Abstract
Background
Facioscapulohumeral muscular dystrophy (FSHD) is an autosomal dominant disease associated with contraction of arrays of tandem 3.3-kb units (D4Z4) on subtelomeric 4q. Disease-linked arrays usually have fewer than 11 repeat units. Equally short D4Z4 arrays at subtelomeric 10q are not linked to FSHD. The newly described 4qA161 haplotype, which is more prevalent in pathogenic 4q alleles, involves sequences in and near D4Z4.
Methods
We developed two new assays for 4qA161, which are based upon direct sequencing of PCR products or detecting restriction fragment length polymorphisms. They were used to analyse single nucleotide polymorphisms (SNPs) indicative of 4q161 alleles.
Results
All (35/35) FSHD patients had one or two 4qA161 alleles (60% or 40%, respectively). In contrast, 46% (21/46) of control individuals had no 4qA161 allele (p<10−4), and 26% had homozygous 4qB163 alleles.
Conclusions
Our results from a heterogeneous population are consistent with the previously described association of the 4qA161 haplotype with FSHD, but a causal association with pathogenesis is uncertain. In addition, we found that haplotype analysis is complicated by the presence of minor 10q alleles. Nonetheless, our sequencing assay for the 4qA161allele can enhance molecular diagnosis of FSHD, including prenatal diagnosis, and is simpler to perform than the previously described assay.
INTRODUCTION
Facioscapulohumeral muscular dystrophy (FSHD) is an enigmatic autosomal dominant disease affecting about 1 out of 15 000 people.1 2 The immediate genetic defect is a shortened array of tandem 3.3-kb repeats (D4Z4) at 4q35 (figure 1A).3 However, the mechanism by which contracted D4Z4 arrays on 4q cause muscular dysfunction is still unclear.4–6 In addition, it is unclear why contraction of subtelomeric 4q D4Z4 arrays is pathogenic but not that of 99% identical D4Z4 arrays at subtelomeric 10q, especially in view of the high 4q/10q homology ~40 kb proximal and distal to the arrays.4 By the age of about 20 years, most individuals who have only 1–10 copies of the repeat at one subtelomeric 4q (instead of the normal 11–100 copies at both subtelomeric 4q) exhibit symptoms of this progressively debilitating and painful disease.1 2 7 There is no efficacious treatment.
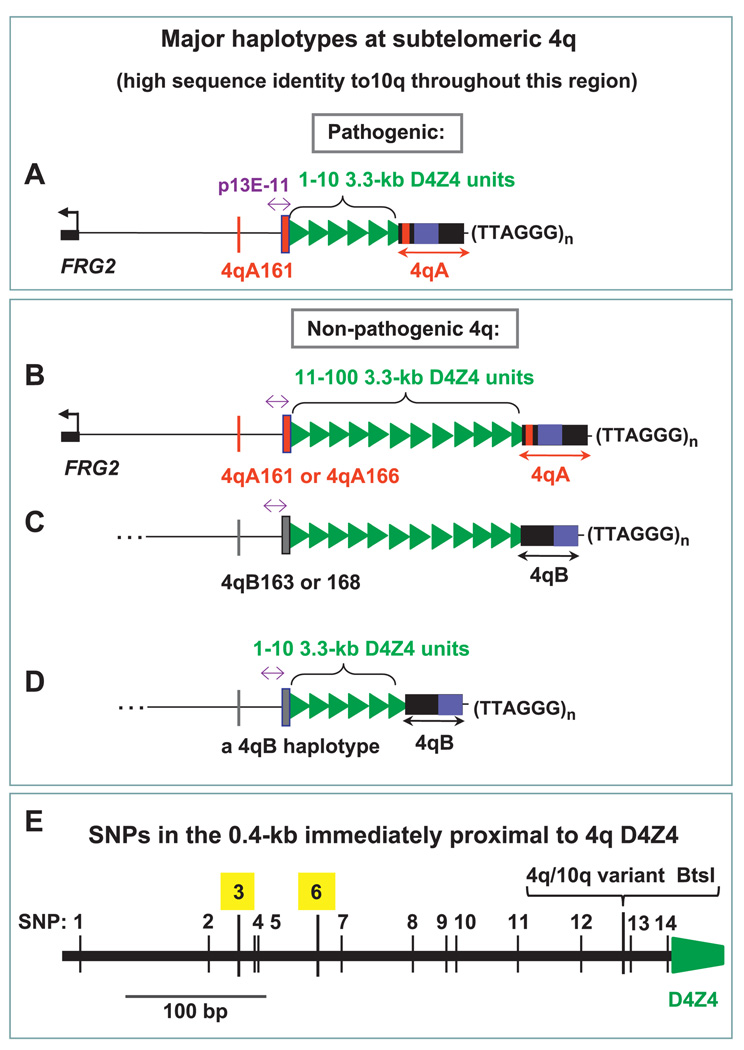
Figure 1.
Cartoon of the major 4q haplotypes. The D4Z4 array consists of variable numbers of 3.3 kb units (green triangles) ~25–33 kb from the telomeric (TTAGGG)n (distal end sequence derived from van Geel et al19 and Genome Reference Consortium h37 10qA and 4qB sequences). Proximal to D4Z4 is a SSLP subregion (thin red or grey line) and SNP-rich 0.4-kb subregion (thick red or grey line); the 0.8-kb p13E-11 subregion (purple double-headed arrow) is indicated. Not included are less frequent haplotypes with unreported sequences.20 23 32 4qA and 4qB alleles have almost equal prevalence at 4q. The predominant D4Z4-distal allele at 10q is very similar to 4qA and called 10qA.19 4qA and 10qA, but not 4qB, contain an ~3-kb subregion of 68-bp tandem repeats (red bar) (A–D). Dense SNPs in the 0.4-kb region adjacent to D4Z4. SNP3 and SNP6 are highlighted. The BtsI site that distinguishes the main 10q allele (BtsIS) and 4q alleles (BtsIR) is shown (E).
