Abstract
Mycobacterium abscessus, Mycobacterium bolletii, and Mycobacterium massiliense (Mycobacterium abscessus sensu lato) are closely related species that currently are identified by the sequencing of the rpoB gene. However, recent studies show that rpoB sequencing alone is insufficient to discriminate between these species, and some authors have questioned their current taxonomic classification. We studied here a large collection of M. abscessus (sensu lato) strains by partial rpoB sequencing (752 bp) and multilocus sequence analysis (MLSA). The final MLSA scheme developed was based on the partial sequences of eight housekeeping genes: argH, cya, glpK, gnd, murC, pgm, pta, and purH. The strains studied included the three type strains (M. abscessus CIP 104536T, M. massiliense CIP 108297T, and M. bolletii CIP 108541T) and 120 isolates recovered between 1997 and 2007 in France, Germany, Switzerland, and Brazil. The rpoB phylogenetic tree confirmed the existence of three main clusters, each comprising the type strain of one species. However, divergence values between the M. massiliense and M. bolletii clusters all were below 3% and between the M. abscessus and M. massiliense clusters were from 2.66 to 3.59%. The tree produced using the concatenated MLSA gene sequences (4,071 bp) also showed three main clusters, each comprising the type strain of one species. The M. abscessus cluster had a bootstrap value of 100% and was mostly compact. Bootstrap values for the M. massiliense and M. bolletii branches were much lower (71 and 61%, respectively), with the M. massiliense cluster having a fuzzy aspect. Mean (range) divergence values were 2.17% (1.13 to 2.58%) between the M. abscessus and M. massiliense clusters, 2.37% (1.5 to 2.85%) between the M. abscessus and M. bolletii clusters, and 2.28% (0.86 to 2.68%) between the M. massiliense and M. bolletii clusters. Adding the rpoB sequence to the MLSA-concatenated sequence (total sequence, 4,823 bp) had little effect on the clustering of strains. We found 10/120 (8.3%) isolates for which the concatenated MLSA gene sequence and rpoB sequence were discordant (e.g., M. massiliense MLSA sequence and M. abscessus rpoB sequence), suggesting the intergroup lateral transfers of rpoB. In conclusion, our study strongly supports the recent proposal that M. abscessus, M. massiliense, and M. bolletii should constitute a single species. Our findings also indicate that there has been a horizontal transfer of rpoB sequences between these subgroups, precluding the use of rpoB sequencing alone for the accurate identification of the two proposed M. abscessus subspecies.
Mycobacterium abscessus is a rapidly growing mycobacterium (RGM) that causes a wide spectrum of disease in humans, including chronic lung disease, skin and soft-tissue disease, and disseminated disease (12, 14, 53). M. abscessus lung disease mostly develops in subjects with underlying lung disorders (e.g., cystic fibrosis [CF] or prior mycobacterial infection) or Lady Windermere syndrome (10, 19, 29, 35, 40, 41, 45, 46). M. abscessus is also a leading cause of sporadic and epidemic cases of skin and soft-tissue RGM infections after surgery or following the use of contaminated syringes and needles (21, 22, 39). Several large outbreaks of skin and soft-tissue infection have been reported following the injection of adrenal cortex extract, mesotherapy, tattooing, and piercing (6, 8, 15, 24, 52, 58).
The Mycobacterium massiliense and Mycobacterium bolletii species were characterized in the early 2000s, and both are closely related to M. abscessus (100% identity of 16S rRNA sequences) (2, 5). Therefore, M. abscessus (now M. abscessus sensu lato) is comprised of three species: M. abscessus sensu stricto (for simplicity, M. abscessus sensu stricto here will be referred to as M. abscessus), M. massiliense, and M. bolletii. Although information about the pathogenic effects of M. massiliense and M. bolletii in humans still is scarce, several recent studies report that they cause a spectrum of diseases similar to those associated with M. abscessus. However, there may be some differences between the three species. Zelazny et al. have reported that M. massiliense is more frequently present in the respiratory tract of younger patients with preexisting lung disease than M. abscessus (59). Differences also have been reported in the susceptibility patterns of the three species (13, 14, 54, 55). For example, M. massiliense was reported to be susceptible to doxycycline, whereas M. abscessus and M. bolletii are not (5), although this finding has not been confirmed by others (32, 52).
M. massiliense and M. bolletii were characterized as new species distinct from M. abscessus on the basis of their rpoB sequences (>3% sequence divergence) (2, 5). Partial rpoB sequencing is now the gold standard for the molecular identification of the three species (3, 4, 16, 38, 48). However, recent studies have highlighted the inaccuracy of single-target sequencing, including rpoB sequencing, for distinguishing between M. abscessus, M. massiliense, and M. bolletii (18, 31, 36, 59). Zelazny et al. found that the partial sequencing of rpoB, hsp65, and secA led to inconsistent results in 7 of 42 clinical isolates; most of these seven isolates had an M. abscessus rpoB sequence and M. massiliense hsp65 and secA sequences, and they clustered with the M. massiliense type strain in repetitive sequence-based PCR (rep-PCR) and pulsed-field gel electrophoresis (PFGE) (59). We reported similar data in a study of a panel of 59 clinical isolates by the partial sequencing of rpoB, hsp65, and sodA (36). Target genes yielded discordant results in 15 isolates, which had interspecific composite patterns (e.g., isolates with a rpoB sequence 100% identical to the M. abscessus type sequence and an hsp65 sequence 100% identical to the M. massiliense type sequence). The identification of these isolates was substantially improved by the partial sequencing of five housekeeping gene sequences, indicating the value of a multilocus sequencing approach (26, 36).
A panel of M. abscessus, M. massiliense, and M. bolletii strains recently has been studied by biochemical tests, high-performance liquid chromatography (HPLC), drug susceptibility testing, PCR restriction enzyme analysis of the hsp65 gene (PRA-hsp65), rpoB and hsp65 gene sequencing, the restriction fragment length polymorphism (RFLP) analysis of the 16S rRNA gene, and DNA-DNA hybridization (34). The clinical isolates studied and the type strains could not be separated, and DNA-DNA hybridization showed more than 70% interstrain relatedness. The authors thus proposed a revision of the taxonomic status of M. abscessus, M. massiliense, and M. bolletii, whereby the three species are in fact a single species (M. abscessus) and two subspecies (M. abscessus subsp. abscessus and M. abscessus subsp. massiliense) (34).
