Abstract
A behavioral memory’s lifetime represents multiple molecular lifetimes, suggesting the necessity for a self-perpetuating signal. One candidate is DNA methylation, a transcriptional repression mechanism that maintains cellular memory throughout development. We found that persistent, gene-specific cortical hypermethylation is induced in rats by a single, hippocampus-dependent associative learning experience and pharmacologic inhibition of methylation one month after learning disrupted remote memory. We propose that the adult brain utilizes DNA methylation to preserve long-lasting memories.
In recent years, neuroscience has gained a relatively deep understanding of how memories are formed. In stark contrast, is our limited understanding of how these same memories are maintained1–4. Previously, we demonstrated that hippocampal DNA methylation is critical for memory formation5. However, these hippocampal changes are transient, returning to basal levels within one day of learning. Here we used contextual fear conditioning to explore the possibility that cortical DNA methylation supports long-lasting memories. Contextual fear memory persists in rodents for many months, during which time the memory transitions from “recent” to “remote”. This change is thought to represent system consolidation, in which control over the memory shifts from the hippocampus (HPC) to a long-term dependence on the dmPFC (anterior cingulate [ACC] and prelimbic cortices)6–7.
We began by examining cortical DNA methylation of three memory-associated genes with large, GC-rich CpG islands (Egr1/zif268 [zif], reelin [Reln] and calcineurin [CaN]) using methylated DNA immunopreciptiation (MeDIP)8 (see Supplementary Methods). A subgroup of animals were tested to confirm the presence of fear memory at 7 days (context (C): 4.3% ± 2.1, shock (S): 2.3% ± 0.8, context + shock (CS): 41.1% ± 3.2; F(2, 14) = 91.35, P ≤ 0.001).
The immediate early gene zif was demethylated in all groups at all time points examined (Supplementary Fig. 1a). Thus, environmental stimuli are broadly capable of altering the methylation state of zif in the dmPFC. In contrast, Reln, a positive regulator of memory9, was hypermethylated only in trained animals within an hour of training. Levels shifted toward controls at later time points (Supplementary Fig. 1b). Methylation of the phosphatase and memory suppressor, CaN10, was not affected shortly after training. However, within 1 day of training, this gene also demonstrated learning-specific hypermethylation that persisted (Fig. 1a). We focused on CaN as a proxy for persistent methylation likely present on many genes after learning. Moreover, CaN’s pattern of methylation raises the intriguing possibility of communication between the HPC and dmPFC during the initial 24 hours after learning, as part of a system consolidation process that involves changes to cortical DNA methylation over time.
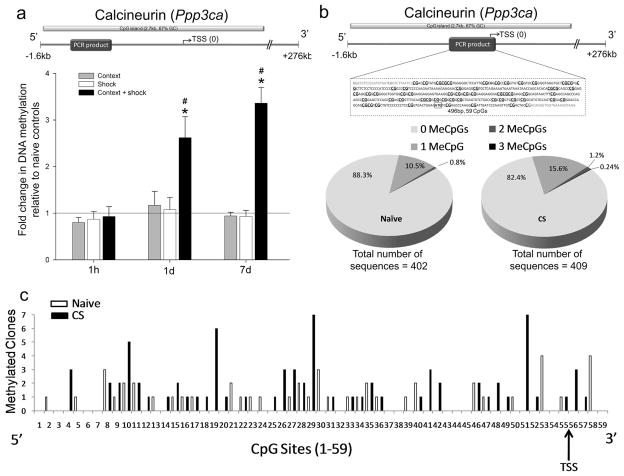
Figure 1. Learning induces persistent DNA methylation of calcineurin in the prefrontal cortex.
a, CaN CpG island analyzed and primer location. CaN is hypermethylated in CS animals at 1d and 7d (no retrieval test; 1h: F(3, 18) = 0.27, 1d: F(3, 19) = 5.73, 7d: F(3, 25) = 33.52). * post hoc P ≤ 0.05. CaN’s hypermethylation is significantly greater at 1 and 7d (F(2, 20) = 13.96, # P ≤ 0.01). See Supplementary Table 1 for N’s. b, Amplicon analyzed by BSP (transcription start site – TSS) at 7d. Charts depict number of alleles with 0, 1, 2 or 3 methylated CpGs in the amplicon. c, Single allele cytosine methylation for the 59 CpGs located in the amplicon at 7d (N = 4 per group). Error bars indicate s.e.m.
