Abstract
Transgenic technologies can provide important animal models for studying drug-metabolizing enzymes. Our overall aim was to generate versatile cell and animal systems that exhibited varying levels of cytochrome P450 oxidoreductase (POR) activity, more accurately modelling the human population for pharmacological and toxicology studies. Towards this goal we evaluated RNA-interference constructs designed for use in vitro and in vivo for reducing POR activity in hepatocytes. This study clearly demonstrates that both POR protein level and reductase activity can be significantly knocked down in Hepa-1 cells in vitro, while highlighting the difficulty in predicting knockdown efficiency in transgenic animals. The high levels of embryonic lethality observed, and inability to produce multi-copy transgenic animals indicates that high levels of shRNA expression may be detrimental to embryonic development.
Keywords: RNA-interference, drug metabolism, lentivirus vectors, liver metabolism, transgenic mice
INTRODUCTION
The metabolism of drugs leading to functional modification or degradation occurs primarily in the liver. This process represents an important determinant of the duration and intensity of the pharmacological action of drugs (Thummel et al, 1997), with outcomes ranging from complete detoxification to the production of metabolites that are more toxic than the original drug. Phase I reactions can either activate or inactivate a drug through a variety of biochemical reactions, with the majority of oxidative events occurring through the cytochrome P450 (CYP) family of enzymes (Nebert et al, 1996). Phase II reactions often function on Phase I generated metabolites, and involve drug-conjugating enzymes that usually result in detoxification (Sheenan et al, 2001).
The liver represents a major site of Phase I drug metabolism through the function of the CYP enzymes (Nebert et al, 1996). The significant variability in drug clearance and clinical response observed between individuals often reflects differences in CYP activity (Ingelman-Sundberg, 2004). The CYP monoxygenases represent a large family of genes that are broadly classified into two groups (Nelson, 1999); those involved in drug metabolism and xenobiotic clearance, which are present at variable levels in different individuals, and those required for specific biological pathways including cholesterol biosynthesis, retinoic acid metabolism and steroid biosynthesis. All CYP enzymes are activated by NADPH: Cytochrome P450 Oxidoreductase (POR; E.C.1.6.2.4). POR catalyses the transfer of electrons from NADPH to CYP enzymes and in the absence of POR, CYP enzymes are essentially inactive. The interaction of drugs with CYP enzymes, and subsequent alteration of CYP activity, is a major source of adverse drug reactions.
Transgenic technologies can provide important animal models for studying drug-metabolizing enzymes, specifically, animals can be engineered to enable studies on the regulation of enzymatic expression and the impact of variable levels of enzyme activity on pharmacological action and associated toxicity. The multiple genes that encode CYP enzymes, and their overlapping functions, make the analysis of gene knockout studies difficult (Henderson et al, 2003). An alternative approach has been to knockout POR, thereby affecting all CYP activity. The POR-null mutation in mice is embryonic lethal (Shen et al, 2002; Wu et al, 2005), but conditional knockout strategies have shown the value of this approach to understanding drug metabolism (Henderson et al, 2003; Finn et al, 2007). Mice lacking hepatic POR are viable and fertile, but completely devoid of liver CYP activity, and are unable to metabolize testosterone, acetaminophen and pentobarbital (Henderson et al 2003). With the aim to develop transgenic strategies providing intermediate POR activities, in this study we have evaluated RNA-interference (RNAi) approaches to knockdown POR activity in the mouse liver.
MATERIALS AND METHODS
Vector construction
shRNA vectors were obtained from Sigma expressing various short hairpin sequences under the control of the human ubiquitous U6 pol-III promoter.
Target sequences:
shPOR1: 5′CCTGACCTACTGGTTCATCTT
shPOR2: 5′CGGAGGCACATCCTAGCCATT
shPOR3: 5′GCATCTAATGCACCTGGAATT
shPOR4: 5′CGGGAAGGAACATTATTGTAT
shPOR5: 5′GCTCGAAATATGGCCAAAGAT
These vectors contained a puromycin resistance gene under the control of a PGK promoter. This was removed using Nsi I and Bam HI, the backbone was blunted and a blunt ended eGFP fragment was ligated into the plasmid using T4 DNA ligase.