Multilocus sequence analysis (MLSA) is a phylogenetic analysis of multiple internal fragments of genes that are ubiquitous to the studied taxon, present as a single copy within the genome, and are not subject to selective pressure (27). MLSA defines an isolate by the sequences obtained from the internal fragments of several housekeeping genes. The usual approach in bacterial taxonomy is to concatenate the sequences of several (typically six to eight) housekeeping genes. The concatenated sequences then are used to assess clustering patterns among large numbers of strains within a genus or part of a genus (17, 23, 27, 28). This approach has been used successfully to delineate microbial species (i.e., well-resolved clusters) within various taxonomic groups, including groups of highly recombinant bacteria, like Neisseria spp. (37). Conversely, MLSA also allows the assignment of unknown strains to species clusters, and this can be performed via the internet, opening the way to electronic taxonomy (11).
In the present study, we developed an MLSA scheme and applied it to a large collection of M. abscessus sensu lato strains. The data obtained with this approach were compared to those obtained by rpoB sequencing to (i) help clarify the taxonomic status of M. abscessus, M. massiliense, and M. bolletii and (ii) evaluate the accuracy of rpoB as a molecular identification target.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
We studied 120 isolates of M. abscessus sensu lato recovered from France (n = 97), Germany (n = 6), Switzerland (n = 7), and Brazil (n = 10) between 1997 and 2007. One hundred nineteen of the isolates were from clinical samples, and one was from the environment (isolated from a sewer in Brazil). The sample origin was known for 90 of the clinical isolates: 79 (87.8%) were from respiratory samples (57 CF subjects, 22 non-CF subjects) and 11 (12.2%) were from other samples (9 from skin and soft tissue, 1 from pericardium, and 1 from a hip prosthesis). All of the clinical isolates were from unrelated cases. One clinical isolate was from a nonsporadic case, having also been recovered during a recent outbreak of M. massiliense skin and soft-tissue disease after a laparoscopic procedure in Brazil (15, 33). The type strains M. abscessus CIP 104536T (ATCC 19977T), M. massiliense (CIP 108297T), and M. bolletii (CIP 108541T) also were included in the strain collection. Bacterial strains were stored at −70°C using cryopreservation beads and were grown on sheep blood agar at 37°C for 4 days prior to use.
rpoB sequencing and rpoB-based identification.
Mycobacterial DNA was extracted using Tris-EDTA, lysozyme, and proteinase K as described previously (36). A 940-bp fragment of the rpoB gene was amplified by PCR using AmpliTaq gold polymerase (Applied Biosystems, Courtaboeuf, France) with the primers MYCOF1 and MYCOR2 (Table 1). Dideoxy sequencing was carried out on both strands using a BigDye Terminator cycle sequencing kit (Applied Biosystems) with the same primers. Sequencing products were purified by gel filtration (Bio-gel P100; Bio-Rad, Marnes-la-Coquette, France) and were run on a 3700 DNA analyzer (Applied Biosystems). The rpoB sequences trimmed to 752 bp (3) were compared to the corresponding rpoB sequences from the reference type strains (http://www.ncbi.nlm.nih.gov/).
Table 1.
Primers used for PCR and sequencing
| Gene | Primer name | Primer sequence | Amplified fragment (bp) | Tm (°C) | Reference or source |
|---|---|---|---|---|---|
| argH | ARGHF | 5′-GACGAGGGCGACAGCTTC-3′ | 629 | 60 | 36 |
| ARGHSR1 | 5′-GTGCGCGAGCAGATGATG-3′ | 58 | |||
| cya | ACF | 5′-GTGAAGCGGGCCAAGAAG-3′ | 647 | 58 | 36 |
| ACSR1 | 5′-AACTGGGAGGCCAGGAGC-3′ | 60 | |||
| gdhA | GDHAF | 5′-GTCAGTGCCCCGATCGCT-3′ | 582 | 60 | This study |
| GDHASR1 | 5′-GGCTCTCGGAGTACGTCGA-3′ | 60 | |||
| glpK | GLPKSF1 | 5′-AATCTCACCGGCGGTGTC-3′ | 609 | 58 | 36 |
| GLPKSFR2 | 5′-GGACAGACCCACGATGGC-3′ | 60 | |||
| gnd | GNDF | 5′-GTGACGTCGGAGTGGTTGG-3′ | 634 | 62 | 36 |
| GNDSR1 | 5′-CTTCGCCTCAGGTCAGCTC-3′ | 62 | |||
| murC | MURCSF1 | 5′-CGGACGAAAGCGACGGCT-3′ | 607 | 60 | 36 |
| MURCSR2 | 5′-CCAAAACCCTGCTGAGCC-3′ | 58 | |||
| pgm | PGMSF1 | 5′-CCATTTGAACCCGACCGG-3′ | 596 | 60 | This study |
| PGMSR2 | 5′-GTGCCAACGAGATCCTGCG-3′ | 66 | |||
| pknA | PKNAF | 5′-CAGGTGGACCTCGGACATG-3′ | 493 | 62 | This study |
| PKNASR1 | 5′-AACCAGGCGCCCACCATC-3′ | 60 | |||
| pta | PTASF1 | 5′-GATCGGGCGTCATGCCCT-3′ | 720 | 60 | This study |
| PTASR2 | 5′-ACGAGGCACTGCTCTCCC-3′ | 66 | |||
| purH | PURHSF1 | 5′-CGGAGGCTTCACCCTGGA-3′ | 634 | 64 | This study |
| PURHSR2 | 5′-CAGGCCACCGCTGATCTG-3′ | 60 | |||
| rpoB | MYCOF1 | 5′-TCCGATGAGGTGCTGGCAGA-3′ | 940 | 68 | This study |
| MYCOR2 | 5′-ACTTGATGGTCAACAGCTCC-3′ | 68 |
PCR amplification and sequencing of 10 housekeeping genes in addition to rpoB.