We next used bisulfite sequencing with subcloning (BSP) to map the cytosine-specific methylation changes in CaN 7 days after training. Similar to our results in Figure 1a, we found that only the CS animals had significant CaN methylation (Supplementary Fig. 2). We then expanded our analysis, focusing on the naïve and CS groups, to obtain a more detailed map. There was no evidence of single sequences with substantially increased methylation (Fig. 1b) and methylated CpGs were randomly distributed across the amplicon (Fig. 1c). Additionally, the resulting single-allele sequences indicated very low levels of methylation in the amplified region (Fig. 1b–c), consistent with reports of typical methylation levels in CpG islands11 and memory’s sparse encoding pattern. Analysis of the additional clones confirmed that CaN hypermethylation was specific to the CS group (naïve: 0.08 ± 0.01, CS: 0.25 ± 0.04; F(1, 7) = 12.91, P ≤ 0.05).
To determine if the lasting cortical methylation reflects associative learning, we gave animals pre-training injections of the NMDA receptor antagonist MK-801. A subgroup of animals, tested 7 days post-training, confirmed the ability of NMDA receptor antagonism to interfere with acquisition of a fear memory12 (veh: 59.4% ± 5.3, MK: 19.0% ± 2.7; F(1, 13) = 50.22; P ≤ 0.001). MK-801 also prevented the 7 day dmPFC CaN (veh: 1.24 ± 0.2, MK: 0.64 ± 0.1; F(1, 13) = 50.22; P ≤ 0.001) and Reln hypermethylation, without affecting zif (Supplementary Fig. 3a), providing further support that the CaN and Reln hypermethylation is a specific response to associative environmental signals.
Frankland et al. have previously investigated what effect inactivation of the ACC, a subregion of the dmPFC, has on fear memory retrieval at various post-training time points6. ACC inactivation at 18 and 36 days (remote memory), but not 1 or 3 days post-training (recent memory), interfered with retrieval. This suggests that system consolidation occurs between 3 and 18 days and further suggests that the cortical DNA methylation events we observe during the first week post-training (Fig. 1) are appropriately timed to participate in the initial incorporation of a memory trace in the cortex. We next infused the NMDA receptor antagonist APV directly into the dorsal HPC (CA1) immediately before training. APV not only interfered with learning (veh: 51.5% ± 11.7, APV: 12.3% ± 6.1; F(1, 9) = 9.73; P ≤ 0.05), but also blocked the CaN (veh: 2.11 ± 0.2, APV: 0.39 ± 0.1; F(1, 13) = 50.22; P ≤ 0.001) and Reln (Supplementary Fig. 3b–c) methylation present in the dmPFC 7 days after training, indicating that a single hippocampus-dependent learning experience is sufficient to drive lasting, gene-specific methylation changes in the cortex. These findings present a tangible marker that can be used for a learning-induced dialogue between the HPC and dmPFC.
In order to support memory persistence, cortical DNA methylation would need to be long-lasting. Therefore, we examined the persistence of fear memory (C: 0% ± 0, S: 1.3% ± 1.3, CS: 46.1% ± 8.3; F(2, 15) = 29.35, * P ≤ 0.001) and the methylation status of these genes 30 days after training. We observed robust methylation of CaN in CS animals (Fig. 2a, Supplementary Fig. 4a) well into the time period that memories are thought to become dependent upon this region. In addition, the rapid methylation observed in the HPC5 versus sustained methylation seen here in the dmPFC is consistent with system consolidation7. In accordance with DNA methylation’s role as a transcriptional repressor, we found CaN mRNA (Fig. 2b, Supplementary Fig. 4b) and protein (Fig. 2c) were specifically reduced in CS animals following retrieval of the 30 day old memory.