The near identity of 4q and 10q subtelomeric D4Z4 arrays complicates molecular diagnosis,8 which is necessary for an unambiguous determination of disease status. The sizes of these 4q and 10q macrosatellites can be determined by pulsed field gel electrophoresis (PFGE) of DNA cleaved outside the arrays and at 4q-specific or 10q-specific D4Z4 restriction sites9 or by linear gel electrophoresis (LGE).10 The standard hybridisation probe for molecular diagnosis of FSHD, p13E-11 (figures 1A and 2A), detects both 10q and 4q arrays. Although ~10% of 10q arrays are short, a pathogenic D4Z4 array on 10q has never been reported.9 11 About3%of controls and of FSHD patients have deletions of the p13E-11 region, which necessitates using a D4Z4 hybridisation probe.12 13 At least 10% of the population have one or more 10q-type, BlnI-sensitive (BlnIS) repeat units on 4q (almost always heterogeneous arrays) or 4q-type, BlnI-resistant (BlnIR) units on 10q (usually homogeneous arrays11 14). This necessitates careful analysis to determine whether the short D4Z4 array is on 4q. Moreover, about 10–30% of patients have sporadic (non-familial) FSHD, and about 25% of these are somatic mosaics with respect to the size of the D4Z4 array; this is best detected by PFGE, rather than LGE.15–18
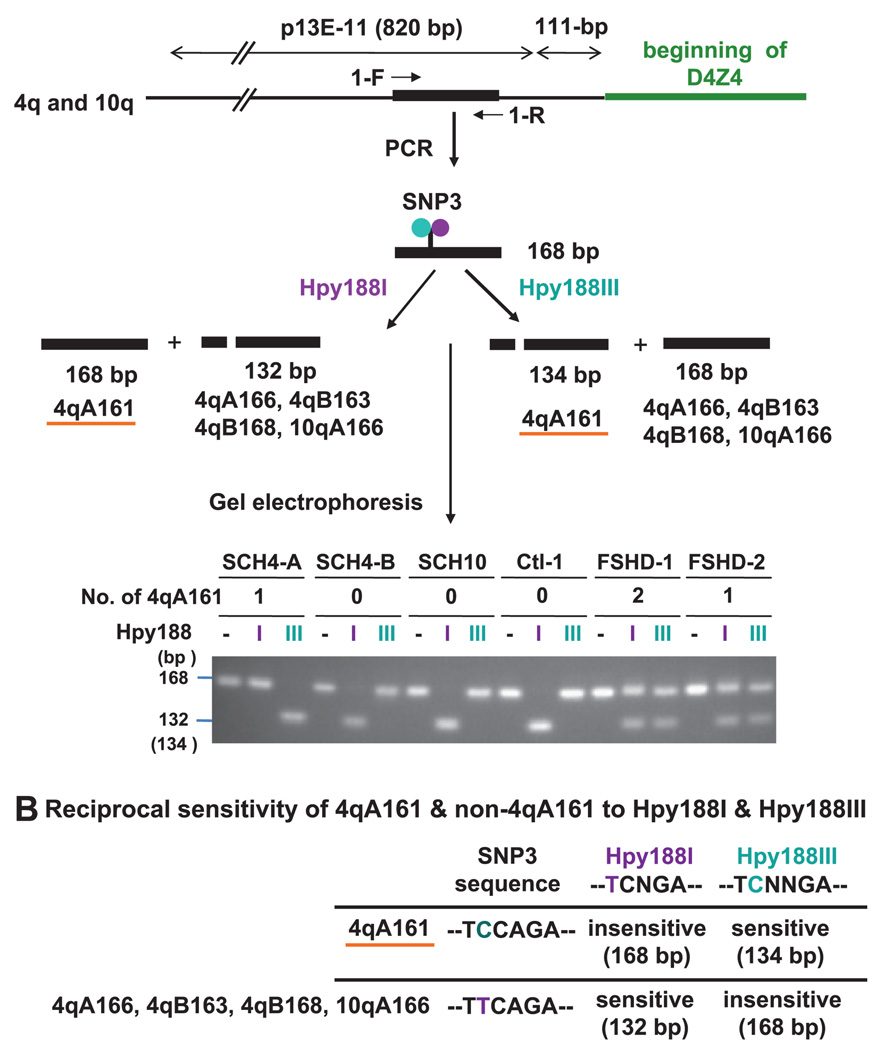
Figure 2.
Testing for the 4qA161 haplotype by differential digestion with Hpy188I and Hpy188III. Genomic DNAs and human-rodent somatic cell hybrids (GM14193, GM11687, and GM10926 containing chr4, chr4, and chr10, respectively; Coriell Institute) were analysed for overlapping Hpy188I (purple) or Hpy188III (blue) sites. Whether one or two 4qA161 alleles were present was subsequently determined by the sequencing assay. GM11448 and GM10115 containing chr4 were negative for 4qA161 (not shown) (A). The Hpy188I or Hpy188III sites are indicated for the forward strand in the SNP-rich region of 4qA161, the other main 4qA alleles, and the predominant 10qA allele (B).
An additional complication is that the 4q D4Z4 distal region is composed of two allelic variants, 4qA and 4qB (figure 1B vs 1C).19 In controls, the frequencies of 4qA and 4qB are almost equal, but short pathogenic D4Z4 arrays on 4q35 have been associated only with 4qA20 21 (figure 1A vs 1D). This has also been demonstrated in studies with large percentages of sporadic cases20 21). Moreover, in studies of over a thousand individuals in the general population, 17 unrelated controls had short D4Z4 arrays (ie, less than 11 repeat units, including arrays with only five repeat units) on 4qB alleles and no signs of FSHD.22 23
In addition to the 4qA/4qB subdivision, there are at least 17 different haplotypes at subtelomeric 4q24 based upon the following: the distal 4qA/B variation and less frequent variants; the simple sequence length polymorphisms (SSLP) ~3 kb proximal to D4Z4 (figure 1A–D, thin red or grey lines); a region rich in single nucleotide polymorphisms (SNPs) adjacent to the proximal end of D4Z4 (figure 1A–D, thick red or grey lines; figure 1E) and the D4Z4 SNP at the beginning of the D4Z4 array. Lemmers and coworkers23 tested for linkage of these haplotypes to FSHD. They did preparative PFGE on restriction digests, PCR and capillary electrophoresis for detection of variants in the D4Z4-proximal SSLP region; PFGE on separate restriction digests and blot hybridisation for analysis of 4qA/4qB status and LGE for analysis of the SNP in the first D4Z4 repeat. They found that all pathogenic, contracted D4Z4 arrays from 86 tested FSHD patients were linked to the haplotype that they call 4qA161. Importantly, 39% of 4q alleles in controls also had the 4qA161 allele.
We describe the use of PCR-DNA sequencing or a PCR-restriction digestion assay for detection of this haplotype. These assays are much simpler to perform and to interpret than the previously described haplotyping.23 24 We discuss the potential use of haplotype testing in molecular diagnosis. In addition, we have confirmed the much stronger association of the 4qA161 haplotype with FSHD than with controls using, for the first time, a population of patients and controls from the USA. This association was previously reported only in a European population. Given recent findings about the strong effect of ethnicity on subtelomeric 4q haplotypes in control populations,24 these results are an important addition to our understanding of the relevance of 4qA161 haplotypes to clinical testing for FSHD.
