 | A | A′ | |||||||||
| B. garinii | 40 | 1 | 41 | B | B′ | ||||||||
| 3 | 23 | 26 | C | C′ | |||||||||
| B. afzelii | 2 | 6 | 2 | 5 | 1 | 1 | 17 | D | D′ | ||||
| B. valaisiana | 11 | 5 | 1 | 17 | F | F′ | |||||||
| Total | 5 | 69 | 2 | 5 | 11 | 1 | 6 | 1 | 1 | 1 | 102 | ||
DNA extraction.
DNA was extracted by a modification of a method described previously (12). After a 20-min incubation at 37°C, 80 μl of 10% sodium dodecyl sulfate (SDS) was added to the preparation (10 μg in 1 ml of PBS), and the preparation was heated at 65°C for 10 min. Next, 20 μl of RNase (10 mg/ml) was added, and the solution was incubated at 37° for 2 h. Following the addition of 10 μl of proteinase K, the preparation was incubated at 37°C for 2 h. Next, the DNA was extracted twice with equal volumes of phenol and once with an equal volume of chloroform. The DNA was precipitated by the addition of 2 volumes of absolute ethanol. The precipitated DNA was washed with 70% ethanol and was resuspended in Tris-EDTA (TE; pH 8.0).
PCR-RFLP.
The 5S-23S rRNA intergenic spacer was amplified by PCR according to a previous report (7). Primers (primer 1, 5′-GCG GCA GAG TAG GTT ATT-3′; primer 2, 5′-CTA GGC ATT CAC CAT AGA CT-3′) were designed with Oligo 5.0 and were synthesized by Boya Bio Company (Shanghai, China). Each reaction mixture consisted of 0.5 μl of each primer (20 μM), 1 U of Taq DNA polymerase, 250 μM each deoxynucleoside triphosphate (dNTP), 50 mM Tris-HCl (pH 8.3), 40 mM KCl, 1.5 mM MgCl2, and 1 μl of temperate DNA in a final volume of 50 μl. PCR was performed as follows: 1 min at 94°C; 35 cycles of 1 min at 94°C, 45 s at 52°C, and 45 s at 72°C; and a final extension step of 10 min at 72°C. The amplicons were analyzed in 2% agarose gels containing 0.5 μg/ml ethidium bromide. DNA bands were visualized under UV light.
The resulting amplicons were digested with the endonucleases MseI (New England Biolabs) and DraI (Promega) according to the manufacturers' protocols. A total of 10 μl of the PCR mixture containing the amplified fragment was digested with 5 U of either MseI or DraI in a total volume of 20 μl. The restriction fragments were subjected to electrophoresis on 16% (0.1 M Tris-borate-EDTA [TBE]) nondenaturing polyacrylamide gels. Next, the gel was fixed in 100 ml of a solution containing 10% ethanol and 0.5% acetic acid for 45 min and was stained with 200 ml of 0.01 M AgNO3. After 2 h, color was developed with 150 ml of 0.75 M NaOH and 0.1 M methanol, and the reaction was stopped with 5% acetic acid.
MLSA.
Seven loci—rrs, hbb, groEL, recA, fla, ospA, and the rrf-rrl intergenic spacer—were used for MLSA and were amplified under conditions described previously (10). All loci were amplified by a single PCR. The reaction was performed in a final volume of 50 μl, comprising 2× Taq PCR Master Mix (Tiangen Biotech, Beijing, China), 50 μM each primer of a primer pair, and 1 μl of temperate DNA. PCR was performed as follows: 1 min at 94°C; 35 cycles of 1 min at 94°C, 45 s at 52°C, and 45 s at 72°C; and a final extension step of 5 min at 72°C. The products were sequenced by the BGI Company.
Sequence analysis.
The CLUSTAL_X (17) algorithm was used for sequence alignments, and MEGA4 software was used for phylogenetic analyses of both individual and concatenated sequences. Distances were calculated using the neighbor-joining method.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been deposited in GenBank under the following accession numbers: rrs, HQ433589 to HQ433694; fla, HQ433695 to HQ433800; groEL, HQ433801 to HQ433906; hbb, HQ433907 to HQ434012; ospA, HQ434013 to HQ434118; recA, HQ434119 to HQ434224; rrf-rr1, HQ434225 to HQ434330.
