Abstract
The cornea continues to mature after birth to develop a fully functional, refractive and protective barrier tissue. Here we investigated the complex biological events underlying this process by profiling global genome-wide gene expression patterns of the immature postnatal day 10 and seven week-old adult mouse cornea. The lens and tendon were included in the study to increase the specificity of genes identified as up regulated in the corneal samples. Notable similarities in gene expression between the cornea and the tendon were in the mesenchymal extracellular matrix collagen (types I, III, V, VI) and proteoglycan (lumican, decorin and biglycan) genes. Expression similarities in the cornea and lens were limited to certain epithelial genes and the crystallins. Approximately 76 genes were over expressed in the cornea samples that showed basal expression levels in the lens and tendon. Thirty-two of these were novel with no known functions in the cornea. These include genes with a potential role in protection against oxidative stress (Dhcr24, Cdo1, Akr1b7, Prdx6), inflammation (Ltb4dh, Wdr1), ion-transport (Pdzk1ip1, Slc12a2, Slc25a17) and transcription (Zfp36l3, Pdzk1ip1). Direct comparison of the cornea of two ages showed selective up regulation of 50 and 12 genes in the P10 and adult cornea, respectively. Of the up regulated P10 genes several encode extracellular matrix collagens and proteoglycans that are stable components of the adult cornea and their high transcriptional activity at P10 indicate a period of active corneal growth and matrix deposition in the young cornea. Much less is known about the genes selectively over expressed in the adult cornea; some relate to immune response and innervation (Npy), and possibly to electron transport (Cyp24a1, Cyp2f2) and others of yet unknown functions in the cornea (Rgs10, Psmb8, Xlr4)). This study detected expression of genes with known functions in the cornea, providing additional validation of the microarray experiments. Importantly, it identified several novel genes whose functions have not been investigated in the cornea.
Keywords: Gene expression pattern, cornea maturation, postnatal cornea, adult cornea, corneal stroma, lens, tendon, extracellular matrix
Introduction
The cornea is a unique connective tissue that combines tensile strength, protection against microbial infections, transparency and refractive power for correct vision (Hay 1979; Maurice 1957; Hay and Revel 1969; Chakravarti 2001). The corneal epithelium is five to seven cell-layered, where the innermost undifferentiated basal cells continue to divide to provide daughter cells that differentiate as they move to the outer layers to replace sloughed off cells. The stroma consists of a highly organized extracellular matrix of collagen fibrils interspersed with specialized fibroblasts, the keratocytes. The innermost layer of the cornea is a single cell-layered endothelium, which by virtue of its barrier and pump functions regulates hydration of the stroma (Fischbarg and Lim 1974).
While prenatal development of the cornea has been studied extensively (Pei and Rhodin 1970; Kaufman 1992; Sivak and Sivak 2000), postnatal maturation and growth of the cornea remains relatively obscure. During postnatal maturation as the eyelids open, the onset of epithelial stratification is a major event that prepares the cornea against external factors. The stroma also shows adaptive changes after eyelid opening that are still poorly understood. In the mouse after eyelid opening, there is an initial decrease in thickness when endothelial pump-function ensues. The cornea gradually thickens thereafter until day 30 after birth when adult thickness is achieved (Song et al. 2003). The cornea continues to mature and grow after birth, as shown in the cat for 1–2 years, for the first 8 months in dogs (Moodie et al. 2001; Montiani-Ferreira et al. 2003), and at least six months in humans (Portellinha and Belfort 1991). The stroma is metabolically active during this critical maturation stage. Thus, comparing gene expression differences between the postnatal and adult cornea is likely to identify new genes that regulate maturation and development of the cornea and possibly candidate genes for corneal developmental diseases, dysgenesis, sclerocornea and cornea plana types (OMIM, the Johns Hopkins University, http://www.ncbi.nlm.nih.gov/omim/).
Genomic and proteomic approaches have been used to study the cornea in the context of wound healing, infection and complex diseases like keratoconus (Cao et al. 2002; Varela et al. 2002; Zhou et al. 2007; Karring et al. 2004). We have also investigated the gene expression patterns of cultured stromal keratocytes, fibroblasts and myofibroblasts to investigate changes in the corneal stromal cells during injury and healing (Chakravarti et al. 2004). In that study the cornea samples were used to generate baseline expression levels to identify genes differentially expressed in the corneal cell cultures. One study compared gene expressions in the postnatal nine-day old and the adult cornea by SAGE analysis with an emphasis on epithelial gene expression (Norman et al. 2004). To identify cornea-enriched genes they further compared the two cornea libraries with commercially available brain, limb, heart and embryonic stem cell libraries.
To identify genes showing preferential expression in postnatal day 10 and adult cornea of the mouse, here we compared genome-wide gene expression patterns in the cornea with that of the lens and tendon. The cornea shares many similarities with the tendon in its collagenous extracellular matrix (ECM). Both tissues contain a stromal ECM elaborated by mesenchymal cells. We compared gene expression of the cornea with that of the tendon to identify genes beyond those of the collagen-rich stromal connective tissue, but possibly more typical of the cornea. We also derived gene expression patterns of the adult lens as another ocular tissue. The cornea forms a functional unit with the lens to provide a clear refractive unit for optimal vision (Piatigorsky 1998). From a developmental and cellular point of view the cornea is entirely different from the lens. The lens is epithelial by origin, containing a basement membranous lens capsule, epithelium and epithelial lens fiber with no interstitial ECM. By comparing gene expression patterns of the cornea with that of the lens, we wished to identify similarities that may arise from a similar functional need. Our study identified gene expressions shared by the cornea and lens and novel genes expressed more specifically in the cornea. Finally we compared the total gene expression pattern of the P10 directly with that of the adult cornea to identify genes differentially expressed in the immature and the adult stage cornea. The results provide insights into biological processes defining maturation of the corneal stroma and functions of the stroma in maintaining a refractive, optically transparent, barrier tissue.
Materials and Methods
Animals
All mice were of the outbred CD-1 strain and used according to protocols approved by the Johns Hopkins University Animal Care and Use Committee and guidelines provided by ARVO for the use of animals in vision research.
Target RNA preparation and microarray hybridization
Isolation of total RNA from the cornea samples were described previously (Chakravarti et al. 2004). Briefly, the corneas were dissected free of the limbus from postnatal day 10 (P10) and seven-week old (adult) mice. To enrich for stromal contribution, the epithelium and the endothelium were scraped off and the cornea placed in TRIzol (Invitrogen Life Technologies, Carlsbad, CA). The scraping off of the epithelium reduced, but did not abrogate epithelial contribution, as judged by the expression of known epithelial markers. The lens and the flexor digitorum longus (FDL) tendon were removed from seven-week old adult mice for isolation of total RNA. For the adult cornea three independent preparations of total RNA was generated. For all the other tissues two independent preparations of total RNA were used. The adult and the P10 microarrays were used as baselines in our previous study of corneal cells in culture to identify genes differentially expressed by cultured cells of the stroma (Chakravarti et al. 2004). The additional gene expression raw data for the tendon and lens, and the new gene expression analyses using, a) the tendon as baseline and b) gene expression in the P10 compared to the adult cornea as baseline, will be deposited in the Gene Expression Omnibus repository.
The Affymetrix high-density oligonucleotide Murine Genome_U74Av2 arrays (Affymetrix, San Diego, CA), that probe 9824 unique transcripts, were used according to manufacturer's recommendations. The arrays were hybridized to fragmented biotin-labeled cRNA prepared from the target total RNA, washed (Affymetrix GeneChip protocols) and stained (R-Phycoerythrin Streptavidin) in an Affymetrix GeneChip Fluidics Station 400 and then scanned by a HP GeneArray Scanner (Affymetrix).
Data Analysis
The output fluorescence of the scanned images was analyzed first using the Affymetrix Microarray Suite 5.0 software to compute absolute expression, background calculation and detection call (present, absent or marginal). Background signal was between 42 to 55 arbitrary units, while the noise to signal ratio was between 1.42 to 1.78 arbitrary units. The expression data from all nine arrays were further analyzed as follows. The model-based DNA-Chip Analyzer (dChip) software (http://www.dchip.org/) was used to normalize the data from the image files for array-to-array comparison (Li and Wong 2001). This software uses an invariant-set algorithm as a basis for normalization and a model-based expression index to calculate the expression value for each transcript. To identify gene expression patterns that were statistically significantly different between any two given tissue types, we used the following criteria: (1) fold change ≥ 2; (2) difference of mean expression index ≥ 100 arbitrary units; (3) t-statistic, p < 0.05. Gene annotations and functional information on genes were obtained from NCBI (http://www.ncbi.nlm.nih.gov/) and GeneCards (http://www.genecards.org/).
Semi-quantitative RT-PCR confirmation
Total RNA was isolated from the whole eye, cornea, lens, heart, lung, liver, kidney, and colon using Trizol (Invitrogen) reagent and utilized in subsequent RT-PCR experiments. First strand cDNA synthesis was conducted using the SuperScript III First Strand Synthesis System (Invitrogen Life Technologies, Carlsbad, CA) with random hexamers from 1 µg of total RNA. Gene specific primers were designed and theoretically tested using Primer3 Input, http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi/, and MacVector version 7.2. All gene-specific primer pairs (Supplemental Table 2) were obtained from Operon Biotechnologies, Huntsville, AL., spanning at least two exons to produce PCR products between 200 and 300 base pairs within the open reading frame (ORF). β-actin was used as a control.