PATIENTS AND METHODS
Patient data
DNA was obtained from blood samples, fibroblasts, lymphoblasts or myoblasts of FSHD patients, who had been clinically confirmed and molecularly diagnosed. Similar numbers of males and females were represented in the patient and control samples from an ethnically heterogeneous US population. Both patients and controls gave informed consent, and samples were deidentified. Molecular diagnosis was by PFGE on EcoRI/HindIII, EcoRI/BlnI and XapI digests (14 samples)12 or LGE of EcoRI and EcoRI/BlnI samples (21 samples) combined with the dosage test.25 Only LGE samples without evidence of non-standard D4Z4 arrays (ie, without canonical 10q-type repeat units on 4q or vice versa) were used.
SNP assay 1: differential digestion by Hpy188I versus Hpy188III
Genomic DNA (100 ng) was amplified in a 25-µl volume using 0.2 µM primers 1-F (5′-CTGTGGTCATCTCTGCTCCA-3′) and 1-R (5′-TTTGCCTGTGAGTTCGAATG-3′) as follows: 95°C, 2 min; then 45 cycles of 95°C, 20 s; 60°C, 20 s; 72°C, 20 s; followed by 72°C for 5 min. For each set of PCRs, a no-template control was the last sample. Aliquots of the single 168-bp PCR product (10 µl) were digested with 20 U of Hpy188I or of Hpy188III (New England Biolabs) for 3 h at 37°C (40 µl reaction volume). For Hpy188III, first 10 U was added, followed by 5 U after 1 and again after 2 h. Digests were electrophoresed in a 2.5% or 3% agarose gel for 3 h, followed by ethidium bromide staining (1 µg/ml) for 30 min. For about half of the reactions, 0.1 µg of a pUC19 plasmid DNA was added as an internal control to an aliquot of the digest and incubated concurrently. Upon electrophoresis, the internal controls showed complete digestion relative to plasmid DNA in the absence of PCR product.
SNP assay 2: DNA sequencing
In this more informative assay, genomic DNA (100 ng) was amplified in a 50-µl volume using 0.3 µM primers 2-F (5′-CCTATTAAACGTCACGGACA-3′) and Bts-R (5′-CCATCTCCCCTCACACACTT-3′) under the above-described cycling conditions. The single 363-bp product was cycle-sequenced using primer 1-R (BigDye Terminator v3.1; ABI 3730 XL, Genetic Analyzer). A no-template control was routinely verified to contain no product by gel electrophoresis. It was not necessary to purify the PCR product before sequencing because we had a large enough amount of product.
RESULTS
Hpy188I/III restriction fragment length polymorphism (SNP assay 1)
To analyse the association of the 4qA161 haplotype with FSHD, we identified a SNP (figure 1E, SNP3) located 307 bp proximal to D4Z4 (GenBank AF117653, nt 4519) at overlapping recognition sites for Hpy188I and Hpy188III (figure 2A). This assay was designed to provide a quick method to exclude the presence of any 4qA161 alleles. Differential digestion at SNP3 should distinguish the 4qA161 allele from the other reported haplotypes of 4q and 10q (figure 2B) based upon sequencing of at least three alleles per haplotype by Lemmers et al.23 We amplified a sequence that contains SNP3 from somatic cell hybrid (SCH) and genomic DNAs and digested aliquots of the PCR products with Hpy188I or Hpy188III. SCH DNAs were completely sensitive to Hpy188I and resistant to Hpy188III or vice versa, as expected (figure 2A and data not shown).
Of the 11 examined FSHD DNA samples, all had at least one 4qA161 allele, as seen by the undigested band in the Hpy188I digest and a digested band in the Hpy188III digest (figure 2A). In contrast, only 12 out of 21 genomic samples displayed this Hpy188IR/Hpy1888IIIS allele. Samples with only one 4qA161 allele and those with two 4qA161 alleles (as subsequently determined in the sequencing assay, see below) gave both an undigested band and a digested band with each enzyme. This is because the highly homologous 10q allele was amplified and its sequence at SNP3 mimicked non-4qA161 4q alleles. Thus, distinguishing between one and two 4qA161 alleles is not possible in this assay.
Direct sequencing of SNP region (SNP assay 2) and disease relationships
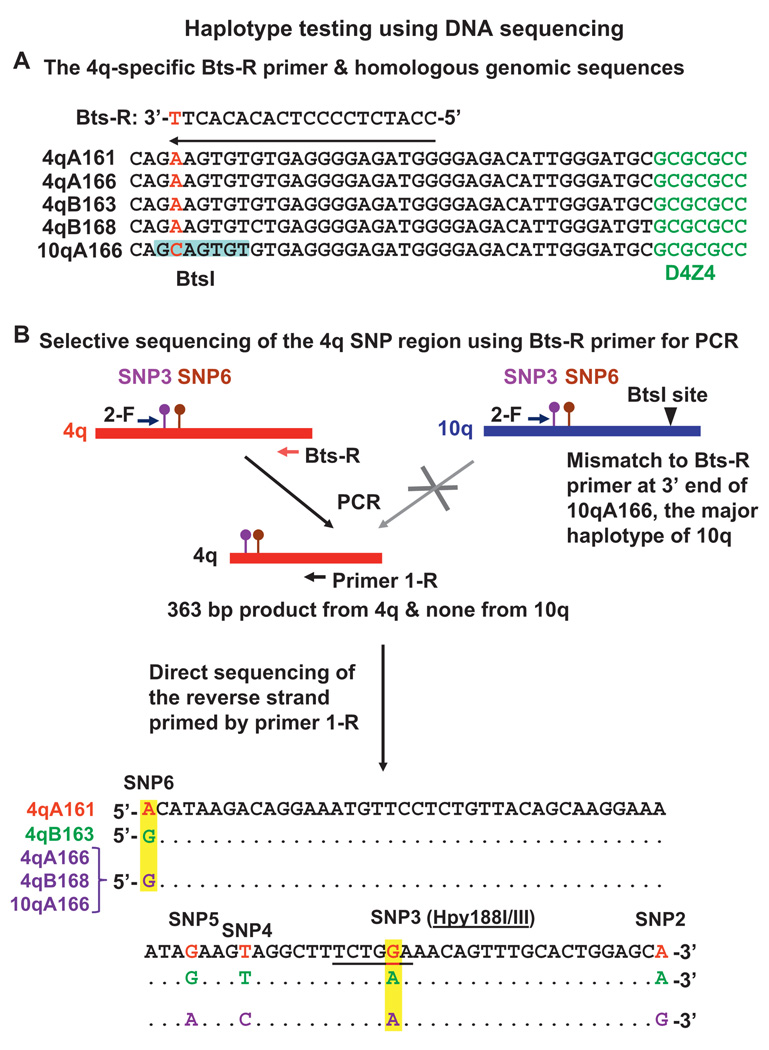
We developed a second assay that provides more detailed information about sequence variations in the D4Z4-proximal region and excludes 10qA166 alleles from amplification. 10qA166 alleles were reported to constitute 96% of 10q alleles.23 Despite ~99% sequence identity in the 41.6-kb region proximal to D4Z4 and within the D4Z4 array, selective amplification of a 363-bp SNP-rich sequence immediately proximal to 4q was achieved with a reverse primer, 1-R. The 3′ end of this primer has a T:C mismatch to 10qA166, the predominant 10q allele23 (figure 3A, SNP13). The 10q alleles in databases (AL845259, AL954635, AY028079, BX294170 and BX005259, GenBank) have the same sequence in this region as 10qA166. As expected, DNA from a SCH containing chr10 (GM10926, Coriell Institute) could not be amplified by these primers, unlike SCH DNA containing chr4.