Figure 2. Cortical DNA methylation persists for at least 30 days.
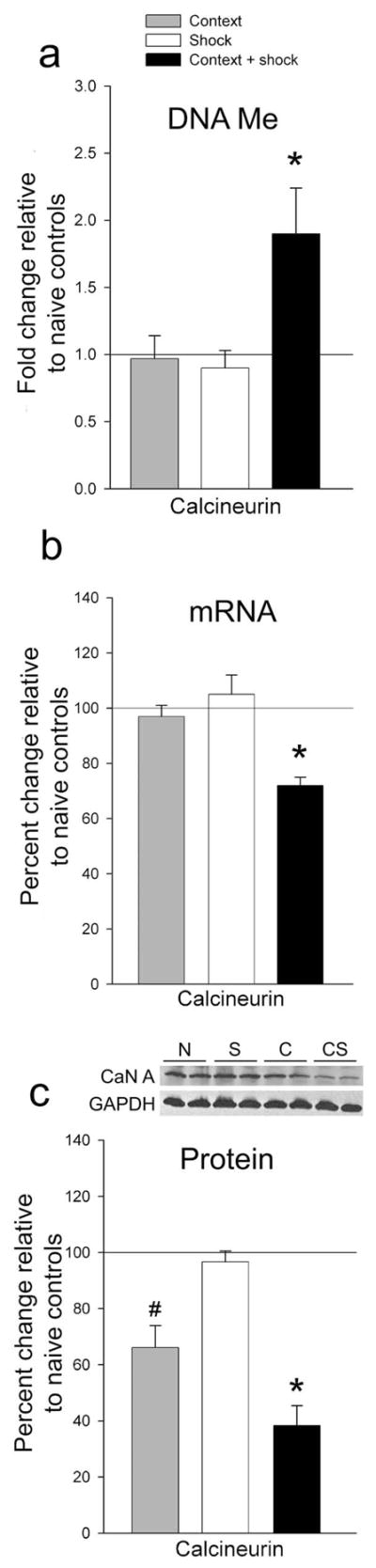
a, 30d after training, CaN is still hypermethylated (F(3, 19) = 4.77).* P ≤ 0.05. b, CaN transcript is decreased in CS at 30d (t5 = −8.36, * P ≤ 0.001; F(2, 15) = 12.72). c, 2h after retrieval test, CaN protein levels are decreased in CS as well (F(3, 30) = 24.31, * P ≤ 0.001; # P ≤ 0.05). See Supplementary Table 2 for N’s.
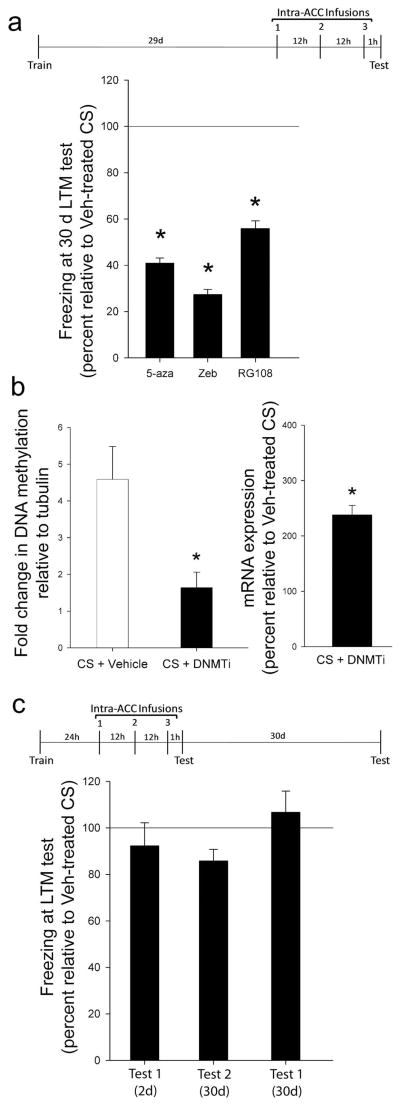
Finally, to determine if DNA methylation is necessary for the maintenance of a remote memory, we inhibited the enzymes responsible for introducing and maintaining cytosine methylation, DNA methyltransferases (DNMT), 30 days after training using 5-azadeoxycytidine (5-aza) or zebularine (zeb). Animals that received intra-ACC infusions of DNMT inhibitors (DNMTi) failed to display normal memory (Fig. 3a, Supplementary Fig. 5), indicating that DNA methylation in the ACC is critical for remote memory stability. However, outside the CNS, both compounds require DNA replication. Because their mechanism is currently unclear in the CNS, we confirmed that a direct DNMT inhibitor, RG108, also interfered with remote memory (Fig. 3a). In addition, DNMTi interfered with the hypermethylation and reduced transcription of CaN (Fig. 3b), confirming its specific effect on DNA methylation.