Immunoblotting
Ten Corneas and lenses were extracted in 0.5 and 1.0 mL, respectively of T-PER (Pierce, Rockford, IL) with protease inhibitor cocktail (Sigma, St Louis, MO). The samples were homogenized and centrifuged at 10,000 × g for 5 minutes and the supernatants were collected. Protein concentrations were estimated by using BCA protein assay kit (Pierce, Rockford, IL) and the final concentrations adjusted to 740 µg/mL. To detect specific proteins and actin as a control, 15 µg and 5 µg of total protein, respectively, were resolved by SDS polyacrylamide gel electrophoresis, and transferred to nitrocellulose membrane. The membrane was blocked with 5% non fat dry milk in 10 mM Tris-HCl (pH 7.4), 20 mM KCl, 150 mM NaCl, 0.05% Tween-20 and probed with antibodies against PDCD4 (Ana Spec, San Jose, CA), prominin-1 (eBioscience, San Diego, CA), plectin-1 (Novus Biologicals, Littleton, CO) and horse radish peroxidase conjugated donkey anti-rabbit (Amersham Biosciences, Piscataway, NJ) as the secondary antibody.
Results
Genome-wide gene expression in the cornea, lens and tendon
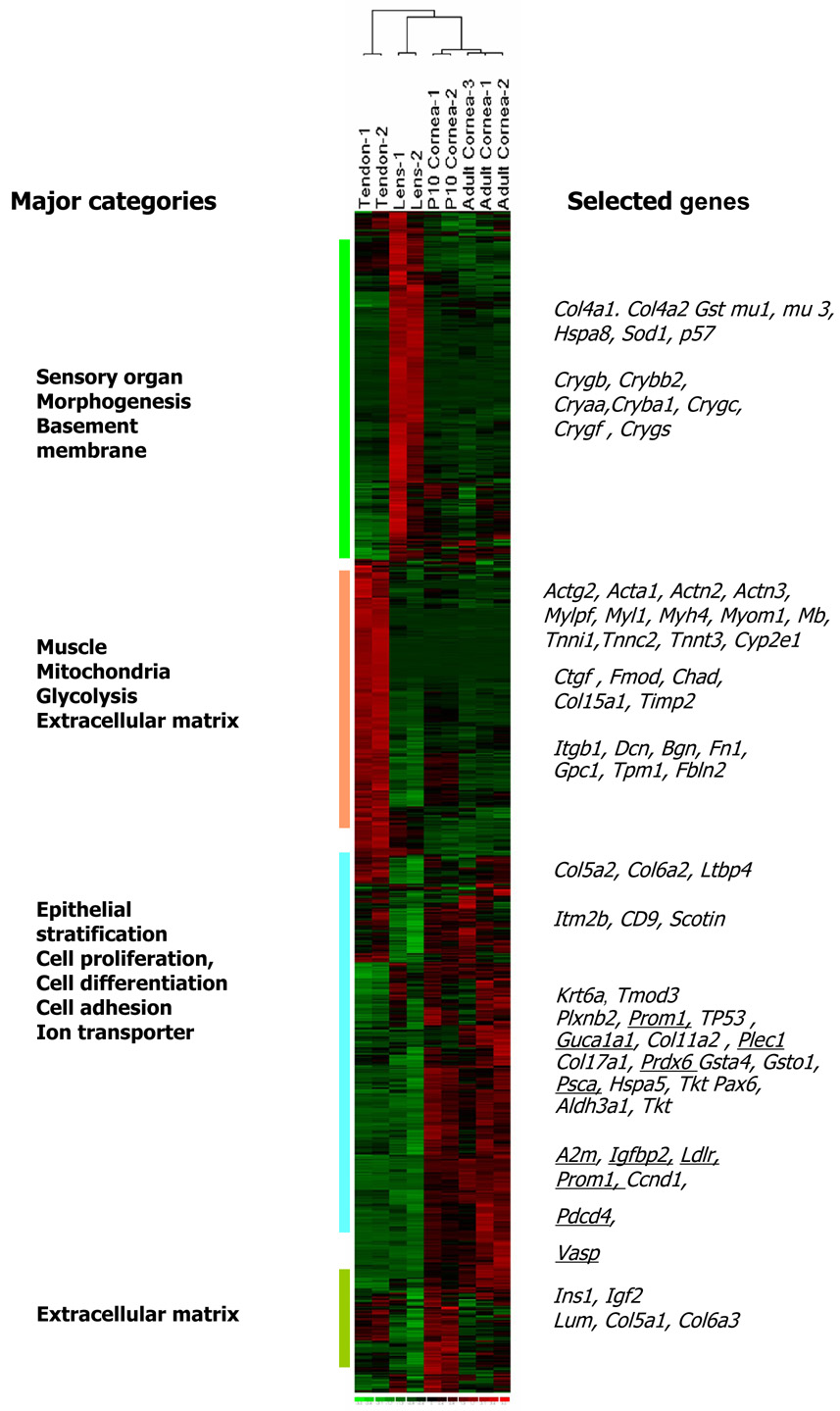
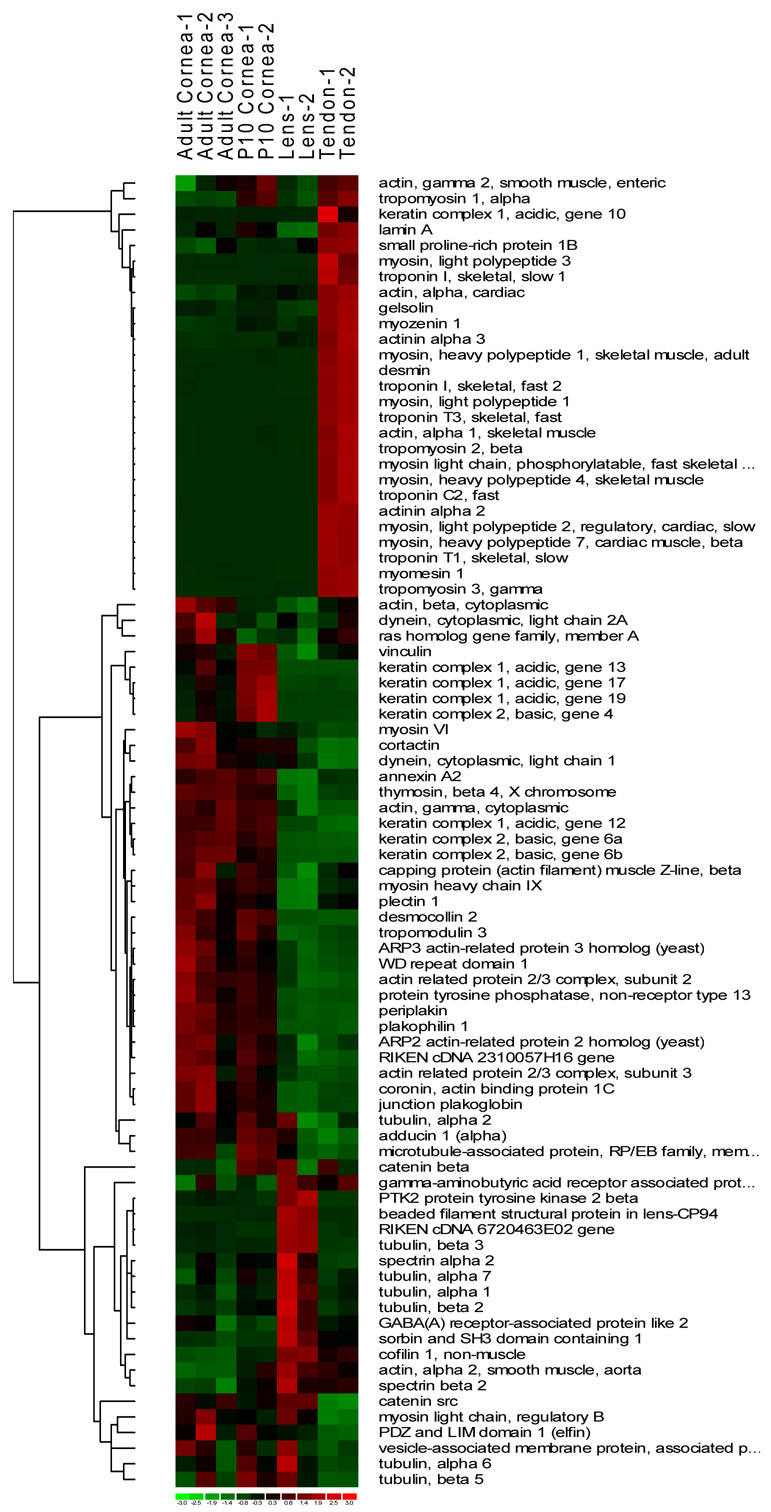
Approximately 45% of all genes tested were expressed in at least one tissue type examined. A fourth of these (1087 genes) were present at high levels (≥ 500 expression index) in at least one of the tested tissue samples (Fig.1). A complete list of these genes and their expression levels are presented in Supplemental Table 1. The complete dataset is available at NCBI Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo. The heat image of expression levels for the 1087 genes show three major areas of tissue-biased expression (Fig.1). Genes preferentially expressed in the cornea included epithelial stratification and differentiation, desmosomal, hemidesmosomal, epithelial, and endothelial cell junction-related genes, connexin 43 (Gja1/Cxc43), gap junction beta2 (Gjb2/Cxc26), desmocollin2 (DSc2), collagen type XVII, alpha 1(Col17a1), cytoskeletal genes, keratin 17, 19 (Krt17,Krt19), tropomodulin 3 (Tmod3), steroid and sterol metabolism (Hmgcs1, Ldlr, Sc4mol, Dhcr24) and transcription factor genes (Tcfap2a, Pax6). Because the epithelium is the most metabolically active segment of the cornea attempts were made to minimize its contribution to the transcript pool by scraping off the epithelial layers. Nevertheless the total RNA contained some epithelial transcripts. Additional over expressed genes in the cornea included those encoding soluble metabolic enzymes that may contribute to corneal transparency and refractivity, such as aldehyde dehydrogenase and transketolase (Aldh3a, Tkt) (Jester et al. 1999).