Figure 3.
Testing for the 4qA161 haplotype by sequencing through SNP3 and SNP6 on the reverse strand. Sequence alignment of the 4qA alleles and 10qA166 allele illustrates the 4q-specific nature of PCR primer Bts-R. The BtsI site overlapping the primer in 10qA166 is highlighted (A). PCR with Bts-R as one of the primers followed by sequencing the reverse strand allows identification of the 4qA161haplotype. As described in the text, digestion with BtsI to eliminate 10qA166 amplicons before PCR is not necessary (B).
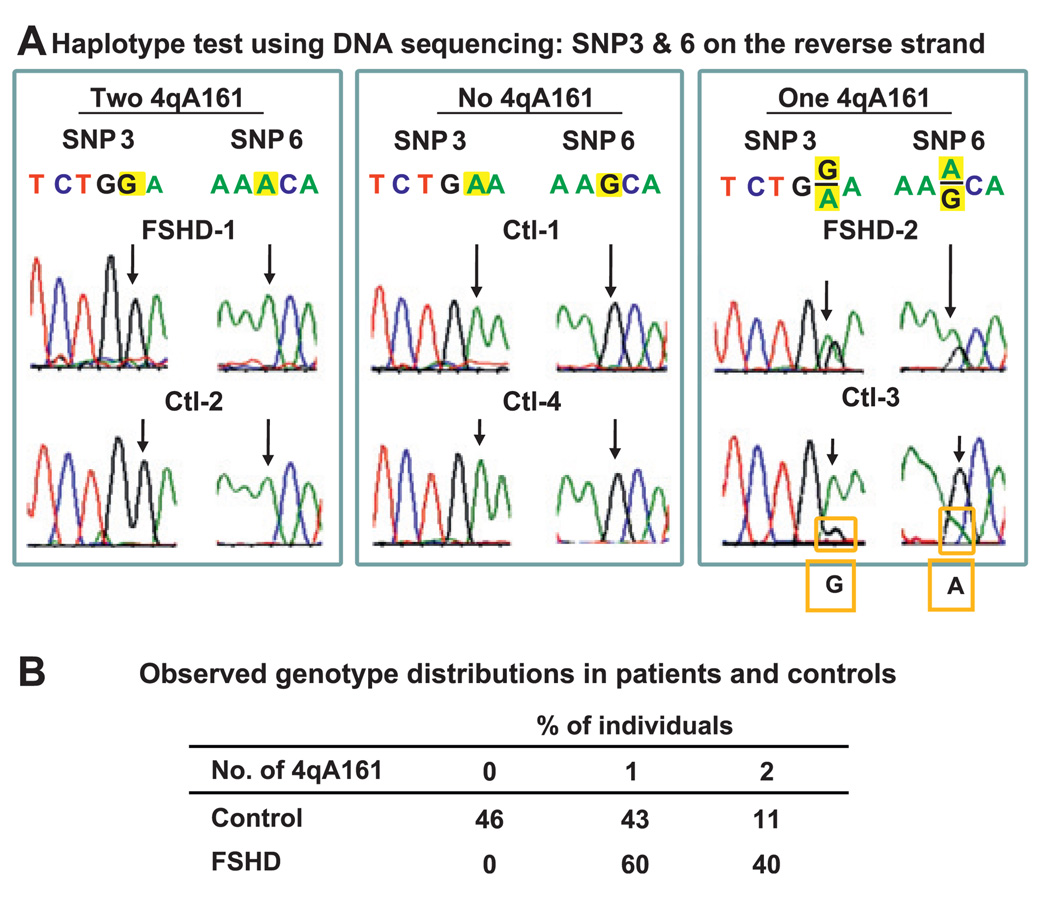
Sequencing the reverse strand of the 363-bp product allowed identification of 4qA161 alleles from a G at SNP3 and an A at SNP6 (figures 3B and 4A). All 35 FSHD samples had at least one 4qA161 allele (figure 4B). In contrast, 21 of the 46 control samples had only A at SNP3 and only G at SNP6, indicating no 4qA161 allele. Approximately 33% of 4q alleles in controls and 70% of alleles in FSHD patients were 4qA161 (see below). Similarly, Lemmers et al23 reported that 4qA161 constituted ~39% of 4q alleles in 100 controls based on analysis involving the SSLP ~3 kb proximal to SNP3 and SNP6. Genome-wide haplotype data26 indicated that the frequency of G at SNP3 (4qA161) was 34–41% in European or Asian controls (86–118 individuals in each of three populations; dbSNP build 130). Our data indicate a highly significant difference (p<10−4, Fisher exact test) between FSHD patients and controls in the frequency of individuals with no 4qA161 allele.
Figure 4.
Results from the DNA sequencing test for 4qA161. Sequencing data from representative samples containing two 4qA161 alleles, none or one (A). Genotype frequencies from our assays of 35 FSHD patients and 46 controls (B).
Ten FSHD and four control samples had only G at SNP3 and A at SNP6, indicating two 4qA161 alleles. Thirteen FSHD and six control samples had approximately equal heights of the G and A peaks at SNP3 and SNP6, signifying one 4qA161 allele and one heterologous 4q allele (eg, FSHD-2, figure 4A). Unexpectedly, eight FSHD and 14 control samples had G:A ratios at SNP3 that differed several fold and had correspondingly skewed A:G ratios at SNP6 (eg, Ctl-3, figure 4A). Similar results were obtained from replicate PCRs. Testing of these samples in a modified Hpy188I/III digestion assay for SNP3 using nested PCR to eliminate 10qA166 alleles gave concordant results to those from the sequencing assay, even for the above-mentioned samples (Supplementary figure 1). The skewed G:A ratios were probably the result of minor, but not rare, alleles of subtelomeric 10q, which were amplified because they completely matched the Bts-R primer, unlike 10qA166 alleles (figure 3A). It was originally reported that 10q alleles other than 10qA166 constitute only 4% of all 10q alleles in a European population,23 but our data from an American population indicates that the percentage is approximately fivefold higher, consistent with a more recent report from the same group.24 We conclude that the 14 samples with G:A ratios at SNP3 of about 1:2 and the eight samples with ratios of about 1:3 to 1:4 contain one 4qA161 allele, one heterologous 4q allele and one or two unusual 10q alleles mimicking a non-4qA161 allele at 4q. In addition, the five samples that had G:A ratios at SNP3 of 2:1 should contain one unusual 10q allele other than 10qA166, as well as two 4qA161 alleles, thereby increasing the number of samples that were identified as homozygous for 4qA161. By taking into account the contribution of minor, but not rare, 10q alleles as above, a sample can be more accurately evaluated as to whether it has one or two 4qA161 alleles.