Fragments from 10 housekeeping genes were amplified using the sets of primers shown in Table 1: argH (argininosuccinate lyase), cya (adenylate cyclase), gdhA (glutamate dehydrogenase), glpK (glycerol kinase), gnd (6-phosphogluconate deshydrogenase), murC (UDP N-acetylmuramate-l-Ala ligase), pgm (phosphoglucomutase), pknA (serine/threonine protein kinase), pta (phosphate acetyltransferase), and purH (phoshoribosylaminoimiazolcarboxylase ATPase subunit). The genes gdhA, glpK, murC, pknA, pta, and purH have been widely used in multilocus sequence typing (MLST) schemes developed for Gram-positive bacteria (e.g., Streptococcus spp., Enterococcus spp., Staphylococcus spp., and Bacillus spp.) (7, 37, 49). The gene products for argH, cya, gnd, and pgm have been used previously in the multilocus enzyme electrophoresis (MLEE) analysis of mycobacteria (20, 56, 57, 60). Amplification was performed using 25 μl of ReddyMix PCR master mix (Thermo Fisher Scientific Inc.) and 1 μl of each primer (10 pmol). The dideoxy sequencing of the amplified gene fragments was carried out on both strands with the Big Dye Terminator cycle sequencing kit (Applied Biosystems), using the same primers as those for amplification. Sequencing products were purified and analyzed with an ABI 3700 DNA analyzer as described above. The sequences were aligned and trimmed to defined start and end positions using BioEdit version 7.0.5.3 (25). Gene sequences from the three type strains were used as reference species sequences. Reference M. abscessus sequences of the 10 housekeeping genes were obtained from the whole-genome sequence of M. abscessus CIP 104536T (http://www.ncbi.nlm.nih.gov) (accession number NC_010397) (43). We recently determined the sequences of the argH, cya, glpK, gnd, and murC genes in the reference strains M. bolletii CIP 108541T and M. massiliense CIP 108297T (36).
Phylogenetic analyses.
Unrooted individual gene trees and trees obtained using concatenated sequences were generated using the neighbor-joining method with 1,000 bootstrap replications. Trees were drawn to scale, with branch lengths representing the inferred evolutionary distances. The evolutionary distances were computed using the maximum composite likelihood method (51), and the data units were the number of base substitutions per site. Codon positions were in frame, and there was a total of 4,071 bp in the final data set. The neighbor-joining method and MEGA 4 software were used for the phylogenetic analysis of the sequence data (50).
Nucleotide sequence accession numbers.
Sequences of the pgm, pta, and purH gene fragments from the reference strains M. bolletii CIP 108541T and M. massiliense CIP 108297T were determined in this study and submitted to the National Center for Biotechnology Information (NCBI) website (accession numbers HM371394, HM371395, and HM371396 for M. bolletii and HM371391, HM371392, and HM371393, for M. massiliense).
RESULTS
Selection of eight housekeeping genes eligible for MLSA.
Ten sets of primers (Table 1) were designed to amplify and sequence internal fragments of 10 housekeeping genes on a first panel, including 10 clinical isolates and the three M. abscessus sensu lato type strains. Fragments of the expected size were amplified from 100% of the strains for eight genes: argH, cya, glpK, gnd, murC, pgm, pta, and purH. The gdhA and pknA genes were eliminated from the subsequent MLSA study: in several strains, the amplification of gdhA did not yield fragments of the expected size, and the sequencing of the amplification products showed phage sequences; for pknA, we repeatedly obtained nonspecific amplification products even with the use of different primer pairs.
The eight selected housekeeping genes (argH, cya, glpK, gnd, murC, pgm, pta, and purH) are scattered throughout the genome of M. abscessus CIP 104536T and are at least 76 kb away from each other (42).
Polymorphism of the genes included in the MLSA scheme.
Internal fragments from the eight housekeeping genes selected for MLSA were amplified, and their nucleotide sequences were determined for the 120 isolates and the three type strains. Fragment sizes varied from 480 to 549 bp, with G+C contents of between 62.4 and 68.2% (Table 2). Polymorphic sites for each gene fragment were least frequent in pta (n = 24) and most frequent in argH (n = 43). The glpK and murC fragments had the lowest number of alleles (n = 16), and the cya and pgm fragments had the highest number of alleles (n = 25). All differences between alleles were due to point mutations; neither deletions nor insertions were observed. The ratio between homologous and nonhomologous substitutions was between 0.0029 (cya) and 0.1971 (pgm) with a mean of 0.0769; thus, most of the mutations were silent and did not generate nucleotide substitutions (Table 2). The maximum sequence divergence between the 123 strains was greatest for argH (5.42%) and lowest for pgm (2.02%); divergence exceeded 3% for four genes: argH, cya, gnd, and murC.
Table 2.
Genes studiedc
| Gene(s) | Size of analyzed fragment (bp) | %G+C | No. of polymorphic sites | No. of alleles | dN/dS | Maximum % of nt divergencea | % of nt divergence between type strain sequencesb |
||
|---|---|---|---|---|---|---|---|---|---|
| MabsT versus MmasT | MabsT versus MbolT | MbolT versus MmasT | |||||||
| argH | 480 | 66.8 | 40 | 19 | 0.1529 | 5.47 | 4.58 | 3.54 | 2.92 |
| cya | 510 | 68.2 | 31 | 25 | 0.0029 | 3.62 | 1.96 | 1.96 | 2.75 |
| glpK | 534 | 62.7 | 25 | 16 | 0.0380 | 2.48 | 1.69 | 1.31 | 0.94 |
| gnd | 480 | 65.2 | 33 | 20 | 0.0307 | 4.73 | 2.29 | 4.58 | 3.54 |
| murC | 537 | 68.6 | 39 | 16 | 0.1259 | 4.44 | 2.05 | 3.54 | 3.35 |
| pgm | 495 | 62.2 | 29 | 25 | 0.1900 | 2.02 | 1.41 | 1.41 | 0.81 |
| pta | 486 | 66.1 | 20 | 20 | 0.0110 | 2.74 | 1.85 | 1.65 | 2.26 |
| purH | 549 | 65.6 | 31 | 21 | 0.0502 | 2.98 | 1.82 | 2.00 | 0.18 |
| All MLSA genes | 4,071 | 65.7 | 248 | 79 | ND | 2.85 | 2.19 | 2.43 | 2.01 |
| rpoB | 752 | 65.6 | 43 | 22 | 0.0827 | 4.79 | 3.32 | 4.12 | 1.46 |
Maximum nucleotide divergence found between the 123 strains studied.