Lentivirus production
Self-inactivating (SIN) lentivirus was generated by Fugene-6 (Roche) mediated co-transfection of the three plasmids encoding the required packaging proteins, envelope and viral genome – 12μg psPAX2, 6μg VSV-G and 9μg pLKO-shRNA, repectively – a T150 flask seeded the previous day at 1×107 cells per flask (Al Yacoub et al, 2007). Conditioned medium was removed at 24hr and 48hr, filtered and centrifuged at 7,000×g O/N at 4°C, the resultant pellet was resuspended in 5ml TSSM buffer consisting of 20mM Tris, pH 7.3, 100mM sodium chloride, 10mg/ml sucrose and 10mg/ml mannitol, and concentrated by ultra-centrifuge at 20,000xg for 2hr at 4°C. The viral pellet was then resuspended in 100μl TSSM and aliquoted for storage at -80°C. Virus titres were determined by Polybrene-mediated serial dilution transduction of HT1080 cells and GFP expressing positive colonies were counted after 5 days (Al Yacoub et al, 2007). Concentrated viral titres ranged from 3.8×109 TU/ml to 6×109 TU/ml.
Cell culture and lentivirus transduction
Hepa-1 cells were grown at 37°C (5%, v/v, CO2) in Dubelecos modified Eagle medium (DMEM) supplemented with 10% (v/v) foetal calf serum (FCS, Gibco). Cells were grown to 80-90% confluence before being passaged. Hepa-1 cells were seeded at ∼12000 cells per well in a 24-well plate 24hr prior to transduction with lentivirus at a multiplicity of infection (MOI) of ∼1 in the presence of 8μg/ml polybrene. The cells were then FACS sorted to isolate the cells into which the pro-virus had integrated and maintained as a pooled population for no more than 10 passages.
POR expression and functional assays
Microsomal protein extracts were purified from Hepa-1 cells and mouse tissue using Sigma Endoplasmic Reticulum Extraction kit following the manufacturer's protocol. Protein concentration was determined using BCA protein assay kit (Thermo Scientific) and expression was analysed by western blot employing ECL detection. POR detection was through rabbit CH59 anti-POR polyclonal antibody (kind gift from Colin Henderson, Dundee University: Henderson et al, 2003) and horseradish peroxidise labelled polyclonal goat anti-rabbit antibody (Dako). POR levels were normalised to β-actin using anti-mouse β-actin (Sigma) and goat anti-mouse (Dako) antibodies. The intensity of the bands was then quantified using PhosphorImager and QuantityOne software. The reductase activity of microsomal POR was determined using a colorimetric Cytochrome P450 reductase assay kit (Sigma) following the manufacturer's protocol (Plonne et al, 1999).
Generation of transgenic mice
All animal experimentation was performed after review by Animal Ethics Committee and under UK Home Office Licence. Transgenic mice were generated as described previously (Ritchie et al, 2007). Eggs harvested from super-ovulated C57BL6 X CBA females were injected with a 10x or 1x pulse of ˜50pl of 109 TU/ml1080 virus using standard micro-injection procedure. The virus was injected into the peri-vitellin space using a glass microinjection needle. Following injection the eggs were transferred to M16 medium (Sigma) for incubation at 37°C 5% (v/v) CO2 until day 4.5 where 15-18 developed blastocysts were surgically implanted into a psuedo-pregnant female. The resultant pups were genotyped by Southern blot (Whitelaw et al, 1992) and PCR (Vasey et al, 2008), using either:
eGFP-F: 5′CACATGAAGCAGCACGACTT with
eGFP-R: 5′TCTGGGTGGACAGGTAGTGG; or
HIVpsiF: 5′GAGAGAGATGGGTGCGAGAAG with
HIVpsiR: 5′GCTGTGCGGTGGTCTTACTT
as previously described. Transgenic mice were bred with C57BL6 X CBA F1 stock mice.