For all 14 samples with a SNP3 G:A ratio of 1:0 (two 4qA161 alleles), the presence of only A, T and G at SNPs 2, 4 and 5, respectively, confirmed that there was no contamination with 10q sequences (data not shown). We could also identify the 4qB163 allele from the reported pattern of SNPs23 (figure 3B). Twelve controls (26%) had predominant or unique A, A, T, G and G peaks in the reverse strand at SNPs 2, 3, 4, 5 and 6 (nt 4499, 4519, 4530, 4534 and 4576 of AF117653, GenBank), respectively, indicating homozygosity for 4qB163. Among FSHD patients and controls, 34% and 30%, respectively, were 4qB163/4qA161 heterozygotes (mixed 4qA161 and 4qB163 sequence patterns at SNPs 2–6). In addition, eight controls (9%) had a mixed pattern of 4qB163 SNPs and 4qA166 or 4qB168 SNPs. In summary, we found that ~50% of 4q alleles in controls and 17% in FSHD patients were 4qB163. We also verified the sequences of the non-polymorphic residues in this region that were previously reported.23
Integrating SNP testing into FSHD molecular diagnosis
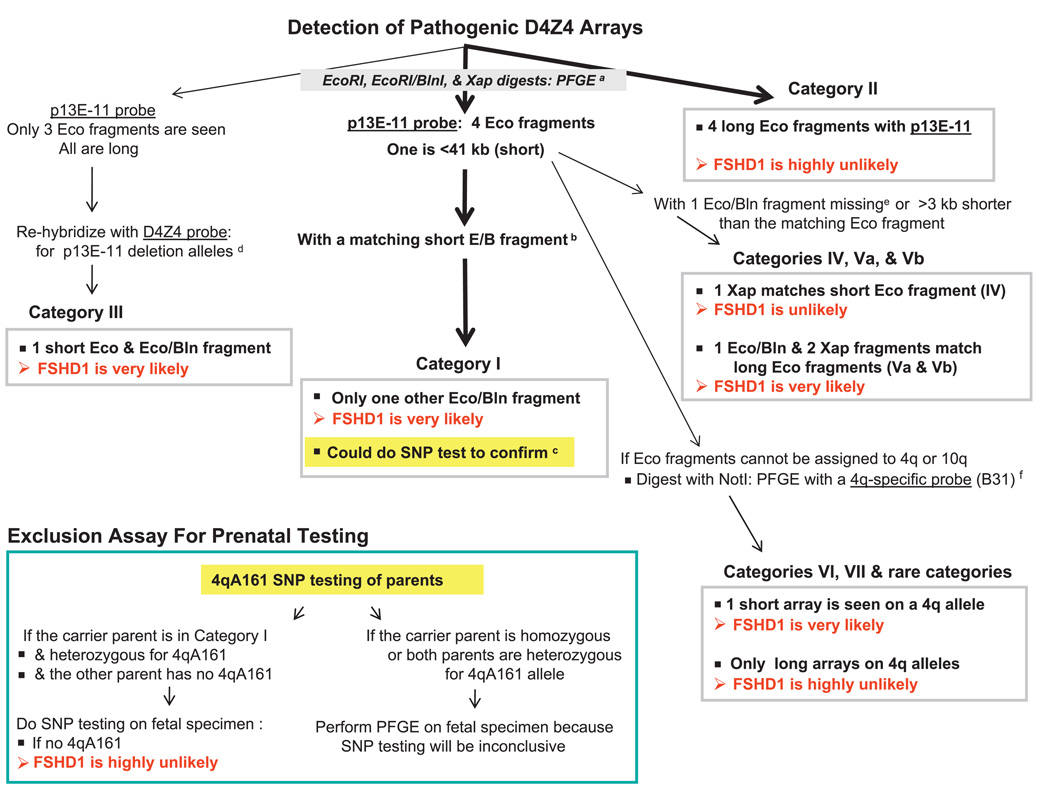
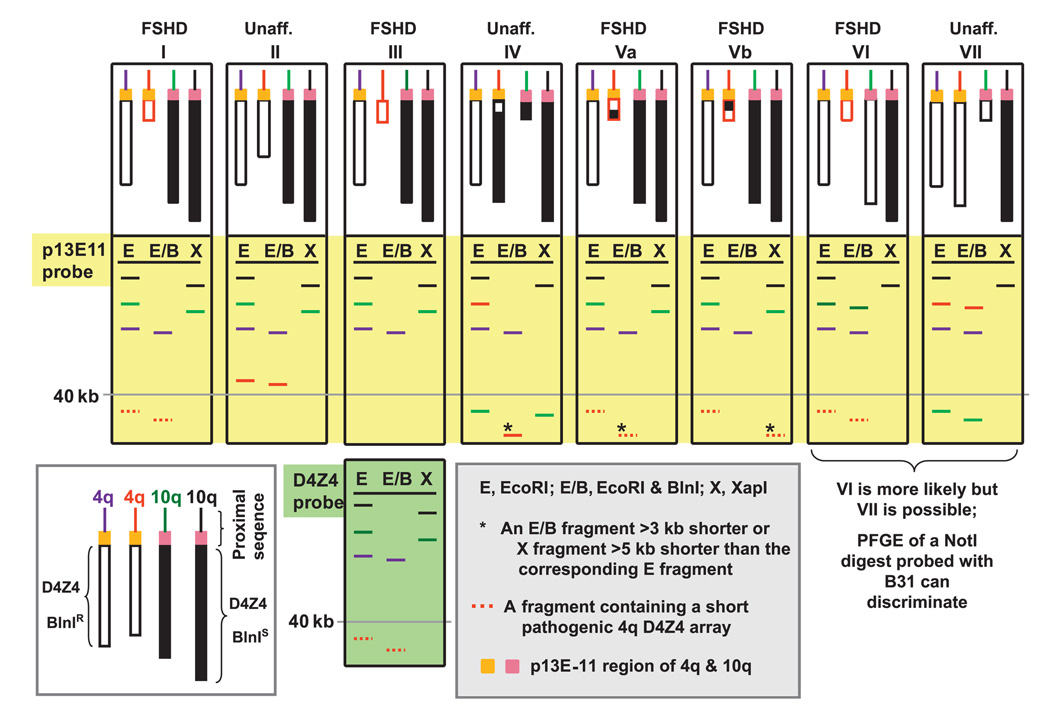
Our recommendations for incorporating 4qA161 SNP testing into molecular diagnosis of FSHD patients and prenatal testing are diagrammed in figure 5 for FSHD linked to a short 4q D4Z4 array (FSHD Type 1A or FSHD1), which is the type of FSHD that we studied. FSHD symptoms without a short D4Z4 array on 4q are termed FSHD Type 1B (or FSHD2), which is ~20 times less prevalent.27 The scheme for molecular diagnosis of FSHD1 is based upon Southern blotting to determine the size of 4q D4Z4 arrays and distinguish them from 10q D4Z4, preferably by PFGE (figure 6). PFGE (or LGE) Southern blot analysis of 4q D4Z4 array size depends on a few restriction site differences between canonical 4q and 10q D4Z4 arrays. These restriction site differences are among the ~1% non-identity between canonical 4q and 10q D4Z4 repeat units.9 28 Sizing of 4q D4Z4 can be complicated by non-standard arrays on 4q containing some of the D4Z4 repeat units as canonical 10q-type arrays and 10q arrays containing 4q-type D4Z4 repeat units (figure 6). Given the complexity of determining the size of D4Z4 arrays on 4q, it is helpful for diagnosis and prenatal testing to have a rapid SNP test as one tool that can be used in the molecular analysis (figure 5).