M. abscessus CIP 104536T, M. massiliense CIP 108297T, and M. bolletii CIP 108541T.
ND, not done; dN/dS, ratio of the rate of nonsynonymous substitutions (dN) to the rate of synonymous substitutions (dS); MabsT, M. abscessus type strain sequence; MbolT, M. bolletii type strain sequence; MmasT, M. massiliense type strain sequence.
rpoB tree and polymorphism of rpoB sequences.
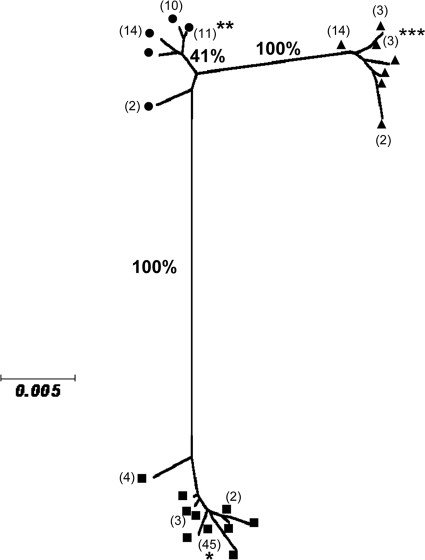
The tree built from the 123 rpoB sequences showed three distinct clusters, each comprising the type strain of one species (Fig. 1). According to Adékambi et al. (2), these three clusters could be equated with the species clusters M. abscessus (59 isolates), M. bolletii (24 isolates), and M. massiliense (37 isolates). The mean sequence divergence within each cluster was extremely low, ranging from 0.36 to 0.41%. The mean divergence between the M. abscessus and M. bolletii clusters was 4.38%, with all values exceeding 3% (extremes were 3.59 to 4.65%), divergence between the M. abscessus and M. massiliense clusters was 3.40%, with some values below 3% (extremes were 2.66 to 3.59%), and that between M. massiliense and M. bolletii was only 1.68%, and all values were below 3% (extremes were 1.33 to 2.13%). The divergence values between type strain sequences were consistent with these differences: 4.12% between M. abscessus CIP 104536T and M. massiliense CIP 108297T, 3.32% between M. abscessus CIP 104536T and M. bolletii CIP 108541T, and 1.46% between M. massiliense CIP 108297T and M. bolletii CIP 108541T (Table 2).
Fig. 1.
Tree constructed from partial rpoB gene sequences. The tree for all studied strains (n = 123) was generated using the neighbor-joining method. Bootstrap support values (%) are indicated for each node. Species assignment of clinical isolates are according to the criteria in Adékambi et al. (2): ■, M. abscessus; •, M. massiliense; ▴, M. bolletii; numbers in parentheses are the numbers of isolates if there are two or more. Type strains: *, M. abscessus CIP 104536T; **, M. massiliense CIP 108297T; ***, M. bolletii CIP 108541T.
Single-gene trees obtained with each of the MLSA genes.
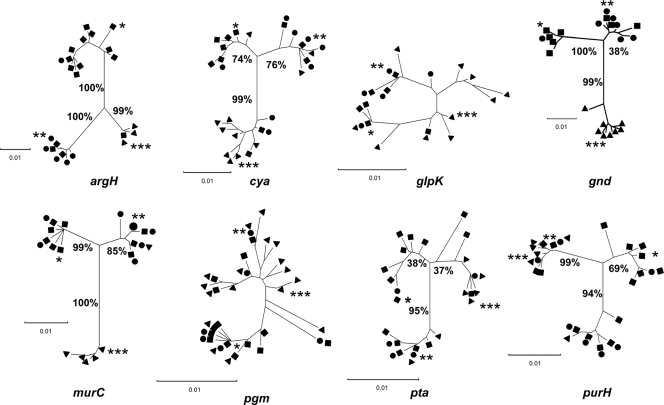
A single-gene tree was built from the sequences for each of the eight genes included in the final MLSA scheme. The trees built using argH, cya, gnd, murC, pta, and purH sequences showed three main branches, each carrying the type strain of one species except for the purH tree (M. massiliense and M. bolletii type strain sequences on the same branch) (Fig. 2). Bootstrap values for the argH and murC genes were >80%. However, there was no tree in which each of the three clusters was made up of strains assigned to a single species by rpoB sequencing (Fig. 2). The resolution obtained with the glpK and pgm sequences was very poor, and this was consistent with the very low polymorphism in these genes (as described above).
Fig. 2.
Trees constructed from the sequences of the eight individual genes included in the final MLSA scheme. The trees for all studied strains (n = 123) were generated using the neighbor-joining method. Bootstrap support values (%) at each of the nodes are indicated only for trees showing well-defined clusters. Species assignment of clinical isolates according to the criteria of Adékambi et al. (3): ■, M. abscessus; •, M. massiliense; ▴, M. bolletii. Type strains:*, M. abscessus CIP 104536T; **, M. massiliense CIP 108297T; ***, M. bolletii CIP 108541T.
The divergence between type strain sequences was highest with argH and gnd and lowest with glpK and pgm. With the argH type sequences, divergence was 4.58% between M. abscessus and M. massiliense and 3.54% between M. abscessus and M. bolletii (3.54%), but it was only 2.92% between M. massiliense and M. bolletii. With the gnd type sequences, divergences were 4.58% between M. abscessus and M. bolletii, 3.54% between M. massiliense and M. bolletii, and only 2.29% between M. abscessus and M. massiliense (Table 2).
Trees obtained with concatenated sequences of the eight MLSA genes.