Fig. 1.
Genome-wide gene expression differences between cornea samples, tendon and lens. Heat image of 1087 genes with ≥ 500 arbitrary expression units in at least one sample type shown in green (below mean), black (mean) and red (above mean). Four major functional groups of genes differentially expressed in the different tissue types are shown on the left margin. Symbols of specific genes as they appear across all samples are shown on the right, and underlined if validated by RT-PCR or immunoblotting.
The major lens genes included sensory organ and morphogenesis related crystallin genes, Crygb, Cryaa, Cryba1, Crygc, Crygf, Crybb2 and Crygs. The tendon gene expression pattern was dominated by muscle and mitochondria/energy pathway related genes, actin (Actg2, Acta1), troponins (Tnni1, Tnnt3, Tnnc2), actinins (Actn2, Actn3) and myosin genes (Myl3, Myl2, Mylpf, Myh7, Myh4). There were some specific overlaps in gene expression between the adult tendon and the P10 cornea. These include genes pertinent to tissue growth and extracellular matrix deposition, insulin1, insulin-like growth factor binding protein 4, several collagens (Col5a1, Col5a2, Col6a2, Col6a3) and collagen fibril associating proteoglycans, decorin, lumican and biglycan (Dcn, Lum and Bgn).
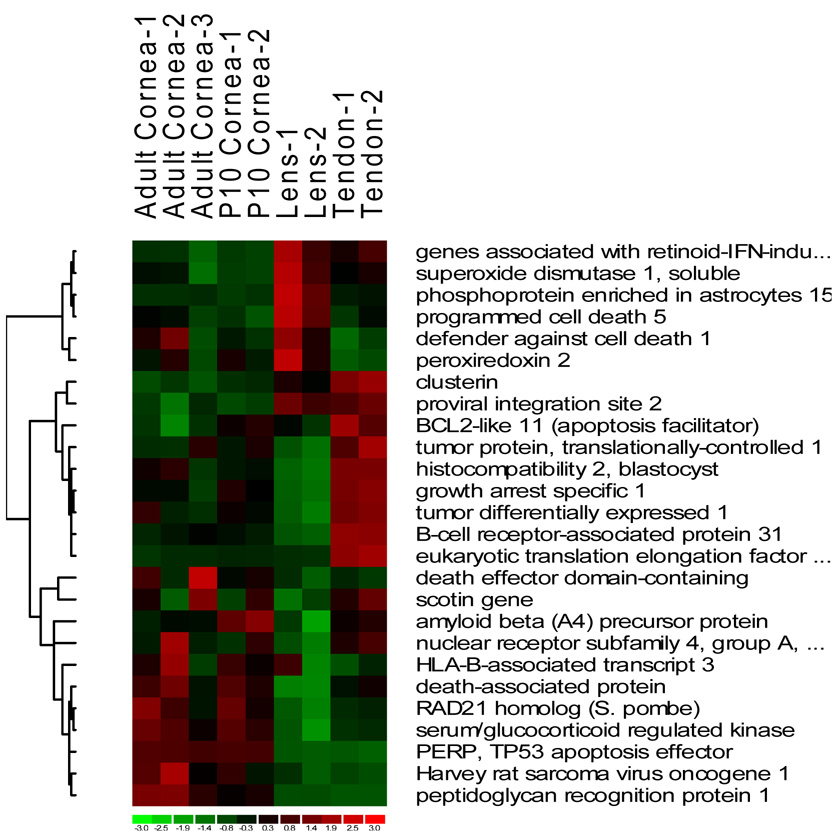
We compared expression patterns in the three tissue types with respect to key functional groups, cell cycle and death, cell adhesion, cytoskeleton, ECM, and immune/defense response (Fig. 2). Overlaps between the cornea and tendon in cell death/cell cycle-related genes include tumor protein translationally controlled (Tpt1), tumor differentially expressed (Tde1/Serinc3) with a possible role in preventing cell death, Gas1 (growth arrest specific 1) regulating mesenchymal cell growth, and Scotin, regulating p53 dependent apoptosis (Fig. 2A). There was little overlap between the cornea and the lens in this functional group.
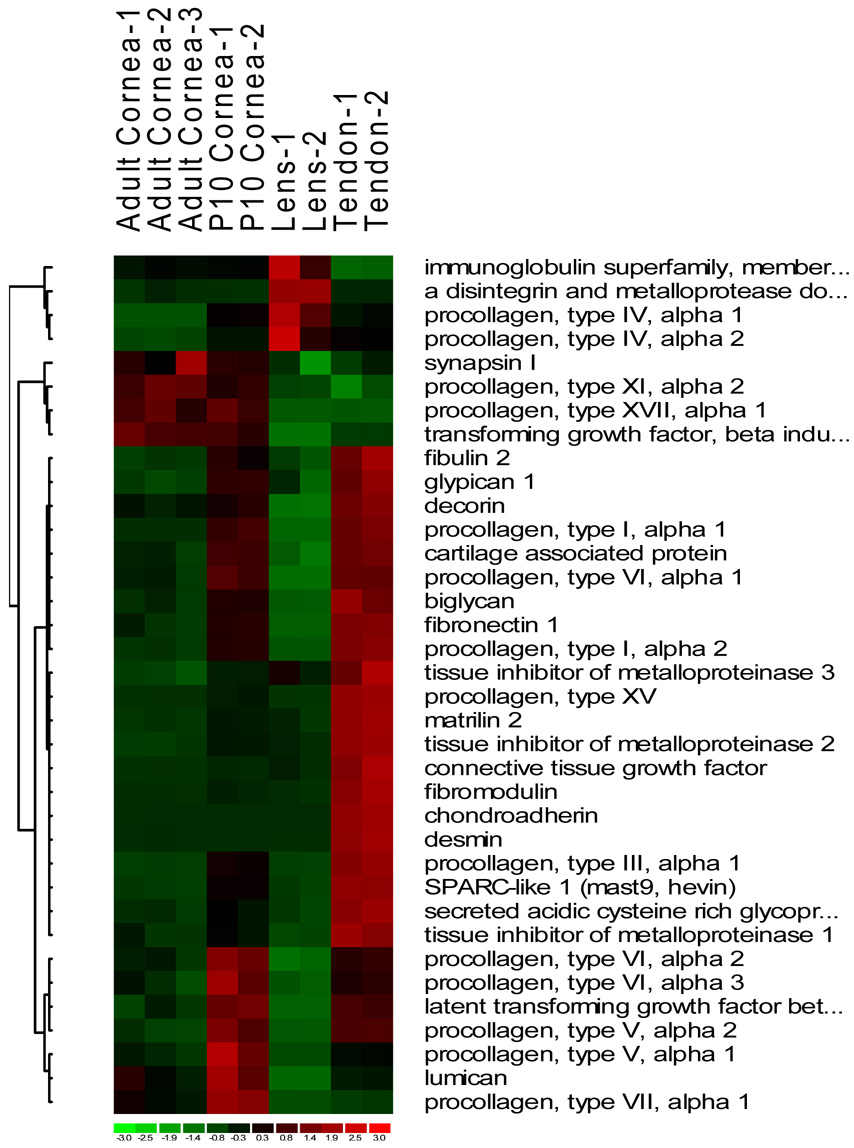
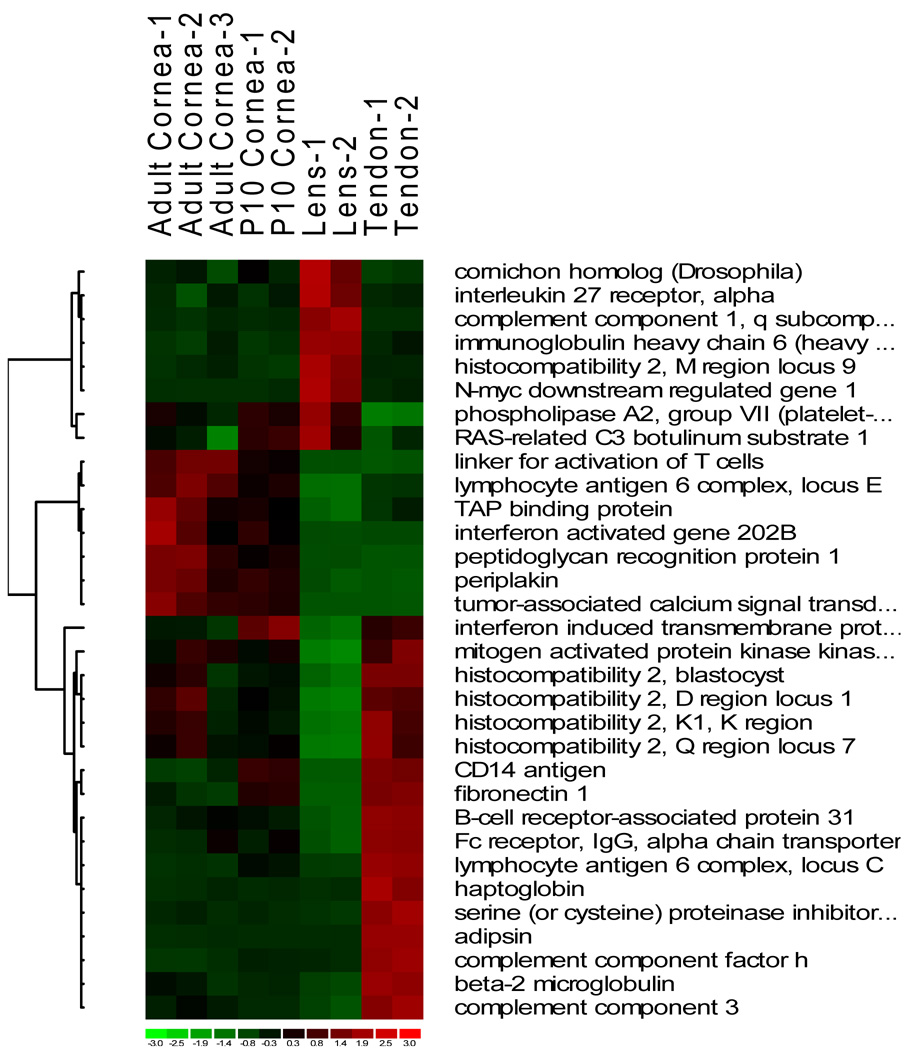
Fig. 2.