Figure 3. Cortical DNA methylation is required for remote memory.
a, Intra-ACC infusions of 5-aza (N’s = 14, 13; F(1, 26) = 11.48), zeb (N’s = 7, 7; F(1, 13) = 21.07) or RG108 (N’s = 10, 10; F(1, 19) = 5.17) 30d after training disrupted remote fear memory. * P ≤ 0.05. b, DNMTi interfered with CS-induced CaN methylation (N’s = 6, 6; F(1,11) = 8.96) and normalized CaN transcript levels (t8 = 8.34, * P ≤ 0.001). c, Intra-ACC infusions of DNMTi 1d after training had no effect on fear memory (N’s = 7, 8; F(1,14) = 0.81, P > 0.05). Same animals also expressed normal fear memory 30 days later (F(1,12) = 0.26, P > 0.05). Experiment was repeated in absence of a test at 2d to confirm lack of damage to ACC. Again, infusions had no effect on fear memory (N’s = 7, 8; F(1,14) = 0.81, P > 0.05). Error bars indicate s.e.m.
Intra-ACC infusions of a DNMTi may produce state-dependent effects or affect the health of cells. To address these potential confounds, we performed a control experiment in which animals received the same infusions 24 hours after training, before the memory had become reliant on the ACC. Both vehicle and DNMTi animals displayed normal fear memory, indicating that DNMTi does not interfere with an animal’s physical ability to display a fear memory (Fig. 3c). Both groups also displayed normal memory 30 days post-training (Fig. 3c), demonstrating that the ACC is of sufficient health to support a memory, such that normal transfer is likely to have occurred during the intervening weeks. These data also suggest that any effect of these drugs on baseline methylation does not preclude the subsequent formation of a remote memory.
Our results indicate that methylation helps to maintain memories and may serve as a marker of the memory trace. This introduces additional, important questions. For instance, is methylation altering a neuron’s basal state, thus altering its response to future stimuli? An example of this might be lowering a neuron’s firing threshold to enable incorporation into the existing cortical network, which transcriptional repression of CaN may accomplish, given its importance in long-term depression13 and interference with long-term potentiation10. An additional possibility is that synaptic proteins and signaling pathways downstream of cellular activation utilize methylation as a mechanism to self-perpetuate through the regulation of their own transcription rate. For example, persistent changes in methylation may support ongoing synthesis of proteins that support input-specific changes to the synapse14–15. These possibilities need not be mutually exclusive. Regardless of the specific answers to these questions, our study demonstrates that cortical DNA methylation is triggered by a learning experience and is a perpetuating signal utilized by the brain to help preserve remote memories. In combination with our previous work5, this study presents evidence that DNA methylation provides the brain with a mechanism enabling it to be dynamic during memory formation, but also stable thereafter, in order to maintain those memories.
Supplementary Material
Supplementary Figure 1. DNA methylation changes in the dmPFC following training for contextual fear conditioning.
Supplementary Figure 2. Bisulfite sequencing of calcineurin’s promoter 7 days after training.
Supplementary Figure 3. NMDA receptor blockade interferes with learning and cortical DNA methylation.