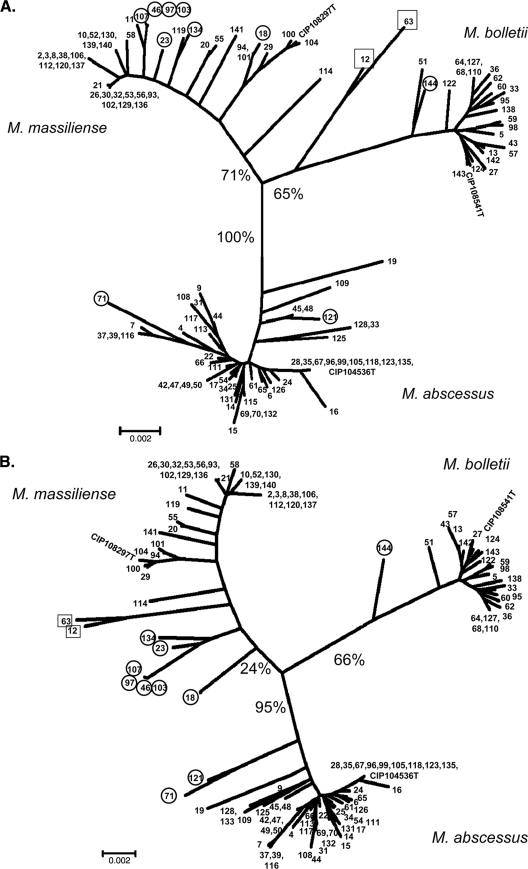
Figure 3A shows the tree obtained for the 123 strains by concatenating the sequences of the eight housekeeping gene fragments (4,071bp). This tree shows three principle clusters, each containing the sequence of only one of the three type strains; it contains, as expected, the M. abscessus, M. massiliense, and M. bolletii clusters. Apart from a few exceptions, the M. abscessus cluster is compact, with a bootstrap value of 100%. The respective bootstrap values for the M. massiliense and M. bolletii branches are much lower at 71 and 65%, respectively. The M. massiliense cluster is more dispersed, with some sequences situated close to the other clusters. One of these sequences is from the type strain CIP 108297T, which is not closely related to most of the other sequences in the cluster. The M. bolletii cluster is more compact, but some of its sequences are close to the main branch point.
Fig. 3.
Trees constructed from concatenated sequences. (A) Concatenated MLSA sequences. (B) Concatenated MLSA + rpoB sequences. The trees for all studied strains (n = 123) were generated by using the neighbor-joining method. Bootstrap support values (%) are indicated for each node. Each isolate is indicated by its number in our collection. CIP type strains also are indicated. Boxes indicate isolates with discordant rpoB-based identification (see Table 3 for further details). Note that isolates 12 and 63 are located on the M. bolletii branch of the MLSA tree (A) and on the M. massiliense branch of the MLSA + rpoB tree (B).
The mean nucleotide divergence values were 0.45% (range, 0 to 1.65%) within the M. abscessus cluster, 0.72% (0 to 1.25%) within the M. massiliense cluster, and 0.64% (0 to 2.06%) within the M. bolletii cluster. The mean divergence values between clusters were 2.17% (range, 1.13 to 2.58%) between the M. abscessus and M. massiliense clusters, 2.37% (1.5 to 2.85%) between the M. abscessus and M. bolletii clusters, and 2.28% (0.86 to 2.68%) between the M. massiliense and M. bolletii clusters. Divergence values for the MLSA-concatenated sequences from type strains were 2.19% between M. abscessus and M. massiliense, 2.43% between M. abscessus and M. bolletii, and 2.01% between M. massiliense and M. bolletii.
Effect of adding the rpoB sequence to the final MLSA scheme.
We studied the effect of adding the rpoB sequence to the concatenated sequences of the eight MLSA genes. The tree obtained (Fig. 3B, MLSA + rpoB tree) was very similar to the tree constructed from the MLSA gene sequences only (MLSA tree). However, the bootstrap value for the M. massiliense branch was even lower (24 versus 71%). With a few exceptions (see isolates 12 and 63), the distribution of isolates into the three groups was very similar in the two trees (Fig. 3A and B).
Discrepancies between MLSA data and rpoB-based identification.
We compared the MLSA and rpoB sequences from each of the 120 isolates using the type strain sequences for reference. The results of this analysis were consistent for 110 isolates and inconsistent for 10 isolates (Table 3). The most frequent inconsistency (n = 7; isolates 18, 23, 46, 97, 103, 107, and 134) was an MSLA sequence with 99.14 to 99.75% identity to the M. massiliense type strain (versus ≤98.03% identity to the M. abscessus and M. bolletii type strains) and an rpoB sequence similar to that of the M. abscessus type strain. Other inconsistencies were an MLSA sequence similar to that of the M. bolletii type strain, an rpoB sequence similar to that of the M. abscessus type strain (isolate 144), an MLSA sequence similar to that of the M. abscessus type strain, and an rpoB sequence similar to that of the M. massiliense type strain (isolates 71 and 121). Four of the isolates with an MLSA sequence similar to that of M. massiliense and an rpoB sequence similar to that of M. abscessus clustered together on the M. massiliense branch of the MLSA tree (Fig. 3, isolates 46, 97, 103, and 107). These four isolates shared identical rpoB sequences.
Table 3.
Isolates with discordant concatenated MLSA sequence and rpoB sequencec
| Isolate no. | % of nt identity with type strain sequence |
|||||
|---|---|---|---|---|---|---|
| MLSA sequencea |
rpoB sequence |
|||||
| MabsT | MbolT | MmasT | MabsT | MbolT | MmasT | |
| 18 | 98.03 | 97.99 | 99.75 | 100 | 95.88 | 96.68 |
| 23 | 97.89 | 97.62 | 99.14 | 100 | 95.88 | 96.68 |
| 46b | 97.69 | 97.86 | 99.19 | 99.34 | 96.28 | 97.34 |
| 97b | 97.69 | 97.86 | 99.19 | 99.34 | 96.28 | 97.34 |
| 103b | 97.69 | 97.86 | 99.19 | 99.34 | 96.28 | 97.34 |
| 107b | 97.67 | 97.84 | 99.16 | 99.34 | 96.28 | 97.34 |
| 134 | 97.84 | 97.57 | 99.34 | 99.73 | 96.14 | 96.81 |
| 144 | 97.96 | 99.31 | 98.01 | 99.87 | 96.01 | 96.68 |
| 71 | 98.85 | 97.89 | 98.18 | 96.68 | 98.54 | 99.73 |
| 121 | 99.04 | 97.35 | 97.94 | 96.68 | 98.54 | 99.73 |
Concatenated MLSA gene sequences (4,071 bp).
Also see Fig. 3.
MabsT, M. abscessus type strain sequence; MbolT, M. bolletii type strain sequence; MmasT, M. massiliense type strain sequence. Boldface numbers indicate discordant species identification by either MLSA or rpoB analysis for a given isolate.