Heat images of differentially expressed genes from four major functional groups, cell death and cell cycle (A), extracellular matrix (B), defense and immune response (C) and cytoskeleton (D).
Compared to the adult cornea the developing P10 cornea shared a greater similarity with the tendon in ECM gene expression patterns (Fig. 2B). Thus, genes for the stromal fibrillar collagens I, III and V, and cell adhesive fibronectin, fibulin, collagen VI and protocadherin were all transcriptionally active in the maturing P10 and in the adult tendon which also undergoes constant remodeling. These proteins are all significant components of the adult cornea. However, due to little turnover in the adult cornea, these are not very active at the transcriptional level. Lum and Dcn, encoding the proteoglycans lumican and decorin, were expressed at high levels in the P10 cornea and tendon, and detectably over expressed in the adult cornea as well*Fug. 2B and supplemental Table 1). Unlike the tendon, the lens is of epithelial origin with no mesenchymal fibroblasts and shows little in common with the cornea with respect to ECM. Beyond the genes for epithelial basement membrane collagen type IV, our microarrays showed little expression of interstitial ECM genes in the lens. Other ECM genes barely expressed in either the lens or the cornea, but expressed at high levels in the tendon were Timp1 and Timp2 (Tissue inhibitor of metalloproteinase), Fmod (Fibromodulin) and Chad (Chondroadherin).
There were 30 differentially expressed genes in the immune response/defense category (Fig. 2C). The cornea samples showed approximately 22% (8/30) similarity in expression pattern with the tendon. A number of MHC (major histocompatibility) class I genes (H2-bl, H2-k1, H2-d1, H2-q7) were expressed at above mean levels in the cornea and tendon. Tapbp (TAP binding protein) regulating antigen processing and presentation was over expressed in the cornea, particularly in the adult group. Only 8 of the defense related genes were expressed at above mean levels in the lens, of which one, Pla2g7 (phospholipase A2, group VII) was also over expressed in the cornea.
With the exception of a few similarities the gene expression pattern of the cytoskeleton group was quite distinctive for the three tissue types (Fig. 2D). Ctnnd1 (Catenin src/cadherin associated protein), an epithelial/ endothelial adherens junction protein with a possible role in leukocyte transmigration was expressed at above mean levels in the lens and the cornea. Epithelial tubulin genes (Tuba1c and Tubb5) were also over expressed in the lens and the cornea. The cornea and the tendon shared over expression patterns for actin (Actg2, Actb), tropomyosin (Tpm1), dynein (Dynlrb1) and vinculin (Vcl) in the cytoskeleton group (Fig. 2D)
Genes over expressed in the cornea compared to the tendon as baseline
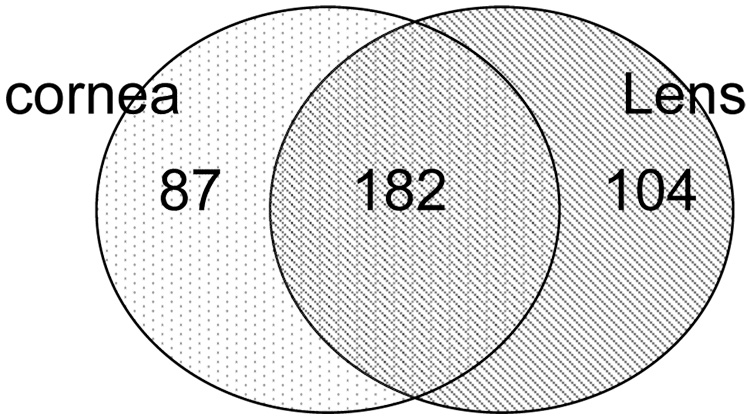
To identify genes expressed in the cornea primarily, we used the expression pattern of the tendon as baseline and selected 531 genes expressed at ≥ 2 fold difference in the cornea and lens. Of these 158 had high basal expression in the tendon and therefore, removed from the analysis to further focus on those that were primarily elevated in the lens and cornea only (Fig. 3). Of the remaining 373 genes, 104 were over expressed in lens; another 182 were expressed in the cornea and lens (the overlapping region in the Venn diagram); these include transketolase (Tkt), aldehyde dehydrogenases (Aldh1a1, Aldh3a1) and certain glutathione S transferases (Gsto1, Gstm1, Gstm3) and crystallin genes (Cryaa, Cryab1, Crygc, Crygf, CrygS). A group of 76 genes (Table 1) and 11 ESTs (not shown) had elevated expression in the cornea with close to background-level expression (100 arbitrary units) in the tendon and lens. These corneal genes include those known to play a role in ocular development (Pax6, Notch3), stem cells, desmosomal and hemidesosomal adhesion, differentiation and stratification (Prom1, Jup, Jam-A/F11r, Plec1, Pkp1, Col17a1), cytoskeletal functions (Krt13, Krt17, Krt19, Krt6b), metabolism (Acly, Adh1, Ces3, Cpd, Fdps) and antioxidative, detoxification properties (Gsta4, Prdx6). Functions of several genes are known based on studies directly on the cornea. However, 32/76 genes are novel and have not been investigated in the cornea. Based on what is known about these genes in other systems, we indicated their speculative role in the cornea. These include several transcription factors (Plac8, Phf7, Zfp36l3), cell cycle regulators (Brd4, Rad21, Rod1) and genes encoding oxidative stress-related proteins (Akr1b7, Cdo1, Prdx6).
Fig. 3.
Genes over expressed in the cornea compared to the tendon as baseline. An initial set of 531 were expressed at ≥ 2 fold in the cornea and lens compared to the tendon as baseline. Of these 158 were taken out because of their high basal expression in the tendon. The distribution of the remaining 373 genes in the cornea (P10 + adult) and lens are shown.
Table 1. Genes differentially expressed in the cornea compared to tendon as baseline.
Genes marked with ~ were novel to the cornea and their functions postulated based on their reported functions in other systems.