RESULTS
Evaluation of shRNA vectors in vitro
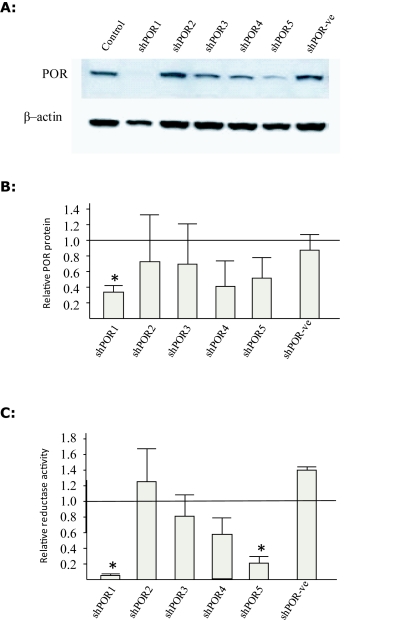
POR knockdown was analysed by western blot in shPOR1 (72%), shPOR4 (55%) and shPOR5 (49%) transduced cell populations although given the variance between replicates only the reduction observed in shPOR1 cells was statistically significant (Figure 1). To determine if the shRNA knockdown of POR protein levels influenced hepatic cellular function we performed a reductase assay (Plonne et al, 1999). The lowest reductase levels were consistently observed for shPOR1 and shPOR5, reflecting the POR expression levels both showing a significant reduction in P450 reductase activity (Figure 1). The relatively high variance observed between replicates could be a consequence of varying numbers of viral integration events between the examined samples. A subsequent experiment, where cells were sorted according to GFP expression, indicated that those cells expressing higher levels of GFP grew slower than those expressing lower levels of GFP and when re-analysed by FACS their GFP expression profile had changed (data not shown). Higher GFP expression was assumed to be as a result of a greater number of integration events, and thus it may be that these cells also express higher quantities of shRNA. This suggests that there may be a significant reduction in growth rate or even toxicity associated with high numbers of integration events and transgene expression, indeed toxicity has previously been reported in cells expressing high levels of shRNAs (Fish et al, 2004). This shift in population may have affected different experimental replicates to a greater or larger degree resulting in a high variance in expression and activity read-outs.
Figure 1.
POR expression and function in Hepa1 cells treated with shPOR lentivirus. Transduction of Hepa-1 cells with one of 6 shRNAs (shPOR1, shPOR2, shPOR3, shPOR4, shPOR5, shPOR-ve) and POR protein determined by western blot (A) with average densitometry data (B) normalized to β-actin protein levels with error bars showing Standard Deviation (n=3). POR protein levels are shown relative to that detected in non-tranfected cells (horizontal line). Cytochrome C reductase activity (C) of microsomal protein with error bars representing the Standard Deviation (n=3). Reductase activity shown relative to that detected in non-tranfected cells (horizontal line).
Evaluation of shRNA vectors in vivo
shRNAs shown to give different levels of knockdown were used to generate transgenic animals. We attempted two separate peri-vitelline injection regimes in an attempt to get various levels of POR knockdown depending on transgene copy number; 1×50pl and 10×50pl of high-titre lentivirus (at least 3.8×109 TU/ml), the outcome of all the injections carried out are summarised in Table 1. Initial development of embryos to blastocysts showed a considerably lower level of development in the 10× injection batch (1×50pl: 241/376 (64%); 10×50pl: 143/375 (38%)) in all viral injections. This indicates that high concentrations of viral particles and/or shRNA expression reduce blastocyst development irrespective of target sequence. Integrational mutagenesis is a recognised problem of using lentiviruses however studies using similar injection strategies showed no significant reduction in development where up to 21 transgene integration events were seen (Lois et al, 2002; Ritchie et al, 2007), indicating that this may be a symptom of high levels of shRNA expression.
Table 1.