Figure 5.
A possible scheme for use of the sequencing SNP assay in clinical testing for FSHD1, which involves short 4q D4Z4 arrays and constitutes ~95% of FSHD cases.27 Analysis paths are shown by arrows in series. Thick arrows indicate most likely outcomes. aCanonical 10q D4Z4 arrays in EcoRI/BlnI digests28 and canonical 4q arrays in XapI digests are reduced to small fragments that electrophorese off the gel.9 PFGE is highly preferable to LGE, for example, PFGE can reveal somatic mosaics having five hybridising EcoRI fragments.17 18 If LGE is used, the so-called dosage test25 should be done to determine if the most proximal D4Z4 repeat unit on both 4q alleles is 4q-type (BlnIR) and on both 10q alleles is 10q-type (BlnIS), and the blot should be re-probed with D4Z4. EcoRI/HindIII digests can be used instead of EcoRI digests to get cleaner bands, but fragments are ~2 kb smaller.12 The maximum size of pathogenic D4Z4 alleles is only an approximate threshold due to incomplete penetrance, especially when EcoRI fragments are 35–41 kb; also, some patients may have very slightly larger D4Z4 arrays.33 bMatching EcoRI/BlnI fragments are ~3 kb shorter and XapI fragments are ~5 kb shorter than the corresponding EcoRI fragment.12 cThe sequencing test is the recommended assay for 4qA161, the predominant haplotype of pathogenic D4Z4 alleles. It can serve as a surrogate for 4qA/4qB testing by PFGE on HindIII digests with 4qA- and 4qB-specific oligonucleotide probes.11 20 22 23 There can be pathogenic non-4qA161 alleles with 10q-type BlnIS D4Z4 repeat units on 4q.32 However, categories I, III and VI in figure 6 do not involve such arrays; therefore, 4qA161 testing would be informative for samples giving these types of PFGE patterns, especially because ~40–50% of controls have no 4qA161 allele. dThe D4Z4 subfragment probe under stringent hybridisation conditions detects fragments with p13E-11 deletions and gives stronger signals than the p13E-11 probe from XapI digests.12 eCategories IV, Va and Vb are non-standard D4Z4 arrays with canonical 10q-type repeat units on 4q or vice versa, as evidenced by a missing EcoRI/BlnI fragment (only one EcoRI/BlnI fragment instead of two, figure 6, Vb) or an EcoRI/BlnI fragment that is less than 3 kb shorter than the corresponding EcoRI fragment (figure 6, IV and Va). fSometimes, the chromosomal origin of a short EcoRI fragment has to be determined by PFGE on NotI digests blot-hybridised to a 4q35-specific probe (B31),13 22 for example, figure 6, category VI versus category VII, and unusual cases with all four arrays beginning with 10q-type D4Z4 units. The NotI fragment length will be the sum of 187 kb and the length of the corresponding EcoRI fragment.
Figure 6.
A cartoon illustrating various categories of subtelomeric 4q and 10q genotypes (I–VII; large white rectangles) analysed by Southern blotting after digestion with EcoRI, EcoRI plus BlnI and XapI (ApoI) and probing with p13E-11 (large yellow rectangles) or D4Z4 (single large light-green rectangle). Non-standard D4Z4 arrays on 4q that contain BlnIS units (black-filled boxes) and those on 10q that contain BlnIR units (white boxes) complicate the determination of whether a short array is present on 4q. These types of D4Z4 arrays with repeat units from chr10 on 4q are almost always mixed with canonical chr4-type units in various patterns, and those from chr4 on 10q are usually homogeneously chr4-type,11 34 as illustrated for categories IV–VII. Other combinations of translocated arrays are possible.35
DISCUSSION
We have developed and applied a rapid method to determine the 4q subtelomeric haplotype, 4qA161, which is associated with FSHD.23 It has been proposed that, of all the 17 discovered haplotypes in this region, the 4qA161 haplotype is responsible for conferring pathogenicity on short 4q arrays and not on short 10q arrays.24 However, there is linkage disequilibrium in this subtelomeric region24 which, coupled with the haplotype-dependent variation in size ranges of D4Z4 arrays, might account for the disease association of the 4qA161 haplotype. We favour the hypothesis that it is the larger chromatin context that determines the 4q specificity of FSHD.5 11 13 29 30 Nonetheless, the association of 4qA161 with FSHD is useful clinically.
The previously reported disease association of 4qA161 with FSHD23 could have been influenced by ethnic factors.24 31 Our analysis of a heterogeneous US population confirmed that all studied FSHD patients had at least one 4qA161 allele, but this was also the case for 54% of controls. Additionally, we demonstrated a much higher frequency of minor 10q alleles than initially reported.23
Our new sequencing assay for the 4qA161 haplotype requires only PCR and sequencing a 363-bp product to analyse two SNPs, which provide redundant, and thereby, confirmatory information. The previously described and much more complicated assay involves a preparative PFGE, PCR and capillary electrophoresis for analysis of the SSLP region; PFGE for 4qA/B determination and LGE for analysis of a SNP in D4Z4.23 24 Analysis of the data involves combining results about small differences in sizes of a heterogeneous array of PCR products and data from several Southern blots. Unlike the previous assay, our assay is not complicated by co-electrophoresis of minor 4q alleles (4qA166 and 4qB166) with the major 10q allele (10qA166).23
It is not yet clear whether patients with sporadic FSHD and with non-standard D4Z4 arrays24 32 have the same strong association of a short D4Z4 array to the 4qA161 haplotype observed in familial cases, which constitute most FSHD patients. It has been reported that there are unusual D4Z4-distal alleles, whose sequences were not published, that are neither 4qA nor 4qB and are linked to short pathogenic D4Z4 arrays at 4q.20 In addition, arrays which have 10-type D4Z4 units on 4q11 can be pathogenic even though they should not be part of a 4qA161-type allele.