| Symbol | Gene Name | Function in the cornea | Adult/td1 | P10/td2 |
|---|---|---|---|---|
| A2m | Alpha-2-macroglobulin | Protease inhibitor | 1095 | 1125 |
| Acly | ATP citrate lyase | ~Lipid synthesis (synthesis of acetyl CoA) | 3 | 3 |
| Actn4 | Actinin alpha 4 | adherens and tight junctions | 9 | 7 |
| Adh1 | Alcohol dehydrogenase 1 (class I) | Detoxification | 211 | 209 |
| Agrn | Agrin | Synaptic transmission | 2 | 5 |
| Aim1 | Absent in melanoma 1(HGNC1 or AIM1) | ~Beta gamma crystallin family member | 24 | 21 |
| Akr1b7 | Aldo-keto reductase family 1, member B7 | ~Oxidative stress protection | 10 | 18 |
| Bat9 | HLA-B-associated transcript 9 | ~HLA-B associated, transcription regulation | 21 | 21 |
| Brd4 | Bromodomain containing 4 | ~Cell cycle regulation | 2 | 2 |
| Capg | Actin filament capping, gelsolin-like | Corneal transparency | 5 | 4 |
| Cdo1 | Cysteine dioxygenase 1, cytosolic | ~Oxidative stress-related | 5 | 6 |
| Ces3 | Carboxylesterase 3 | Peptide processing | 4 | 2 |
| Cldn7 | Claudin 7 | Epithelial tight junction | 824 | 335 |
| Clic1 | Chloride intracellular channel 1 | Oxidative stress-related | 4 | 3 |
| Col17a1 | Procollagen, type XVII, alpha 1 | ECM | 37 | 38 |
| Col5a1 | Procollagen, type V, alpha 1 | ECM | 1 | 3 |
| Col7a1 | Procollagen, type VII, alpha 1 | ECM | 2 | 4 |
| Coro1c | Coronin, actin binding protein 1C | ~Actin binding, cytokinesis | 4 | 3 |
| Cpd | Carboxypeptidase D | Peptide processing | 3 | 4 |
| Ctsh | Cathepsin H | Protease | 3 | 5 |
| Cyfip1 | Cytoplasmic FMR1 interacting protein 1 | Regulation of actin cytoskeleton | 4 | 3 |
| Dhcr24 | 24-dehydrocholesterol reductase | ~Oxidative stress-related | 8 | 4 |
| Dsc2 | Desmocollin 2 | Epithelial differentiation | 169 | 196 |
| Elf3 | E74-like factor 3 | Epithelial differentiation | 26 | 16 |
| JAM-A | F11 receptor | Tight junction | 7 | 6 |
| F3 | Coagulation factor III | Blood vessel dev | 11 | 19 |
| Fdps | Farnesyl diphosphate synthetase | ~Biosynthesis | 63 | 98 |
| Fxyd3 | FXYD domain-containing | ~Na, K ATPase regulator | 7 | 5 |
| Gjb2 | Gap junction membrane channel beta 2 | Gap junction | 14 | 14 |
| Gsta4 | Glutathione S-transferase, alpha 4 | Detoxification | 32 | 34 |
| Hspa1a | Heat shock protein 1A | Protein chaperone | 119 | 69 |
| Itgb4 | Integrin beta 4 | epithelial migration and healing | 29 | 22 |
| Jup | Junction plakoglobin | epithelial migration and healing | 13 | 12 |
| Krt13 | Keratin complex 1, acidic, gene 13 | Epithelial cytoskeletal | 39 | 74 |
| Krt17 | Keratin complex 1, acidic, gene 17 | ~Epithelial cytoskeletal | 7 | 19 |
| Krt19 | Keratin complex 1, acidic, gene 19 | Epithelial cytoskeletal | 16 | 61 |
| Krt6b | Keratin complex 2, basic, gene 6b | ~Epithelial cytoskeletal | 33 | 21 |
| Ldlr | Low density lipoprotein receptor | Lipid metabolism | 6 | 3 |
| Lmo7 | LIM domain only 7 | ~Protein-degradation pathway | 6 | 6 |
| Ltb4dh | Leukotriene B4 12-hydroxydehydrogenase | ~Inflammation regulation | 45 | 25 |
| Mal | myelin and lymphocyte protein, T-cell | Lipid raft, innate immune response | 38 | 29 |
| Mknk2 | MAP kinase-interacting serine/threonine kinase 2 | ~G protein-coupled receptor kinase | 22 | 14 |
| Myo6 | Myosin VI | Cytoskeleton | 6 | 2 |
| Ncoa4 | Nuclear receptor co-activator 4 | ~Receptor tyrosine kinase signaling | 5 | 2 |
| Notch3 | Notch gene homolog 3 (Drosophila) | Cell-fate determination. | 3 | 4 |
| Npy | Neuropeptide Y | Corneal innervation | 4 | 3 |
| Ostf1 | Osteoclast stimulating factor 1 | ~Src-signaling | 24 | 5 |
| Pax6 | Paired box gene 6 | Morphogenesis | 4 | 2 |
| Pdcd4* | Programmed cell death 4 | Growth regulation | 219 | 269 |
| Pdzk1ip1 | PDZK1 interacting protein 1 | ~Na/phosphate transporter | 4 | 3 |
| Perp | PERP, TP53 apoptosis effector | Cell cycle regulation | 20 | 20 |
| Pglyrp1 | Peptidoglycan recognition protein 1 | ~Antimicrobial | 58 | 32 |
| Phf7 | PHD finger protein 7 | ~Transcription factor | 3 | 1 |
| Pkp1 | Plakophilin 1 | Desmosomal protein | 47 | 39 |
| Plac8 | Placenta-specific 8 | ~Transcription factor | 160 | 36 |
| Plec1* | Plectin 1 | Hemidesmosomal protein | 2 | 1 |
| Ppl | Periplakin | ~Epithelial barrier function | 8 | 7 |
| Ppp1r14b | Protein phosphatase 1, regulatory subunit 14B | ~Epithelial cell migration | 3 | 2 |
| Prdx6 | Peroxiredoxin 6 | ~Oxidative stress-related | 5 | 4 |
| Prom1* | Prominin 1 | CD133, Stem cell marker | 55 | 61 |
| Psca | Prostate stem cell antigen | Cell proliferation | 59 | 2 |
| Ptpn13 | Protein tyrosine phosphatase, type 13 | ~Cytoskeleton-plasma membrane interacting | 7 | 6 |
| Rad21 | RAD21 homolog (S. pombe) | ~ Ptpn13Cell cycle regulation | 2 | 2 |
| Rod1 | ROD1 regulator of differentiation 1 | ~Cell differentiation | 3 | 3 |
| Sc4mol | Sterol-C4-methyl oxidase-like | Unkown | 4 | 2 |
| Scel | Sciellin | ~Epithelial stratification/barrier functions | 7 | 8 |
| Sdc1 | Syndecan 1 | Epithelial cell migration | 30 | 23 |
| Serpinb5 | Serine (or cysteine) proteinase inhibitor | Antiproteinase | 20 | 13 |
| Slc12a2 | Solute carrier family 12, member 2 | ~Ion transport | 5 | 4 |
| Slc25a17 | Solute carrier family 25 member 17 | ~Ion transport | 3 | 3 |
| Smarcd2 | SWI/SNF related, matrix associated | ~Notch signaling and development | 4 | 3 |
| Tacstd2 | Tumor-associated calcium signal transducer2 | Cell adhesion/Gelatinous corneal dystrophyIII | 1823 | 1284 |
| Tgm2 | Transglutaminase 2, C polypeptide | deamidation/Detoxification | 5 | 0.4 |
| Txnl5 | Thioredoxin-like 5 | Oxidative stress protection | 5 | 4 |
| Wdr1 | WD repeat domain 1 | ~neutrophil functions | 3 | 2 |
| Zfp36l3 | Zinc finger protein 36, C3H type-like 1 | ~response to growth factors | 3 | 3 |
Confirmed by immunoblotting (see Figure 5).
Function in the cornea predicted from studies in other systems.
Mean gene expression in adult cornea versus mean gene expression in tendon
Mean gene expression in P10 cornea versus mean gene expression in tendon
Genes differentially expressed in the adult and P10 cornea
To identify genes that may have potentially important roles in the postnatal maturation of the cornea, or in the maintenance of the adult cornea, we compared the expression of 4000 genes (remaining after removing those expressed at the background level) in the cornea of two ages. Only 50 genes were selectively up regulated in the P10 cornea compared to the adult, while another 12 genes were up regulated in the adult (Table 2). Genes over expressed in the P10 cornea included those regulating cell growth (Cri1/ Eid-1, Cdknc1, Emp2, Pten, Peg3), extracellular matrix synthesis (Postn, Col1a1, Col1a2, Col3a1, Col4a1, Col4a2, Col6a2, Col8a1, Col14a1, Eln), and corneal innervation (Sema4a, Nrp1). Genes over expressed in the adult cornea include mitochondrial respiration-related genes Cyp24a1 and endoplasmic reticular Cyp2f2. However, not much is known about either in the context of the cornea or the whole eye. Psca is another gene that is highly over expressed in the adult eye, a similar observation was reported in an earlier SAGE analysis of the cornea (Norman et al. 2004). Its function in the cornea is not known.
Table 2. Fold change in gene expression in the P10 compared to the adult cornea.
Genes differentially expressed in the adult versus postnatal day 10 cornea. A total of 50 genes over expressed in the P10 cornea appear as positive numbers and 12 over expressed in the adult cornea as negative numbers.