Summary of blastocyst transfers, pregnancies, birth rates and transgenesis. The efficiency of development to blastocyst stage, through gestation to birth varied between viruses and injection regime. Several preganncies did not come to term (* denotes pups reabsorbed, consumed at birth or died within 3 days of birth) and those that did showed low levels of transgenesis.
| Virus | Injection regime | Blastocyst transfers(×18) | Pregnancies (<10days) | Litters | Pups | GFP | Transgenic |
|---|---|---|---|---|---|---|---|
| shPOR1 | 10x | 2 | 1 | 1 | /* | /* | / |
| shPOR1 | 1x | 5 | 3 | 3 | 11 | 11 | 7 |
| shPOR4 | 10x | 5 | 4 | 4 | /* | /* | / |
| shPOR4 | 1x | 3 | 2 | 2 | 0 | 0 | 0 |
| shPOR-ive | 10x | 4 | 1 | 1 | 4* | 4 | 4 |
| shPOR-ive | 1x | 4 | 3 | 3 | 18 | 0 | 1 |
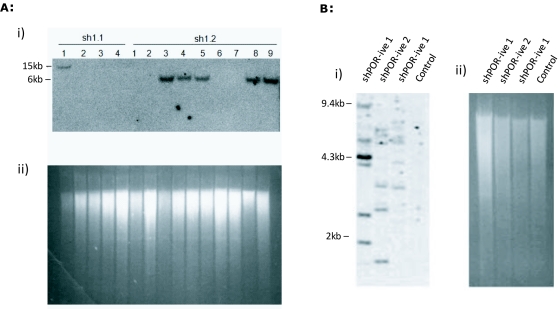
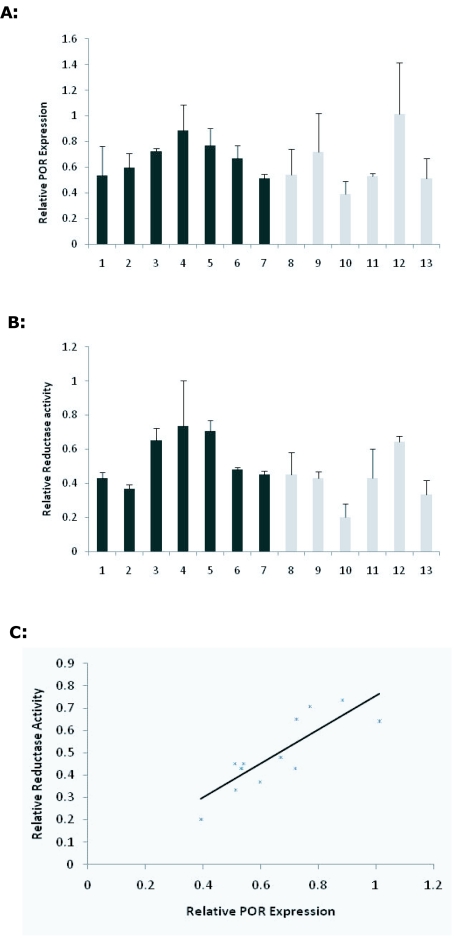
1×50pl injection of shPOR1 into developing zygotes yielded three founder transgenic mice that were identified as positive by PCR of tail clip biopsy DNA, Southern blot analysis showed each positive contained only one copy of the shPOR1 transgene (data not shown); offspring were bred from two of the founder animals and genotyped by Southern blot (Figure 2A) and PCR. Liver microsomal protein extracts from the off-spring were analysed by western blot and Cytochrome P450 reductase assay (Figure 3). A clear relationship between POR expression and reductase activity was observed in these mice (Figure 3C) but, overall, no significant difference in POR expression or reductase activity was observed when compared to wild-type (WT) littermates.
Figure 2.
Southern blot analysis of transgenic pups. The mating of shPOR1.1 and shPOR1.2 founder animals with WT females gave two litters of which 6 were transgenic and they all contained one copy of the transgene (A). Multicopy transgene integration was observed in dead off-spring generated from a 10× 50pl injection of 5.8×109 TU/ml shPOR-ve lentivirus (B). These animals were the only pups to show multiple integration events and died within three days of birth. SybrSafe image of digested DNA (ii) shows efficient digestion of DNA and similar loading of samples.
Figure 3.
POR expression in shRNA1 transgenic mice. Microsomal protein extracts were taken from liver samples of 21 day old pups and POR protein levels determined by Western blot with average densitometry data normalized to β-actin protein levels of three separate blots with Standard Deviation error bars (A). Cytochrome C reductase activity (B) of microsomal protein with Standard Deviation error bars from three repeats of each assay. Non-transgenic samples 1-5 and transgenic samples 6-11. The correlation between protein expression and function is shown (C) where expression is plotted against function.