RESULTS
PCR-RFLP analysis.
The 5S-23S rRNA intergenic spacer fragment was amplified from all isolates and was digested by using both MseI and DraI. The results showed that the pattern of 1 strain isolated from a Caprolagus sinensis bladder in Hunan province corresponded to that of B. burgdorferi sensu stricto, and the pattern of 17 isolates from Guizhou province corresponded to that of B. valaisiana. The other patterns obtained were similar to those of B. garinii and B. afzelii (Table 1).
MLSA.
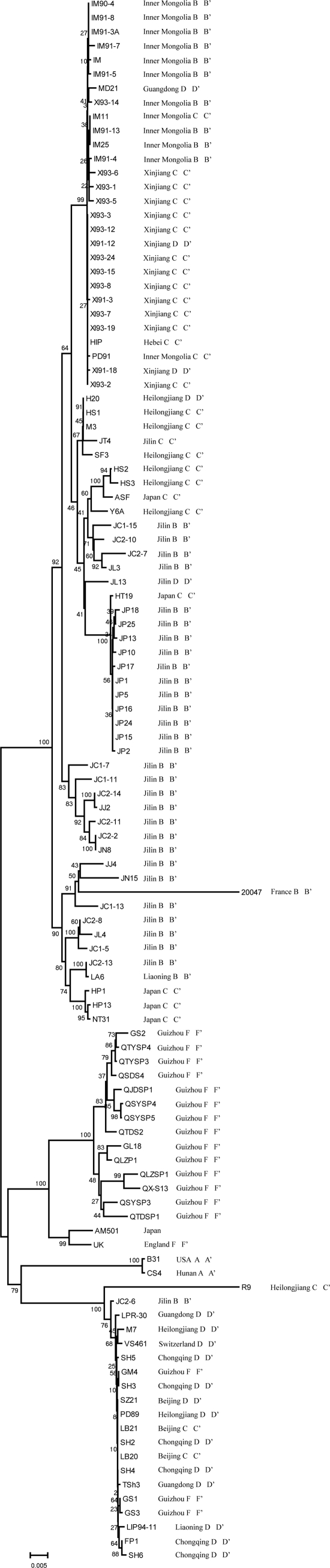
In the phylogenetic tree for the concatenated sequences of the seven loci, the isolates were classified into four clusters: B. burgdorferi sensu stricto, B. garinii, B. afzelii, and B. valaisiana. However, the MLSA results differed slightly from those of PCR-RFLP. Two isolates (LB20 and LB21) from Beijing were distantly related to isolates from Chongqing, which belonged to the B. afzelii cluster. Isolate JL13 from Jilin was classified in the B. garinii cluster by MLSA and in the B. afzelii cluster by PCR-RFLP; isolates GM4, GS1, and GS2 from Guizhou were classified in the B. afzelii cluster by MLSA and the B. valaisiana cluster by PCR-RFLP (Fig. 1).
Fig. 1.
Phylogenetic tree for the concatenated sequences of the seven loci.
Distribution of B. burgdorferi sensu lato in China.
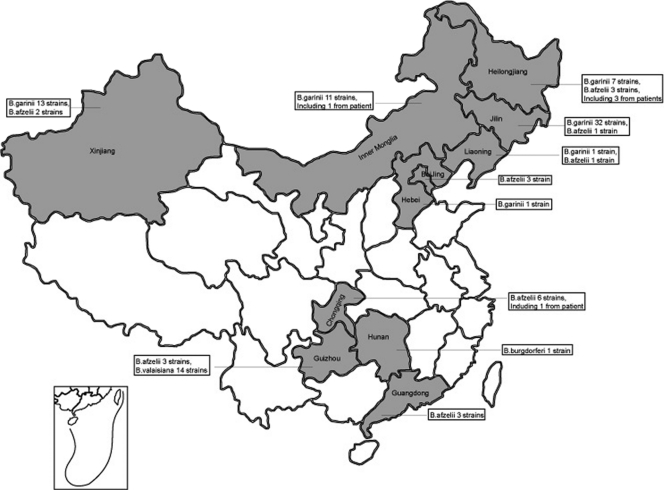
Our results showed that B. burgdorferi was isolated only from Hunan Province and that B. garinii was distributed in Jilin, Heilongjiang, Liaoning, Inner Mongolia, Xinjiang, and Hebei. B. afzelii was distributed in Jilin, Heilongjiang, Liaoning, Xinjiang, Beijing, Chongqing, Guangdong, and Guizhou. B. valaisiana was distributed in Guizhou Province (Fig. 2).