| Symbol | Gene | Function | *Fold change |
|---|---|---|---|
| 5730454B08Rik | RIKEN cDNA 5730454B08 gene | Unknown | 3.8 |
| Anpep | Alanyl (membrane) aminopeptidase | Proteolysis (angiogenesis, antimicrobial) | 2.6 |
| Cd14 | CD14 antigen | Innate immune response | 2.6 |
| Cd63 | CD63 antigen | Growth regulation | 2.8 |
| Cdkn1c | Cyclin-dependent kinase inhibitor 1C (P57) | Cell growrth regulation | 9.0 |
| Clic4 | Chloride intracellular channel 4 (mitochondrial) | Apoptosis (p53 mediated) | 3.4 |
| Col14a1 | Procollagen, type XIV, alpha 1 | ECM (collagen fibril organization) | 6.4 |
| Col1a1 | Procollagen, type I, alpha 1 | ECM (fibrillar collagen) | 2.8 |
| Col1a2 | Procollagen, type I, alpha 2 | ECM (fibrillar collagen) | 3.8 |
| Col3a1 | Procollagen, type III, alpha 1 | ECM (fibrillar collagen) | 55.6 |
| Col4a1 | Procollagen, type IV, alpha 1 | ECM (basement membrane collagen) | 4.6 |
| Col4a2 | Procollagen, type IV, alpha 2 | ECM (basement membrane collagen) | 3.2 |
| Col6a2 | Procollagen, type VI, alpha 2 | ECM cell adhesive | 2.5 |
| Col8a1 | Procollagen, type VIII, alpha 1 | ECM (cell adhesive collagen) | 6.2 |
| Cri1 | CREBBP/EP300 inhibitory protein 1 | Transcription Factor | 3.1 |
| Ctsk | Cathepsin K | Proteolysis | 2.8 |
| Cyp2a4 | Cytochrome P450, family 2, subfamily a, polypeptide 4 | Electron transport | 3.3 |
| Ddr2 | Discoidin domain receptor family, member 2 | (collagen-receptor for dendritic cells | 5.0 |
| Eln | Elastin | ECM (tissue elastomeric properties) | 6.3 |
| Emp2 | Epithelial membrane protein 2 | Cell proliferation/apoptosis | 3.1 |
| Fbn1 | Fibrillin 1 | ECM (connective tissue microfibril) | 3.0 |
| Fgf2 | Fibroblast growth factor receptor 2 | Angiogenesis | 3.2 |
| Fndc1 | Fibronectin type III domain containing 1 | ECM (cell adhesive) | 2.4 |
| Fstl1 | Follistatin-like 1 | Cell proliferation/differentiation | 4.6 |
| Ggh | Gamma-glutamyl hydrolase | Proteolysis (exipeptidase) | 2.8 |
| Gpc3 | Glypican 3 | Cell growth and/or maintenance | 2.8 |
| Gtl2 | Maternally expressed untranslated mRNA | Unknown | 4.4 |
| H0D4S114 | DNA segment, human D4S114 | Unknown | 3.8 |
| Ifitm3 | Interferon induced transmembrane protein 3 | Immune response | 2.8 |
| Igfbp4 | Insulin-like growth factor binding protein 4 | Cell growth regulation | 2.7 |
| Itm2a | Integral membrane protein 2A | Cell differentiation | 5.0 |
| Krt1-19 | Keratin complex 1, acidic, gene 19 | Cytoskeleton (intermediate filament) | 3.9 |
| Ltbp4 | Latent transforming growth factor beta binding protein 4 | TGF beta signaling | 3.2 |
| Mfap2 | Microfibrillar-associated protein 2 | Connective tissue microfibril | 4.3 |
| Nrp1 | Neuropilin | Chemorepulsive axon guidance | 4.0 |
| Peg3 | Paternally expressed 3 | Transcription factor neuronal death via p53/Bax | 8.4 |
| Pitpnb | Phosphotidylinositol transfer protein, beta | Lipid-mediated signaling | 3.6 |
| Postn | Periostin, osteoblast specific factor | Collagen fubrl assembly (Akt/PKB ) | 2.9 |
| Ppic | Peptidylprolyl isomerase C | Protein folding | 4.1 |
| Prss11 | Protease, serine, 11 (Igf binding) | regulates IGF release and availability | 3.2 |
| Pten | Phosphatase and tensin homolog | Cell cycle regulator (tumor suppressor) | 7.1 |
| Rest | RE1-silencing transcription factor | Unknown | 4.9 |
| Rnase4 | Ribonuclease, RNase A family 4 | Monocyte differentiation | 3.2 |
| Sema4a | Semaphorin 4A | Chemorepulsive axon guidance | 4.3 |
| Serpinh1 | Serine/cysteine proteinase inhibitor, clade H, member 1 | Proteinase inhibitor | 3.1 |
| Sf3b1 | Splicing factor 3b, subunit 1 | RNA processing/gene regulation | 5.5 |
| Sfrs7 | Splicing factor, arginine/serine-rich 7 | RNA processing/gene regulation | 3.3 |
| Slc1a4 | Solute carrier family 1member 4 | Glutamate/neutral amino acid transporter | 2.8 |
| Sparcl1 | SPARC-like 1 (mast9, hevin) | ECM (serine protease inhibitor) | 4.0 |
| Zfp148 | Zinc finger protein 148 | Transcription factor (apoptosis regulation) | 2.7 |
| 2300003P22Rik | RIKEN cDNA 2300003P22 gene | Unknown | −4.7 |
| Cyp24a1 | Cytochrome P450, family 24, subfamily a, polypeptide 1 | Electron transport | −12.3 |
| Cyp2f2 | Cytochrome P450, family 2, subfamily f, polypeptide 2 | Electron transport | −4.3 |
| Expi | Extracellular proteinase inhibitor | proteinase inhibitor | −3.6 |
| Guca1a | Guanylate cyclase activator 1a (retina) | Calcium binding (photoreceptor activity) | −3.4 |
| Mknk2 | MAP kinase-interacting serine/threonine kinase 2 | Cell proliferation/differentiation | −3.1 |
| Npy | Neuropeptide Y | Cell migration, anti-angiogenic | −4.8 |
| Plac8 | Placenta-specific 8 | Immune response | −4.5 |
| Psca | Prostate stem cell antigen | Cell growth or immune response | −32.8 |
| Psmb8 | Proteosome (macropain) subunit, beta type 8 | Antigen processing | −3.4 |
| Rgs10 | Regulator of G-protein signalling 10 | Unknown | −2.6 |
| Xlr4 | X-linked lymphocyte-regulated 4 | Unknown | −4.0 |
Mean gene expression in P10/Adult cornea
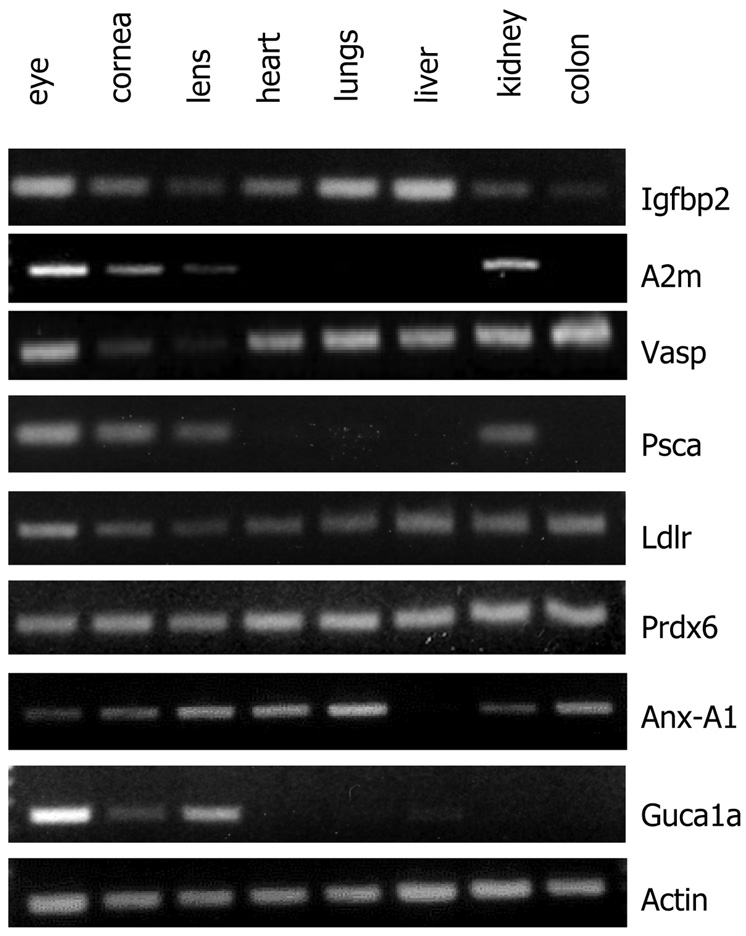
Validation of expressions of selected genes by RT-PCR
Semi-quantitative RT-PCR analyses were used to test expressions of selected genes in total RNA from whole eyes, cornea, lens and a panel of other organs and tissues not tested by microarray gene expression profiling. Our goal was to confirm expression of genes in the cornea and lens identified by the microarray experiments and to determine their extent of expression in other organs and tissues. Therefore we selected a few genes whose detection in the cornea was relatively novel, and by microarray appeared to have high expression in the cornea. The microarrays indicated high levels of Igfbp2 transcript in the P10 and adult cornea and low levels in the lens (Fig. 1 and Supplemental Table 1). Our RT-PCR results on Igfbp2 confirmed high expression in the cornea, but also strong expression in the heart, lungs, liver, and weaker, but measurable expression in the lens, kidney and colon. The RT-PCR results also confirmed microarray expression of A2m, Vasp, and Psca in the cornea. The A2m transcript was also detected in whole eyes, cornea, lens and kidney. By microarray the Psca transcript was detected at high levels in the adult cornea only, and by RT-PCR we detected it in total RNA from whole eyes, adult cornea, lens and kidney. In addition, the RT-PCR results confirmed microarray-detected Ldlr, Prdx6 and Anxa1 in the cornea and showed their wide spread expression in other tissues as well. Guca1a is known to be expressed in the retina and associated with cone dystrophy; its transcript was detected in the microarray profiles at above 500 arbitrary units in the adult cornea and at basal levels in the P10 (Table 2). We tested its expression by RT-PCR and detected it in total RNA from whole eyes and also from the cornea and lens. Whether it is measurably expressed at the protein levels in the lens and cornea remains to be seen.