DISCUSSION
Both the molecular identification and the taxonomy of M. abscessus, M. massiliense, and M. bolletii currently are based upon the partial sequencing of rpoB (2, 3, 16). However, several studies recently have questioned the effectiveness of sequencing only the rpoB gene for the molecular identification of these species (36, 59). More recently, doubts have been raised about the taxonomic classification of M. massiliense and M. bolletii (34). For the first time for this group of species, we used an MLSA scheme with the aim of resolving these issues. We compared the results obtained from using this scheme to those obtained from the partial sequencing of rpoB.
The study of our collection of isolates with the chosen MLSA scheme (eight housekeeping gene sequences, 4,071 bp) clearly shows the existence of three principal groups, with each containing the type strain of one of the three species (M. abscessus CIP 104536T, M. massiliense CIP 108297T, and M. bolletii CIP 108541T). At first sight, these groups can be termed M. abscessus, M. massiliense, and M. bolletii. However, although the M. abscessus branch is robust, the M. bolletii and M. massiliense branches have relatively low bootstrap values (65 and 71%, respectively) and a diffuse appearance, particularly in the M. massiliense branch. Furthermore, the current M. massiliense type strain (CIP 108297T) does not cluster closely with the more tightly grouped isolates in the M. massiliense branch.
Sequence analysis of the fragment of the rpoB gene, described by Adékambi as the gold standard for the molecular diagnosis of RGM infection (3), also was very informative. Our study is the first to use this approach with a large and diverse collection of isolates. We found that the isolates can indeed be divided into three distinct groups, each clustered around one of the three type strains. However, the divergence between the groups casts doubts about the extent to which these groups are different species according to the criteria of Adékambi (i.e., >3% rpoB sequence divergence between two RGM species) (2, 5). This was most marked for the M. massiliense and M. bolletii groups and for the M. massiliense and M. bolletii type strains. The most different rpoB sequences in the M. massiliense and M. bolletii groups strains diverged by only 2.13%, and the divergence between the rpoB sequences of the M. massiliense and M. bolletii type strains (not reported by Adékambi et al. [2, 5]) was only 1.46%. Therefore, according to the criteria of Adékambi et al., M. massiliense and M. bolletii do not constitute different species (3).
The divergence of rpoB between M. massiliense/M. bolletii and M. abscessus similarly raises doubts about whether M. massiliense and M. bolletii are species distinct from M. abscessus. The divergence of the rpoB sequences between the M. abscessus and M. massiliense type strains and between the M. abscessus and M. bolletii type strains were more than 3% (4.12 and 3.32%, respectively). However, in our collection, the divergence threshold of 3% was exceeded between the M. abscessus and M. bolletii groups (values from 3.59 to 4.65%) but not between the M. abscessus and M. massiliense groups (values from 2.66 to 3.59%). As M. massiliense and M. bolletii appear not to be entirely separate species, we compared the M. massiliense/M. bolletii group to the M. abscessus group: the divergence values were between 2.66 and 4.79%. Therefore, the combined M. massiliense/M. bolletii group and the M. abscessus group are not entirely separate species. These findings indicate the need for the complete revision of the current taxonomic classification of M. abscessus sensu lato; as recently proposed by Leao et al. (34), M. abscessus, M. massiliense, and M. bolletii should comprise a single species (M. abscessus).
The other objective of our study was to evaluate the use of rpoB gene sequencing for the molecular identification of isolates and discrimination between the different groups of M. abscessus sensu lato. Several recent studies have questioned the reliability of approaches based solely on rpoB sequencing (31, 34, 47, 48). This is because of the possible horizontal transfer of the rpoB gene between the different groups of M. abscessus sensu lato, especially from the M. abscessus group to the M. massiliense group. Our comparative analysis of the data for rpoB and MLSA confirmed that such horizontal transfer has occurred: almost 10% of the isolates studied had discordant MLSA and rpoB sequences. The most frequent situation was isolates having an rpoB sequence belonging to the M. abscessus group and an MLSA sequence belonging to the M. massiliense group. Thus, rpoB sequencing can identify the revised M. abscessus species (formerly M. abscessus sensu lato) but cannot discriminate 100% between M. abscessus, M. massiliense, and M. bolletii. We recently have shown that other potential targets for the molecular identification of RGM strains, such as hsp65 and sodA, also can be horizontally transferred within the M. abscessus sensu lato groups (36). We also found evidence of lateral transfer events involving the recently proposed target secA (unpublished data). If it were useful to distinguish between the M. abscessus, M. massiliense, and M. bolletii groups, because of clinical or epidemiological features specific to the infectious agent, for example, it would be valuable to identify other targets or combinations of targets to overcome the problem of horizontal gene transfer between these groups. Our team currently is addressing this issue.
This study provides information relevant to the phylogeny of M. abscessus sensu lato. We show the existence of three groups, M. abscessus, M. bolletii, and M. massiliense. Of these, the M. massiliense group seems to have emerged most recently. Our findings also reveal substantial horizontal gene transfer between the three groups, with a particularly marked flow of rpoB from M. abscessus to M. massiliense. Some transfer from M. abscessus to M. massiliense may be recent, as a small subgroup of isolates in the M. massiliense group (obtained from respiratory samples in France and Brazil) had an rpoB sequence that was almost 100% identical to the M. abscessus CIP 104536T sequence. Although the three groups are not entirely separate species, our data suggest that each group evolves in its own way, maintaining a certain cohesion, and that genetic material is exchanged between the groups, most likely via phages (30, 43, 44). The fact that they exchange genetic material indicates that they share similar biotopes, but their distinctness indicates a degree of specialization of each group. However, almost all of the isolates that we tested were from clinical samples; few M. abscessus sensu lato isolates from the environment are available (see below). We therefore cannot totally exclude sampling bias toward human pathogenic strains.
The analysis of the M. abscessus CIP 104536T genome by our group has improved our understanding of the microorganism's natural lifestyle. Genes in the M. abscessus CIP 104536T genome also are found in bacteria living in soil or aquatic environments, probably in close contact with plants. However, M. abscessus also contains a large number of genes known to be involved in intracellular parasitism, suggesting that it may have evolved to escape free-living amoebas (1, 5, 9); this would explain why M. abscessus is rarely isolated from soil or water, although there is general agreement that it lives in such environments. We have launched an extensive investigation of the presence of RGM in the water treatment systems in the Paris region, a region where respiratory infections involving M. abscessus are particularly prevalent in at-risk populations. Our MLSA approach will be valuable for comparing the population structures of M. abscessus sensu lato isolates obtained in this survey to those of clinical strains.