As described in the Introduction, the DNA analysis of the sizes of D4Z4 arrays at 4q in the diagnosis of FSHD is complicated by several factors that need to be considered to avoid false positive or false negative conclusions. The DNA sequencing haplotype analysis described here is a useful additional tool that detects whether there are no 4qA161 alleles or one or two of these alleles. In about 40% of the FSHD population, the 4qA161 sequencing assay will demonstrate that both 4q alleles are 4qA. Therefore, use of the sequencing haplotype assay in primary diagnostic testing will eliminate the need for 4qA/B Southern blot analysis in the same percentage of cases. However, gel electrophoresis (preferably PFGE, figure 6) will still need to be done to look for a short pathogenic 4q because the vast majority of the general population with two 4qA161 alleles does not have the pathogenic short 4q D4Z4 and, hence, is unaffected. When only one 4qA161 allele and one short 4q D4Z4 array are found, linking the short array to the 4qA161 allele requires either a family study or physical resection of a gel band containing the shortened D4Z423 and retesting the purified DNA for 4qA161. In an initial screen of patients where a diagnosis of FSHD is clinically unlikely, either the above-described sequencing or restriction assays for 4qA161 could be used as a probable exclusion test20 32 when the absence of any 4qA161 alleles is being scored. These assays for 4qA161 will also be useful to exclude FSHD during prenatal diagnosis of families in which the affected parent had been shown to have a single, pathogenic 4qA161 allele and the partner has none.
Supplementary Material
Acknowledgements
We thank Drs. Rabi Tawil (University of Rochester), V. Vedanarayanan (University of Mississippi Medical Center, Jackson, MS) and Steven A. Moore (The University of Iowa), as well as the patients for the FSHD samples and Steve Moore, for critical reading of the manuscript.
Funding This research was supported in part by the National Institutes of Health (NS04885 to M.E). Some of the cultured cells from FSHD patients were provided by the Tissue and Cell Culture Repository at The University of Iowa, a component of the Iowa Wellstone Muscular Dystrophy Cooperative Research Center, (NIH NS053672).
Footnotes
An additional figure is published online only. To view this file please visit the journal online (http://jmg.bmj.com).
Competing interests None.
Provenance and peer review Not commissioned; externally peer reviewed.
REFERENCES
- 1.Lunt P. Facioscapulohumeral Muscular Dystrophy: Diagnostic and Molecular Aspects. In: Deymeer F, editor. Neuromuscular Diseases: From Basic Mechanisms to Clinical Management. Vol 18: Monogr. Clin. Neurosci. Basel: Karger; 2000. pp. 44–60. [Google Scholar]
- 2.Pandya S, King WM, Tawil R. Facioscapulohumeral dystrophy. Phys Ther. 2008;88:105–113. doi: 10.2522/ptj.20070104. [DOI] [PubMed] [Google Scholar]
- 3.van Deutekom JC, Wijmenga C, van Tienhoven EA, Gruter AM, Hewitt JE, Padberg GW, van Ommen GJ, Hofker MH, Frants RR. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 4.van der Maarel SM, Frants RR. The D4Z4 repeat-mediated pathogenesis of facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2005;76:375–386. doi: 10.1086/428361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu X, Tsumagari K, Sowden J, Tawil R, Boyle AP, Song L, Furey TS, Crawford GE, Ehrlich M. DNaseI hypersensitivity at gene-poor, FSH dystrophy-linked 4q35.2. Nucleic Acids Res. 2009;37:7381–7393. doi: 10.1093/nar/gkp833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng W, de Greef JC, Chen YY, Chien R, Kong X, Gregson HC, Winokur ST, Pyle A, Robertson KD, Schmiesing JA, Kimonis VE, Balog J, Frants RR, Ball AR, Jr, Lock LF, Donovan PJ, van der Maarel SM, Yokomori K. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Upadhyaya M, Maynard J, Rogers MT, Lunt PW, Jardine P, Ravine D, Harper PS. Improved molecular diagnosis of facioscapulohumeral muscular dystrophy (FSHD): validation of the differential double digestion for FSHD. J Med Genet. 1997;34:476–479. doi: 10.1136/jmg.34.6.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrlich M. Exploring hypotheses about the molecular etiology of FSHD: loss of heterochromatin spreading and other long-range interaction models. In: Cooper DN, Upadhyaya M, editors. FSHD Facioscapulohumeral Muscular Dystrophy: Molecular Cell Biology & Clinical Medicine. New York, NY: BIOS Scientific Pub.; 2004. pp. 253–276. [Google Scholar]
- 9.Lemmers RJL, de Kievit P, van Geel M, van der Wielen MJ, Bakker E, Padberg GW, Frants RR, van der Maarel SM. Complete allele information in the diagnosis of facioscapulohumeral muscular dystrophy by triple DNA analysis. Ann Neurol. 2001;50:816–819. doi: 10.1002/ana.10057. [DOI] [PubMed] [Google Scholar]
- 10.Lemmers RJ, van der Wielen MJ, Bakker E, Padberg GW, Frants RR, van der Maarel SM. Somatic mosaicism in FSHD often goes undetected. Ann Neurol. 2004;55:845–850. doi: 10.1002/ana.20106. [DOI] [PubMed] [Google Scholar]
- 11.Rossi M, Ricci E, Colantoni L, Galluzzi G, Frusciante R, Tonali PA, Felicetti L. The facioscapulohumeral muscular dystrophy region on 4qter and the homologous locus on 10qter evolved independently under different evolutionary pressure. BMC Med Genet. 2007;8:8. doi: 10.1186/1471-2350-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrlich M, Jackson K, Tsumagari K, Camano P, Lemmers RJFL. Improved hybridization analysis of D4Z4 repeat arrays linked to FSHD. Chromosoma. 2007;116:107–116. doi: 10.1007/s00412-006-0080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deak KL, Lemmers RJ, Stajich JM, Klooster R, Tawil R, Frants RR, Speer MC, van der Maarel SM, Gilbert JR. Genotype-phenotype study in an FSHD family with a proximal deletion encompassing p13E-11 and D4Z4. Neurology. 2007;68:578–582. doi: 10.1212/01.wnl.0000254991.21818.f3. [DOI] [PubMed] [Google Scholar]