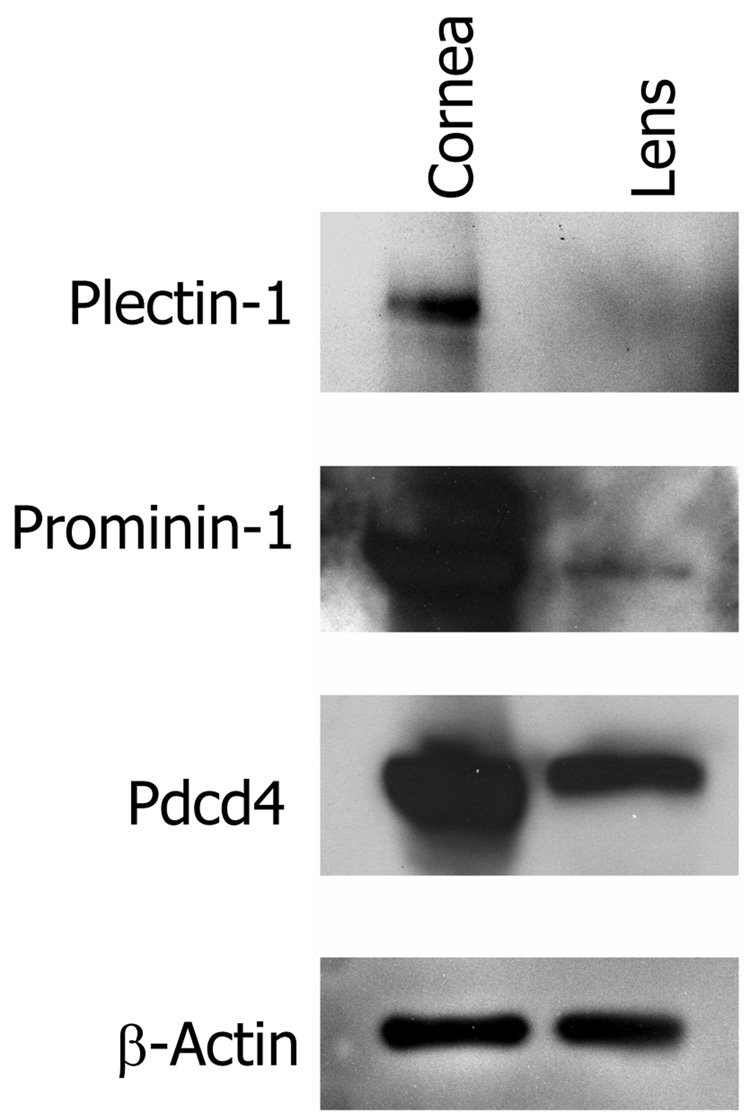
Validation of selected proteins by immunoblotting
We selected three genes that were identified as over expressed in the adult and P10 cornea by microarray, for further identification of the encoded proteins by immunoblotting (Fig. 5). These included Plectin-1 (Plec1), Programmed cell death 4 (Pdcd4) and Prominin-1 (Prom1). The primary reasons for selecting these three genes were a) availability of antibodies and b) their relative novelty to the cornea. All three proteins were detected at high levels in the cornea. Plectin-1 was not present in the lens preparations, whereas prominin-1 was detected at low levels in the lens as well. The Pdcd4 protein was present at high levels in the cornea and at moderately high levels in the lens.
Fig. 5.
Detection of Plectin-1, Prominin-1 and Pdcd 4 by immunoblotting. Total protein was extracted from isolated adult cornea and lens and selected proteins detected using Protein-specific primary antibodies and horse radish peroxidase conjugated donkey anti-rabbit secondary antibody.
Discussion
We analyzed gene expression patterns of the young and adult cornea compared to tendon and lens to gain insight into corneal maturation and biological processes that define the cornea. Genes differentially expressed in the cornea, were related to epithelial stratification, protection against oxidative stress, detoxification and antimicrobial activities. The cornea, particularly at the developing P10 stage showed similarities with the tendon in over expressions of ECM genes encoding fibrillar collagens and proteoglycans indicating common mesenchymal origin of these two tissues. Additional overlaps included expression of mitochondrial, cytoskeletal, and immune function-related genes. Similarities in gene expression between the cornea and the lens were limited to several crystallins, certain detoxification genes, and a few epithelial and basement membrane genes.
Direct comparison of the immature P10 to the adult cornea, not surprisingly, showed a large overlap in their gene expression patterns. Thus, only 50 transcripts were differentially over expressed in the P10 compared to the adult cornea while even fewer genes (12 to be specific) were selectively up regulated in the adult. Genes selectively over expressed in the immature cornea indicate three broad categories of biological processes underlying postnatal development of the cornea. These include cell proliferation, expansion of the extracellular matrix and corneal innervation. In the adult cornea the corneal basal cells proliferate rapidly, the stromal keratocytes are quiescent (G0 stage) and proliferate upon corneal injury (Francesconi et al. 2000). The endothelial cells are also quiescent, arrested at the G1 stage (Joyce et al. 1996). In the immature cornea, cells proliferate in the epithelial, stromal and endothelial layer. The balance between cell proliferation and differentiation may be attained by a number of genes, some promoting and others restricting cell proliferation (Zieske et al. 2004). Cyclin D1 (Ccnd1), a positive regulator of cell cycle was elevated in the adult and young cornea. Cri1 (alias Eid-1) encodes a transcription factor that binds the Rb1 and a co-activator, Ep300 (MacLellan et al. 2000). Its up regulation in the P10 cornea is suggestive of an effort towards exiting from cell cycle and passage to differentiation. Also elevated in the P10 cornea was Cdknc1 (p57), an inhibitor of cyclin-dependent kinase that restricts cell proliferation. Pten, expressed at high levels in the P10 may restrict cell survival functions of Akt1 and may also serve to restrict cell proliferation in the immature cornea. An earlier study detected Pten in the corneal endothelium and suggested its role in contact inhibition of endothelial cells (Chen et al. 2005). Emp2 may mediate trafficking of diverse proteins such as α6β1 integrin and MHC class I to lipid raft microdomains and function in epithelial membrane cycling (Wadehra et al. 2003). Gpc, encoding a cell surface heparan sulfate proteoglycan, glypican, is expressed in various embryonic tissues and known to regulate tissue growth (Cano-Gauci et al. 1999). Its over expression in the maturing P10 cornea suggests a cell growth regulatory role for glypican during postnatal maturation of the cornea. Gpc functions may be mediated via IGF-II, and Igf2 was elevated in P10 corneas (Supplemental Table 1). A recent study also reported IGF-II in the bovine cornea and its role in regulating stromal keratocyte proliferation (Musselmann et al. 2008). Igfbp2, encoding an IGF binding protein that modulates IGF levels and its autocrine and paracrine functions was also found to be over expressed in the cornea of both ages by microarray and RT-PCR. Thus IGF mediated pathways may be involved in guiding postnatal maturation and growth of the cornea. Prominin-1 or CD133, a stem cell marker, was detected n the adult and P10 cornea gene expression patterns and presence of the protein was further confirmed in corneal protein extracts. Two recent studies identified this marker in the corneal stroma and a subset of corneal keratocytes (Perrella et al. 2007; Thill et al. 2007). While prominin-1 is known as a hematopoietic stem cell marker, its expression at such high level in the cornea suggests that it may be a marker for the subset of keratocytes that are derived from bone marrow progenitor cells. It also raises the possibility that in the adult mouse cornea, a majority of the keratocytes are derived from bone marrow progenitor cells and not the neural crest. This is also consistent with our previous study that identified close overlaps in gene expression between the corneal keratocytes and bone marrow derived monocyte/macrophage expressed genes (Chakravarti et al. 2004).
The corneal stroma is an ECM rich tissue that develops rapidly during postnatal maturation of the cornea. ECM and stroma building-related gene expressions were evident in the P10 cornea. Ltbp4, involved in the secretion and activation of TGFβ in the immature cornea may regulate diverse TGF functions from regulating cell proliferation to activation of ECM genes. Follistatin, known to modulate TGFβ family activin-signaling, may regulate keratocyte differentiation (You and Kruse 2002). Also up regulated in the P10 cornea were several basement membrane and interstitial ECM collagen genes (Col1a1, Col1a2, Col3a1, Col4a1, Col4a2 and Col6a2) and basement membrane fibrillin (Fbn1). Another interstitial ECM gene Postn, shown recently to regulate collagen assembly was also up regulated in the postnatal cornea (Norris et al. 2007; Snider et al. 2008). The collagens are clearly present in the adult cornea but there is very little turnover which may explain low transcript levels in the adult cornea. Collagen type III has been studied in the context of developing tendon and increased synthesis is coincident with developing collagen fibrils (Birk and Mayne 1997). Our expression data showed Collagen type III to be the one that is most over expressed in the immature cornea. Over expressed Fibulin, biglycan and collagen VI in the P10 cornea and tendon may also represent a dynamic and immature interstitial ECM in these tissues. The genes encoding collagen associating proteoglycans, lumican and decorin were all over expressed at high levels in both the P10 and adult cornea. These proteoglycans may have a higher turnover rate in the cornea for constant fine tuning of the extracellular matrix. In our earlier gene expression study of the corneal stromal keratocytes, fibroblasts and myofibroblasts, the stromal collagen and proteoglycan transcripts were elevated in the stromal fibroblasts and myofibroblasts underscoring their role in matrix turnover and wound repair (Chakravarti et al. 2004). Keratocan was the only other small leucine-rich repeat proteoglycan transcript that was detectable in the corneal total RNA preparations and not in the primary fibroblast and myofibroblast cultures (Chakravarti et al. 2004; Chakravarti 2006).