Fig. 2.
Provincial distribution of B. burgdorferi sensu lato strains in China (inset: map of South China Sea).
DISCUSSION
A total of 102 Borrelia burgdorferi sensu lato strains were analyzed in this study. The distribution of these strains has geographic multiplicity, a diversity of origins, and representativeness of genospecies. Among the strains, five isolates were obtained from patients. PD91 (Inner Mongolia), Y6A (Heilongjiang), PD89 (Heilongjiang), and FP1 (Chongqing) were isolated from patient blood; R9 (Heilongjiang) was isolated from the cerebrospinal fluid (CSF) of a patient.
Four genospecies—B. burgdorferi sensu stricto, B. garinii, B. afzelii, and B. valaisiana—were isolated from 11 provinces in China. Amost all of the B. garinii strains were isolated from Ixodes persulcatus in northern China. B. afzelii strains were isolated from I. persulcatus in northern China and Haemaphysalis bispinosa in southern China. The findings indicate that I. persulcatus is the main vector of Lyme disease in northern China and that H. bispinosa may serve as one of the vectors of Lyme disease in southern China. These results are in agreement with those reported previously (16, 18, 22). According to our research and previous reports, there are two foci in China: the northern China focus and the southern China focus. The northern China focus is characterized by I. persulcatus as its main vector. It is also characterized by the presence of only two Borrelia species: B. afzelii and B. garinii. However, the southern China focus is more complex. H. bispinosa and Ixodes granulatus may serve as the main vectors, and at least three Borrelia species are involved: B. afzelii, B. valaisiana, and B. sinica (5, 15, 18, 19, 22).
Analysis of the 17 isolates from Guizhou by MLSA revealed that 14 isolates were distantly related to B. valaisiana; these have been named Borrelia yangtze sp. nov. (1). In addition, we found three isolates (GM4, GS1, and GS2) that were more related to the isolates from Chongqing and Guangdong, which were classified as B. afzelii. B. afzelii was not previously found in Guizhou Province; these findings suggest that human cases may also be present in Guizhou.
One very interesting finding of our research was the isolation of B. burgdorferi from Caprolagus sinensis in Hunan Province. B. burgdorferi is present in both Europe and North America (4, 6, 9). However, this species has not been found in Japan or Korea until now (3). Conversely, isolates of B. burgdorferi have been isolated from several different rodents in Taiwan (12). The isolate from Hunan Province is the first B. burgdorferi strain in the mainland of China, but additional studies are necessary to determine whether other vertebrates harbor B. burgdorferi in China. The identification of its vector is also of crucial importance for determining whether infection of humans by this species can occur in China.
In conclusion, our research gave a first insight into the diversity and distribution of B. burgdorferi strains in different provinces of China. The results also provide evidence that measures for the control and prevention of Lyme disease in China are advisable.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the project “Network surveillance technology of pathogens” of the National Key Programme of Mega Infectious Diseases (2008ZX10004-008).
We thank Chen Chen and Haiyin Wang of the Bioinformatics Department, National Institute for Communicable Disease Control and Prevention, for help in submitting DNA sequences to GenBank.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 24 November 2010.