- 14.van der Maarel SM, Frants RR, Padberg GW. Facioscapulohumeral muscular dystrophy. Biochim Biophys Acta. 2007;1772:186–194. doi: 10.1016/j.bbadis.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Zatz M, Marie SK, Passos-Bueno MR, Vainzof M, Campiotto S, Cerqueira A, Wijmenga C, Padberg G, Frants R. High proportion of new mutations and possible anticipation in Brazilian facioscapulohumeral muscular dystrophy families. Am J Hum Genet. 1995;56:99–105. [PMC free article] [PubMed] [Google Scholar]
- 16.Zatz M, Marie SK, Cerqueira A, Vainzof M, Pavanello RC, Passos-Bueno MR. The facioscapulohumeral muscular dystrophy (FSHD1) gene affects males more severely and more frequently than females. Am J Med Genet. 1998;77:155–161. [PubMed] [Google Scholar]
- 17.Tonini MM, Lemmers RJ, Pavanello RC, Cerqueira AM, Frants RR, van der Maarel SM, Zatz M. Equal proportions of affected cells in muscle and blood of a mosaic carrier of facioscapulohumeral muscular dystrophy. Hum Genet. 2006;119:23–28. doi: 10.1007/s00439-005-0100-2. [DOI] [PubMed] [Google Scholar]
- 18.van der Maarel SM, Deidda G, Lemmers RJ, van Overveld PG, van der Wielen M, Hewitt JE, Sandkuijl L, Bakker B, van Ommen GJ, Padberg GW, Frants RR. De novo facioscapulohumeral muscular dystrophy: frequent somatic mosaicism, sex-dependent phenotype, and the role of mitotic transchromosomal repeat interaction between chromosomes 4 and 10. Am J Hum Genet. 2000;66:26–35. doi: 10.1086/302730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Geel M, Dickson MC, Beck AF, Bolland DJ, Frants RR, van der Maarel SM, de Jong PJ, Hewitt JE. Genomic analysis of human chromosome 10q and 4q telomeres suggests a common origin. Genomics. 2002;79:210–217. doi: 10.1006/geno.2002.6690. [DOI] [PubMed] [Google Scholar]
- 20.Thomas NS, Wiseman K, Spurlock G, MacDonald M, Ustek D, Upadhyaya M. A large patient study confirming that facioscapulohumeral muscular dystrophy (FSHD) disease expression is almost exclusively associated with an FSHD locus located on a 4qA-defined 4qter subtelomere. J Med Genet. 2007;44:215–218. doi: 10.1136/jmg.2006.042804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemmers RJ, de Kievit P, Sandkuijl L, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet. 2002;32:235–236. doi: 10.1038/ng999. [DOI] [PubMed] [Google Scholar]
- 22.Lemmers RJ, Wohlgemuth M, Frants RR, Padberg GW, Morava E, van der Maarel SM. Contractions of D4Z4 on 4qB subtelomeres do not cause facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2004;75:1124–1130. doi: 10.1086/426035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemmers RJ, Wohlgemuth M, van der Gaag KJ, van der Vliet PJ, van Teijlingen CM, de Knijff P, Padberg GW, Frants RR, van der Maarel SM. Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2007;81:884–894. doi: 10.1086/521986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemmers RJ, van der Vliet PJ, van der Gaag KJ, Zuniga S, Frants RR, de Knijff P, van der Maarel SM. Worldwide Population Analysis of the 4q and 10q Subtelomeres Identifies Only Four Discrete Interchromosomal Sequence Transfers in Human Evolution. Am J Hum Genet. 2010;86:364–377. doi: 10.1016/j.ajhg.2010.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Maarel SM, Deidda G, Lemmers RJ, Bakker E, van der Wielen MJ, Sandkuijl L, Hewitt JE, Padberg GW, Frants RR. A new dosage test for subtelomeric 4;10 translocations improves conventional diagnosis of facioscapulohumeral muscular dystrophy (FSHD) J Med Genet. 1999;36:823–828. [PMC free article] [PubMed] [Google Scholar]
- 26.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Greef JC, Lemmers RJ, van Engelen BG, Sacconi S, Venance SL, Frants RR, Tawil R, van der Maarel SM. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum Mutat. 2009;30:1–11. doi: 10.1002/humu.21091. [DOI] [PubMed] [Google Scholar]
- 28.Deidda G, Cacurri S, Piazzo N, Felicetti L. Direct detection of 4q35 rearrangements implicated in facioscapulohumeral muscular dystrophy (FSHD) J Med Genet. 1996;33:361–365. doi: 10.1136/jmg.33.5.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masny PS, Bengtsson U, Chung SA, Martin JH, van Engelen B, van der Maarel SM, Winokur ST. Localization of 4q35.2 to the nuclear periphery: is FSHD a nuclear envelope disease? Hum Mol Genet. 2004;13:1857–1871. doi: 10.1093/hmg/ddh205. [DOI] [PubMed] [Google Scholar]
- 30.Lemmers RJ, Osborn M, Haaf T, Rogers M, Frants RR, Padberg GW, Cooper DN, van der Maarel S, Upadhyaya M. D4F104S1 deletion in facioscapulohumeral muscular dystrophy: phenotype, size, and detection. Neurology. 2003;61:178–183. doi: 10.1212/01.wnl.0000078889.51444.81. [DOI] [PubMed] [Google Scholar]
- 31.Matsumura T, Goto K, Yamanaka G, Lee J, Zhang C, Hayashi YK, Arahata K. Chromosome 4q;10q translocations; Comparison with different ethnic populations and FSHD patients. BMC Neurol. 2002;2:7. doi: 10.1186/1471-2377-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemmers RJL. Genotyping of the SSLP, 2008. http://www.urmc.rochester.edu/fields-center/protocols/LeidenFSHDGenotyping.cfm.
- 33.Butz M, Koch MC, Muller-Felber W, Lemmers RJ, van der Maarel SM, Schreiber H. Facioscapulohumeral muscular dystrophy. Phenotype-genotype correlation in patients with borderline D4Z4 repeat numbers. J Neurol. 2003;250:932–937. doi: 10.1007/s00415-003-1116-y. [DOI] [PubMed] [Google Scholar]
- 34.de Greef JC, Frants RR, van der Maarel SM. Epigenetic mechanisms of facioscapulohumeral muscular dystrophy. Mutat Res. 2008;647:94–102. doi: 10.1016/j.mrfmmm.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buzhov BT, Lemmers RJ, Tournev I, Dikova C, Kremensky I, Petrova J, Frants RR, van der Maarel SM. Genetic confirmation of facioscapulohumeral muscular dystrophy in a case with complex D4Z4 rearrangments. Hum Genet. 2005;116:262–266. doi: 10.1007/s00439-004-1237-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.