Supplementary Figure 4. DNA methylation and transcription in the cortex 30 days after training.
Supplementary Figure 5. Inhibition of DNA methylation in the ACC.
Supplementary Table 1. N’s for Figure 1 and Supplementary Figure 1 MeDIP
Supplementary Table 2. N’s for Figure 2 and Supplementary Figure 4 MeDIP and mRNA analyses
Acknowledgments
The authors would like to thank Omar S. Ahmed for his technical assistance, as well as Mei Han of the UAB Genomics Core Facility. This work was supported by the NIMH, NINDS, NIA, NIDA, American Health Assistance Foundation, the Evelyn F. McKnight Brain Research Foundation and Philip Morris.
Footnotes
Author Contributions C.A.M. and J.D.S. conceived of the project. C.A.M., G.R. and J.D.S. designed the experiments. G.R. contributed to assay development. C.A.M., C.F.G., J.A.W., R.R.P., I.M.R., A.H., M.D.R. and C.R.Y. performed the experiments. C.A.M. wrote the manuscript. G.R. and J.D.S. edited the manuscript.
References
- 1.Roberson ED, Sweatt JD. Learn & Mem. 1999;6:381–388. [PMC free article] [PubMed] [Google Scholar]
- 2.Crick F. Nature. 1984;312:101. doi: 10.1038/312101a0. [DOI] [PubMed] [Google Scholar]
- 3.Holliday R. J Theor Biol. 1999;200:339–341. doi: 10.1006/jtbi.1999.0995. [DOI] [PubMed] [Google Scholar]
- 4.Sacktor TC. Prog Brain Res. 2008;169:27–40. doi: 10.1016/S0079-6123(07)00002-7. [DOI] [PubMed] [Google Scholar]
- 5.Miller CA, Sweatt JD. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 6.Frankland PW, Bontempi B, Talton LE, Kaczmarek L, Silva AJ. Science. 2004;304:881–883. doi: 10.1126/science.1094804. [DOI] [PubMed] [Google Scholar]
- 7.Dudai Y. Annu Rev Psychol. 2004;55:51–86. doi: 10.1146/annurev.psych.55.090902.142050. [DOI] [PubMed] [Google Scholar]
- 8.Weber M, et al. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 9.Weeber EJ, et al. J Biol Chem. 2002;277:39944–39952. doi: 10.1074/jbc.M205147200. [DOI] [PubMed] [Google Scholar]
- 10.Malleret G, et al. Cell. 2001;104:675–686. doi: 10.1016/s0092-8674(01)00264-1. [DOI] [PubMed] [Google Scholar]
- 11.Bird AP. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 12.Benvenga MJ, Spaulding TC. Pharmacol Biochem Behav. 1988;30:205–207. doi: 10.1016/0091-3057(88)90445-5. [DOI] [PubMed] [Google Scholar]
- 13.Mulkey RM, Endo S, Shenolikar S, Malenka RC. Nature. 1994;369:486–488. doi: 10.1038/369486a0. [DOI] [PubMed] [Google Scholar]
- 14.Kasai H, Fakuda M, Watanabe S, Hayahi-Takagi A, Noguchi J. TINS. 2010;33:121–129. doi: 10.1016/j.tins.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Kessels HW, Malinow R. Neuron. 2009;61:340–350. doi: 10.1016/j.neuron.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. DNA methylation changes in the dmPFC following training for contextual fear conditioning.
Supplementary Figure 2. Bisulfite sequencing of calcineurin’s promoter 7 days after training.
Supplementary Figure 3. NMDA receptor blockade interferes with learning and cortical DNA methylation.
Supplementary Figure 4. DNA methylation and transcription in the cortex 30 days after training.
Supplementary Figure 5. Inhibition of DNA methylation in the ACC.
Supplementary Table 1. N’s for Figure 1 and Supplementary Figure 1 MeDIP
Supplementary Table 2. N’s for Figure 2 and Supplementary Figure 4 MeDIP and mRNA analyses