Curiously all transgenic mice successfully raised carried only one copy of the transgene (Figure 2); similar studies have shown both higher efficiency of transgenesis and the generation of multicopy transgenic animals using similar quantities of virus (Lois et al, 2002). It is conceivable that in our study, any multicopy animals, which would presumably have higher shRNA expression levels and (based on our in vitro data) reduced POR activity, may have died in utero. Reduced growth rate seen in Hepa-1 cells, reduced blastocyst development and the following observations in the animal studies support this scenario.
In all 10x injections the pups were either re-absorbed or consumed at birth by the mother, in the one litter that survived (shPOR-ve) all the pups had died within 3 days. Where DNA samples were isolated from dead animals, Southern blot data indicated multiple copies of the transgene (Figure 2). The majority of 1x injections led to successful pregnancies although a low level of transgenesis was obtained (8/29; 27.5%) and all transgenic animals only contained one copy of the transgene (Figure 2b). For shPOR lentivirus injections less than half identified pregnancies (at day 10 gestation) resulted in births. If embryonic lethality was occurring it could result from high levels of shRNA (irrespective of target) which has been reported to cause mortality due to competitive inhibition of the endogenous miRNA system (Castanotto et al, 2007; Gimm et al, 2007) or reflect the known phenotypic consequence of significantly reducing POR activity during development (Shen et al 2005).
Limited success has been noted when conjugated antisense oligonucleotides were delivered to the rat liver where a small reduction in POR reductase activity was seen (Venkateswaran et al, 2010). In this study the antisense oligos were delivered using a non-viral delivery system which allowed large doses of the oligonucleotides to be delivered into adult rats, and despite a modest reduction in function, no significant change was seen in the protein expression level. This, together with our findings, suggests that POR is hard to specifically knockdown in vivo using RNAi and antisense technologies, indicating that more factors may be involved than the just previously reported embryonic lethality.
CONCLUSIONS
The overall aim of this study was to generate robust and versatile cell and animal systems that exhibited varying levels of POR activity, more accurately modelling the human population for pharmacological and toxicology studies. Specifically, altering POR activity would allow the role of cytochrome P450 activity on drug metabolism to be determined, enabling identification of bioavailability, efficacy and characterisation of drug metabolites to efficacy and toxicity to be evaluated (Henderson et al, 2003; Finn et al, 2007).
Although this study clearly demonstrates that both POR protein level and reductase activity can be significantly knocked down in Hepa-1 cells in vitro, the data also highlight the difficulty in knockdown of POR in an animal model. We believe that for strategies aiming to deliver gene knockdown in transgenic animals, the prior testing of RNAi constructs in cell systems is not necessarily a reliable indicator of subsequent in vivo activity. This study demonstrates that POR is a difficult target to knock down in vivo and inability to produce multi-copy transgenic animals suggests that a high level of shRNA expression is detrimental to the development of the embryo.
Acknowledgments
We thank Colin Henderson, John Hayes and Douglas Vasey for insightful discussion; and Colin Henderson for reagents. This work was funded through a Biotechnology and Biological Sciences Research Council (BBSRC)-CASE studentship to NM and the European Commission NEST029025 project INTEGRA.
COMPETING INTERESTS
None declared.