ACKNOWLEDGMENTS
We warmly thank Erick Denamur (INSERM U722, Paris, France) for helpful discussions and Cristianne Kayoko Matsumoto for the characterization of the Brazilian isolates.
We thank the association Vaincre la Mucoviscidose for the financial support of this work.
Footnotes
Published ahead of print on 24 November 2010.
REFERENCES
- 1. Adékambi T., Ben Salah S., Khlif M., Raoult D., Drancourt M. 2006. Survival of environmental mycobacteria in Acanthamoeba polyphaga. Appl. Environ. Microbiol. 72:5974–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adekambi T., Berger P., Raoult D., Drancourt M. 2006. rpoB gene sequence-based characterization of emerging nontuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int. J. Syst. Evol. Microbiol. 56:133–143 [DOI] [PubMed] [Google Scholar]
- 3. Adékambi T., Colson P., Drancourt M. 2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 41:5699–5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adékambi T., Drancourt M. 2004. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int. J. Syst. Evol. Microbiol. 54:2095–2105 [DOI] [PubMed] [Google Scholar]
- 5. Adekambi T., et al. 2004. Amoebal coculture of “Mycobacterium massiliense” sp. nov. from the sputum of a patient with hemoptoic pneumonia. J. Clin. Microbiol. 42:5493–5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alvarado-Esquivel C., et al. 2009. Molecular analysis of Mycobacterium isolates from extrapulmonary specimens obtained from patients in Mexico. BMC Clin. Pathol. 9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anonymous 2003. Multilocus sequence typing. Imperial College London, London, United Kingdom [Google Scholar]
- 8. Appelgren P., et al. 2008. Late-onset posttraumatic skin and soft-tissue infections caused by rapid-growing mycobacteria in tsunami survivors. Clin. Infect. Dis. 47:e11–e16 [DOI] [PubMed] [Google Scholar]
- 9. Ben Salah I., Drancourt M. 2010. Surviving within the amoebal exocyst: the Mycobacterium avium complex paradigm. BMC Microbiol. 10:99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bhatt S. P., Nanda S., Kintzer J. S., Jr 2009. The Lady Windermere syndrome. Prim. Care Respir. J. 18:334–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bishop C. J., et al. 2009. Assigning strains to bacterial species via the internet. BMC Biol. 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brown-Elliott B. A., Griffith D. E., Wallace R. J., Jr 2002. Diagnosis of nontuberculous mycobacterial infections. Clin. Lab. Med. 22:911–925 [DOI] [PubMed] [Google Scholar]
- 13. Brown-Elliott B. A., Wallace R. J., Jr 2001. Clarithromycin resistance to Mycobacterium abscessus. J. Clin. Microbiol. 39:2745–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brown-Elliott B. A., Wallace R. J., Jr 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 15:716–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cardoso A. M., et al. 2008. Emergence of nosocomial Mycobacterium massiliense infection in Goiás, Brazil. Microbes Infect. 10:1552–1557 [DOI] [PubMed] [Google Scholar]
- 16. Devulder G., Perouse de Montclos M., Flandrois J. P. 2005. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int. J. Syst. Evol. Microbiol. 55:293–302 [DOI] [PubMed] [Google Scholar]
- 17. Doolittle W. F., Zhaxybayeva O. 2009. On the origin of prokaryotic species. Genome Res. 19:744–756 [DOI] [PubMed] [Google Scholar]
- 18. Duarte R. S., et al. 2009. Epidemic of postsurgical infections caused by Mycobacterium massiliense. J. Clin. Microbiol. 47:2149–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Esther C. R., Jr., Henry M. M., Molina P. L., Leigh M. W. 2005. Nontuberculous mycobacterial infection in young children with cystic fibrosis. Pediatr. Pulmonol. 40:39–44 [DOI] [PubMed] [Google Scholar]
- 20. Feizabadi M. M., Robertson I. D., Cousins D. V., Dawson D. J., Hampson D. J. 1997. Use of multilocus enzyme electrophoresis to examine genetic relationships amongst isolates of Mycobacterium intracellulare and related species. Microbiology 143:1461–1469 [DOI] [PubMed] [Google Scholar]
- 21. Feldman E. M., Ellsworth W., Yuksel E., Allen S. 2009. Mycobacterium abscessus infection after breast augmentation: a case of contaminated implants? J. Plast. Reconstr. Aesthet. Surg. 62:e330–e332 [DOI] [PubMed] [Google Scholar]
- 22. Fisher E. J., Gloster H. M., Jr 2005. Infection with Mycobacterium abscessus after Mohs micrographic surgery in an immunocompetent patient. Dermatol. Surg. 31:790–794 [DOI] [PubMed] [Google Scholar]
- 23. Fraser C., Alm E. J., Polz M. F., Spratt B. G., Hanage W. P. 2009. The bacterial species challenge: making sense of genetic and ecological diversity. Science 323:741–746 [DOI] [PubMed] [Google Scholar]
- 24. Garcia-Navarro X., et al. 2008. Mycobacterium abscessus infection secondary to mesotherapy. Clin. Exp. Dermatol. 33:658–659 [DOI] [PubMed] [Google Scholar]
- 25. Hall T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 26. Hanage W. P., Fraser C., Spratt B. G. 2005. Fuzzy species among recombinogenic bacteria. BMC Biol. 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hanage W. P., Fraser C., Spratt B. G. 2006. Sequences, sequence clusters and bacterial species. Philos. Trans. R Soc. Lond. B Biol. Sci. 361:1917–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hanage W. P., et al. 2005. Using multilocus sequence data to define the pneumococcus. J. Bacteriol. 187:6223–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jönsson B. E., et al. 2007. Molecular epidemiology of Mycobacterium abscessus, with focus on cystic fibrosis. J. Clin. Microbiol. 45:1497–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kenzaka T., Tani K., Nasu M. 2010. High-frequency phage-mediated gene transfer in freshwater environments determined at single-cell level. ISME J. 4:648–659 [DOI] [PubMed] [Google Scholar]