REFERENCES
- 1. Chu C. Y., et al. 2008. Novel genospecies of Borrelia burgdorferi sensu lato from rodents and ticks in southwestern China. J. Clin. Microbiol. 46:3130–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fukunaga M., Okada K., Nakao M., Konishi T., Sato Y. 1996. Phylogenetic analysis of Borrelia species based on flagellin gene sequences and its application for molecular typing of Lyme disease borreliae. Int. J. Syst. Bacteriol. 46:898–905 [DOI] [PubMed] [Google Scholar]
- 3. Lee S. H., et al. 2000. Characterization of Borrelia burgdorferi strains isolated from Korea by 16S rDNA sequence analysis and PCR-RFLP analysis of rrf (5S)-rrl (23S) intergenic spacer amplicons. Int. J. Syst. Evol. Microbiol. 50(Pt. 2):857–863 [DOI] [PubMed] [Google Scholar]
- 4. Margos G., et al. 2008. MLST of housekeeping genes captures geographic population structure and suggests a European origin of Borrelia burgdorferi. Proc. Natl. Acad. Sci. U. S. A. 105:8730–8735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Masuzawa T., et al. 2001. Borrelia sinica sp. nov., a Lyme disease-related Borrelia species isolated in China. Int. J. Syst. Evol. Microbiol. 51:1817–1824 [DOI] [PubMed] [Google Scholar]
- 6. Piesman J. 2006. Strategies for reducing the risk of Lyme borreliosis in North America. Int. J. Med. Microbiol. 296(Suppl. 40):17–22 [DOI] [PubMed] [Google Scholar]
- 7. Postic D., Assous M. V., Grimont P. A., Baranton G. 1994. Diversity of Borrelia burgdorferi sensu lato evidenced by restriction fragment length polymorphism of rrf (5S)-rrl (23S) intergenic spacer amplicons. Int. J. Syst. Bacteriol. 44:743–752 [DOI] [PubMed] [Google Scholar]
- 8. Postic D., Garnier M., Baranton G. 2007. Multilocus sequence analysis of atypical Borrelia burgdorferi sensu lato isolates—description of Borrelia californiensis sp. nov., and genomospecies 1 and 2. Int. J. Med. Microbiol. 297:263–271 [DOI] [PubMed] [Google Scholar]
- 9. Postic D., et al. 1999. Common ancestry of Borrelia burgdorferi sensu lato strains from North America and Europe. J. Clin. Microbiol. 37:3010–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Richter D., et al. 2006. Delineation of Borrelia burgdorferi sensu lato species by multilocus sequence analysis and confirmation of the delineation of Borrelia spielmanii sp. nov. Int. J. Syst. Evol. Microbiol. 56:873–881 [DOI] [PubMed] [Google Scholar]
- 11. Rudenko N., Golovchenko M., Grubhoffer L., Oliver J. H., Jr 2009. Borrelia carolinensis sp. nov., a new (14th) member of the Borrelia burgdorferi sensu lato complex from the southeastern region of the United States. J. Clin. Microbiol. 47:134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shih C. M., Chang H. M., Chen S. L., Chao L. L. 1998. Genospecies identification and characterization of Lyme disease spirochetes of genospecies Borrelia burgdorferi sensu lato isolated from rodents in Taiwan. J. Clin. Microbiol. 36:3127–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Steere A. C. 1989. Lyme disease. N. Engl. J. Med. 321:586–596 [DOI] [PubMed] [Google Scholar]
- 14. Steere A. C., et al. 1977. Erythema chronicum migrans and Lyme arthritis. The enlarging clinical spectrum. Ann. Intern. Med. 86:685–698 [DOI] [PubMed] [Google Scholar]
- 15. Takada N., et al. 1998. Lyme disease spirochetes in ticks from northeastern China. J. Parasitol. 84:499–504 [PubMed] [Google Scholar]
- 16. Takada N., et al. 2001. Lyme disease Borrelia spp. in ticks and rodents from northwestern China. Appl. Environ. Microbiol. 67:5161–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wan K., Zhang Z., Dou G. 1998. Investigation on primary vectors of Borrelia burgdorferi in China. Zhonghua Liu Xing Bing Xue Za Zhi 19:263–266 (In Chinese.) [PubMed] [Google Scholar]
- 19. Wang D. M., et al. 2003. Study on ribotyping of Lyme borreliosis spirochete in Guizhou province. Zhonghua Liu Xing Bing Xue Za Zhi 24:1129–1131 (In Chinese.) [PubMed] [Google Scholar]
- 20. Wang G., van Dam A. P., Schwartz I., Dankert J. 1999. Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin. Microbiol. Rev. 12:633–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wilske B., et al. 1992. Antigenic variation and strain heterogeneity in Borrelia spp. Res. Microbiol. 143:583–596 [DOI] [PubMed] [Google Scholar]
- 22. Zhang Z. F., Wan K. L., Zhang J. S. 1997. Studies on epidemiology and etiology of Lyme disease in China. Zhonghua Liu Xing Bing Xue Za Zhi 18:8–11 (In Chinese.) [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.