REFERENCES
- Al Yacoub N, Romanowska M, Haritonova N, Foerster J. Optimized production and concentration of lentiviral vectors containing large inserts. J Gene Med. 2007;9:579–584. doi: 10.1002/jgm.1052. [DOI] [PubMed] [Google Scholar]
- Castanotto D, Sakurai K, Lingeman R, et al. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nuc Acids Res. 2007;35:5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D, Scherer L. RNA Interference. San Diego, USA: Elsevier Academic Press Inc; 2005. Targeting cellular genes with PCR cassettes expressing short interfering RNAs. pp. 173–183. [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz K, Jopling CL, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Finn RD, McLaren AW, Carrie D, Henderson CJ, Wolf CR. Conditional deletion of cytochrome P450 oxidoreductase in the liver and gastrointestinal tract: A new model for studying the functions of the P450 system. J Pharm Exp Ther. 2007;322:40–47. doi: 10.1124/jpet.107.121780. [DOI] [PubMed] [Google Scholar]
- Fish RJ, Kruithof EKO. Short-term cytotoxic effects and long-term instability of RNAi delivered using lentiviral vectors. BMC Mol Biol. 2004:5–15. doi: 10.1186/1471-2199-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson CJ, Otto DME, Carrie D, et al. Inactivation of the hepatic cytochrome P450 system by conditional deletion of hepatic cytochrome P450 reductase. J Biol Chem. 2003;278:13480–13486. doi: 10.1074/jbc.M212087200. [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M. Pharmacogenetics of cytochrome P450 and its applications in drug therapy: the past, present and future. Trends Pharm Sci. 2004;25:193–200. doi: 10.1016/j.tips.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Li LM, Lin XY, Khvorova A, Fesik SW, Shen Y. Defining the optimal parameters for hairpin-based knockdown constructs. RNA. 2007;13:1765–1774. doi: 10.1261/rna.599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Nelson DR, Coon MJ, et al. The P450 superfamily – Update on new sequences, gene mapping and recommended nomenclature. DNA Cell Biol. 1991;10:1–14. doi: 10.1089/dna.1991.10.1. [DOI] [PubMed] [Google Scholar]
- Nebert DW, McKinnon RA, Puga A. Human drug-metabolizing enzyme polymorphisms: Effects on risk of toxicity and cancer. DNA Cell Biol. 1996;15:273–280. doi: 10.1089/dna.1996.15.273. [DOI] [PubMed] [Google Scholar]
- Nelson DR. Cytochrome P450 and the individuality of species. Arch Biochem Biophys. 1999;369:1–10. doi: 10.1006/abbi.1999.1352. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Cleary M, Silva JM, et al. Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nat Methods. 2004;1:163–167. doi: 10.1038/nmeth1104-163. [DOI] [PubMed] [Google Scholar]
- Plonne D, Cartwright I, Linss W, Dargel R, Graham JM, Higgins JA. Separation of the intracellular secretory compartment of rat liver and isolated rat hepatocytes in a single step using self-generating gradients of iodixanol. Anal Biochem. 1999;276:88–96. doi: 10.1006/abio.1999.4311. [DOI] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotech. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Ritchie WA, Neil C, King T, Whitelaw CB. Transgenic embryos and mice from low titre lentiviral vectors. Transgenic Res. 2007;16:661–664. doi: 10.1007/s11248-007-9102-2. [DOI] [PubMed] [Google Scholar]
- Thummel KE, Kunze KL, Shen DD. Enzyme catalysed processes of first-pass hepatic and intestinal drug extraction. Adv Drug Del Rev. 1997;27:99–127. doi: 10.1016/s0169-409x(97)00039-2. [DOI] [PubMed] [Google Scholar]
- Sangkhathat S, Kusafuka T, Miao JY, et al. In vitro RNA interference against beta-catenin inhibits the proliferation of pediatric hepatic tumors. Int J Oncol. 2006;28:715–722. [PubMed] [Google Scholar]
- Sheehan D, Meade G, Foley VM, Dowd CA. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem J. 2001;360:1–16. doi: 10.1042/0264-6021:3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen AL, O'Leary KA, Kasper CB. Association of multiple developmental defects and embryonic lethality with loss of microsomal NADPH-cytochrome p450 oxidoreductase. J Biol Chem. 2002;277:6536–6541. doi: 10.1074/jbc.M111408200. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci USA. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasey DB, Wolf CR, MacArtney T, Brown K, Whitelaw CB. p21-LacZ reporter mice reflect p53-dependent toxic insult. Toxicol Appl Pharmacol. 2008;227:440–450. doi: 10.1016/j.taap.2007.11.029. [DOI] [PubMed] [Google Scholar]
- Whitelaw CB, Harris S, McLenaghan M, Simons JP, Clark AJ. Position-independent expression of the ovine beta-lactoglobulin gene in transgenic mice. Biochem J. 1992;286:31–39. doi: 10.1042/bj2860031. [DOI] [PMC free article] [PubMed] [Google Scholar]