- 31. Kim H. Y., et al. 2008. Proportion of Mycobacterium massiliense and Mycobacterium bolletii in Korean Mycobacterium chelonae-Mycobacterium abscessus group isolates. J. Clin. Microbiol. 46:3384–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim H. Y., et al. 2007. Outbreak of Mycobacterium massiliense infection associated with intramuscular injections. J. Clin. Microbiol. 45:3127–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leão S. C., et al. 2010. Epidemic of surgical-site infections by a single clone of rapidly growing mycobacteria in Brazil. Future Microbiol. 5:971–980 [DOI] [PubMed] [Google Scholar]
- 34. Leao S. C., et al. 2009. Characterization of mycobacteria from a major Brazilian outbreak suggests a revision of the taxonomic status of members of the Mycobacterium chelonae-abscessus group. J. Clin. Microbiol. 47:2691–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Levy I., et al. 2008. Multicenter cross-sectional study of nontuberculous mycobacterial infections among cystic fibrosis patients, Israel. Emerg. Infect. Dis. 14:378–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Macheras E., et al. 2009. Inaccuracy of single-target sequencing for discriminating species of the Mycobacterium abscessus group. J. Clin. Microbiol. 47:2596–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maiden M. C., et al. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mollet C., Drancourt M., Raoult D. 1997. rpoB sequence analysis as a novel basis for bacterial identification. Mol. Microbiol. 26:1005–1011 [DOI] [PubMed] [Google Scholar]
- 39. Newman M. I., Camberos A. E., Ascherman J. 2005. Mycobacterium abscessus outbreak in US patients linked to offshore surgicenter. Ann. Plast. Surg. 55:107–110 [DOI] [PubMed] [Google Scholar]
- 40. Olivier K. N., et al. 2003. Nontuberculous mycobacteria. I: multicenter prevalence study in cystic fibrosis. Am. J. Respir. Crit. Care Med. 167:828–834 [DOI] [PubMed] [Google Scholar]
- 41. Reich J. M. 2009. Pathogenesis of Lady Windermere syndrome. Am. J. Respir. Crit. Care Med. 179:1165. [DOI] [PubMed] [Google Scholar]
- 42. Ripoll F., et al. 2010. Mycobacterium abscessus chromosome, complete sequence. National Center for Biotechnology Information, Bethesda, MD: http://www.ncbi.nlm.nih.gov/nuccore/CU458896 [Google Scholar]
- 43. Ripoll F., et al. 2009. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One 4:e5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rolain J.-M., et al. 2009. Genomic analysis of an emerging multiresistant Staphylococcus aureus strain rapidly spreading in cystic fibrosis patients revealed the presence of an antibiotic inducible bacteriophage. Biol. Direct. 4:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roux A. L., et al. 2009. Multicenter study of prevalence of nontuberculous mycobacteria in patients with cystic fibrosis in france. J. Clin. Microbiol. 47:4124–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sermet-Gaudelus I., et al. 2003. Mycobacterium abscessus and children with cystic fibrosis. Emerg. Infect. Dis. 9:1587–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shin J.-H., Cho E.-J., Lee J., Yu J.-Y., Kang Y.-H. 2009. Novel diagnostic algorithm using tuf gene amplification and restriction fragment length polymorphism is promising tool for identification of nontuberculous mycobacteria. J. Microbiol. Biotechnol. 19:323–330 [DOI] [PubMed] [Google Scholar]
- 48. Shin J. H., Lee H. K., Cho E. J., Yu J. Y., Kang Y. H. 2008. Targeting the rpoB gene using nested PCR-restriction fragment length polymorphism for identification of nontuberculous mycobacteria in hospital tap water. J. Microbiol. 46:608–614 [DOI] [PubMed] [Google Scholar]
- 49. Spratt B. G. 1999. Multilocus sequence typing: molecular typing of bacterial pathogens in an era of rapid DNA sequencing and the Internet. Curr. Opin. Microbiol. 2:312–316 [DOI] [PubMed] [Google Scholar]
- 50. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 51. Tamura K., Nei M., Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030–11035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Viana-Niero C., et al. 2008. Molecular characterization of Mycobacterium massiliense and Mycobacterium bolletii in isolates collected from outbreaks of infections after laparoscopic surgeries and cosmetic procedures. J. Clin. Microbiol. 46:850–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wallace R. G., Jr., Silcox V., Brown B. A. 1994. Taxonomy of rapidly growing mycobacteria. Clin. Infect. Dis. 18:121–122 [DOI] [PubMed] [Google Scholar]
- 54. Wallace R. J., Jr., Brown-Elliott B. A., Crist C. J., Mann L., Wilson R. W. 2002. Comparison of the in vitro activity of the glycylcycline tigecycline (formerly GAR-936) with those of tetracycline, minocycline, and doxycycline against isolates of nontuberculous mycobacteria. Antimicrob. Agents Chemother. 46:3164–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wallace R. J., Jr., et al. 2001. Activities of linezolid against rapidly growing mycobacteria. Antimicrob. Agents Chemother. 45:764–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wallace R. J., Jr., et al. 1989. Diversity and sources of rapidly growing mycobacteria associated with infections following cardiac surgery. J. Infect. Dis. 159:708–716 [DOI] [PubMed] [Google Scholar]
- 57. Yakrus M. A., et al. 2001. Comparison of methods for Identification of Mycobacterium abscessus and M. chelonae isolates. J. Clin. Microbiol. 39:4103–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yuan J., et al. 2009. Mycobacterium abscessus post-injection abscesses from extrinsic contamination of multiple-dose bottles of normal saline in a rural clinic. Int. J. Infect. Dis. 13:537–542 [DOI] [PubMed] [Google Scholar]
- 59. Zelazny A. M., et al. 2009. Cohort study of molecular identification and typing of Mycobacterium abscessus, Mycobacterium massiliense and Mycobacterium bolletii: a cohort study. J. Clin. Microbiol. 47:1985–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang Y., et al. 2004. Pulsed-field gel electrophoresis study of Mycobacterium abscessus isolates previously affected by DNA degradation. J. Clin. Microbiol. 42:5582–5587 [DOI] [PMC free article] [PubMed] [Google Scholar]