Innervation is another key event during postnatal development of the cornea (Muller et al. 2003). In the P10 cornea we detected expression of Sema4a and Nrp1, encoding semaphorin 4A and neuropilin, respectively. Semaphorin 4A has been studied in the retina (Rice et al. 2004) with no prior reports of its presence in the cornea. Semaphorin 3A and its receptor neuropilin have been reported to play a role in the initial formation of a nerve ring surrounding the cornea (Lwigale and Bronner-Fraser 2007). Sema4A may be a novel regulator of corneal innervation.
Very little is known about the 12 genes (Cy24a1, Cyp2f2, Psca, Plac 8 and others) selectively activated in the adult cornea. Most of these are novel to the cornea field and may be of relevance to immune response, anti-protease activity and anti-angiogenesis; all necessary in maintaining adult corneal health.
Overall, our genome-wide gene expression patterns detected previously known and unknown genes in the cornea. Among genes previously identified in the cornea detected by our microarray were transcription and regulatory genes, Notch3, Pax6, ECM genes Col1, Col5, Lum, Bgn, cornea transparency-related genes (Aldh1a1, Aldh3a1), and others. Novel genes with no known functions in the cornea include Emk2, Aim1, Acly, Coro1, Dhcr24, F3, Lmo7, Ostf1, Zfp36i3 and others. The increasing availability of gene-targeted mutations in the mouse will make it possible for investigators to move rapidly from identification of a gene to its functional study. Thus, functional studies of the newly identified corneal genes will provide a better understanding of the cornea in health and biological processes underlying its injury, infection and inflammation.
Supplementary Material
Gene expression in arbitrary units of 1087 expressed at ≥ 500 arbitrary units in any one sample type.
Gene-specific primer information for semi-quantitative RT-PCR
Fig. 4.
RT-PCR validation of selected genes Igfbp2 (Insulin growth factor binding protein2), A2m (Alpha2 macroglobulin), Vasp (Vesicle associated membrane protein), Psca (prostate stem cell antidgen), Ldlr (Low density lipoprotein receptor), Prdx6 (Peroxiredoxin 6), Anx-A1 (Annexin a1), Guca1a (guanylate cyclase activator 1a). Primer information for each gene is provided in supplemental Table 2.
Acknowledgements
The study was funded by NIH EY11654 (to S.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Birk DE, Mayne R. Localization of collagen types I, III and V during tendon development. Changes in collagen types I and III are correlated with changes in fibril diameter. Eur J Cell Biol. 1997;72:352–361. [PubMed] [Google Scholar]
- Cano-Gauci DF, Song HH, Yang H, McKerlie C, Choo B, Shi W, Pullano R, Piscione TD, Grisaru S, Soon S, Sedlackova L, Tanswell AK, Mak TW, Yeger H, Lockwood GA, Rosenblum ND, Filmus J. Glypican-3-deficient mice exhibit developmental overgrowth and some of the abnormalities typical of Simpson-Golabi-Behmel syndrome. J Cell Biol. 1999;146:255–264. doi: 10.1083/jcb.146.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Wu HK, Bruce A, Wollenberg K, Panjwani N. Detection of differentially expressed genes in healing mouse corneas, using cDNA microarrays. Invest Ophthalmol Vis Sci. 2002;43:2897–2904. [PubMed] [Google Scholar]
- Chakravarti S. The cornea through the eyes of knockout mice. Exp. Eye Res. 2001;73 doi: 10.1006/exer.2001.1055. [DOI] [PubMed] [Google Scholar]
- Chakravarti S. Focus on molecules: keratocan (KERA) Exp Eye Res. 2006;82:183–184. doi: 10.1016/j.exer.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Chakravarti S, Wu F, Vij N, Roberts L, Joyce S. Microarray Studies Reveal Macrophage-like Function of Stromal Keratocytes in the Cornea. Invest Ophthalmol Vis Sci. 2004;45:3475–3484. doi: 10.1167/iovs.04-0343. [DOI] [PubMed] [Google Scholar]
- Chen WL, Harris DL, Joyce NC. Effects of SOV-induced phosphatase inhibition and expression of protein tyrosine phosphatases in rat corneal endothelial cells. Exp Eye Res. 2005;81:570–580. doi: 10.1016/j.exer.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Fischbarg J, Lim JJ. Role of cations, anions and carbonic anhydrase in fluid transport across rabbit corneal endothelium. J Physiol. 1974;241:647–675. doi: 10.1113/jphysiol.1974.sp010676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi CM, Hutcheon AE, Chung EH, Dalbone AC, Joyce NC, Zieske JD. Expression patterns of retinoblastoma and E2F family proteins during corneal development. Invest Ophthalmol Vis Sci. 2000;41:1054–1062. [PubMed] [Google Scholar]
- Hay ED. Development of the vertebrate cornea. Int Rev Cytol. 1979;63:263–322. doi: 10.1016/s0074-7696(08)61760-x. [DOI] [PubMed] [Google Scholar]
- Hay ED, Revel JP. Fine structure of the developing avian cornea. Monogr Dev Biol. 1969;1:1–144. [PubMed] [Google Scholar]
- Jester JV, Moller-Pedersen T, Huang J, Sax CM, Kays WT, Cavangh HD, Petroll WM, Piatigorsky J. The cellular basis of corneal transparency: evidence for 'corneal crystallins'. J Cell Sci. 1999;112:613–622. doi: 10.1242/jcs.112.5.613. [DOI] [PubMed] [Google Scholar]
- Joyce NC, Navon SE, Roy S, Zieske JD. Expression of cell cycle-associated proteins in human and rabbit corneal endothelium in situ. Invest Ophthalmol Vis Sci. 1996;37:1566–1575. [PubMed] [Google Scholar]
- Karring H, Thogersen IB, Klintworth GK, Enghild JJ, Moller-Pedersen T. Proteomic analysis of the soluble fraction from human corneal fibroblasts with reference to ocular transparency. Mol Cell Proteomics. 2004;3:660–674. doi: 10.1074/mcp.M400016-MCP200. [DOI] [PubMed] [Google Scholar]
- Autho . The atlas of mouse development. Academic Press; 1992. [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lwigale PY, Bronner-Fraser M. Lens-derived Semaphorin3A regulates sensory innervation of the cornea. Dev Biol. 2007;306:750–759. doi: 10.1016/j.ydbio.2007.04.012. [DOI] [PubMed] [Google Scholar]
- MacLellan WR, Xiao G, Abdellatif M, Schneider MD. A novel Rb- and p300-binding protein inhibits transactivation by MyoD. Mol Cell Biol. 2000;20:8903–8915. doi: 10.1128/mcb.20.23.8903-8915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice DM. The structure and transparency of the cornea. J. Physiology. 1957;136:263–286. doi: 10.1113/jphysiol.1957.sp005758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiani-Ferreira F, Petersen-Jones S, Cassotis N, Ramsey DT, Gearhart P, Cardoso F. Early postnatal development of central corneal thickness in dogs. Vet Ophthalmol. 2003;6:19–22. doi: 10.1046/j.1463-5224.2003.00196.x. [DOI] [PubMed] [Google Scholar]
- Moodie KL, Hashizume N, Houston DL, Hoopes PJ, Demidenko E, Trembly BS, Davidson MG. Postnatal development of corneal curvature and thickness in the cat. Vet Ophthalmol. 2001;4:267–272. doi: 10.1046/j.1463-5216.2001.00198.x. [DOI] [PubMed] [Google Scholar]
- Muller LJ, Marfurt CF, Kruse F, Tervo TM. Corneal nerves: structure, contents and function. Exp Eye Res. 2003;76:521–542. doi: 10.1016/s0014-4835(03)00050-2. [DOI] [PubMed] [Google Scholar]
- Musselmann K, Kane BP, Alexandrou B, Hassell JR. IGF-II is present in bovine corneal stroma and activates keratocytes to proliferate in vitro. Exp Eye Res. 2008;86:506–511. doi: 10.1016/j.exer.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman B, Davis J, Piatigorsky J. Postnatal gene expression in the normal mouse cornea by SAGE. Invest Ophthalmol Vis Sci. 2004;45:429–440. doi: 10.1167/iovs.03-0449. [DOI] [PubMed] [Google Scholar]
- Norris RA, Damon B, Mironov V, Kasyanov V, Ramamurthi A, Moreno-Rodriguez R, Trusk T, Potts JD, Goodwin RL, Davis J, Hoffman S, Wen X, Sugi Y, Kern CB, Mjaatvedt CH, Turner DK, Oka T, Conway SJ, Molkentin JD, Forgacs G, Markwald RR. Periostin regulates collagen fibrillogenesis and the biomechanical properties of connective tissues. J Cell Biochem. 2007;101:695–711. doi: 10.1002/jcb.21224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei YF, Rhodin JA. The prenatal development of the mouse eye. Anat Rec. 1970;168:105–125. doi: 10.1002/ar.1091680109. [DOI] [PubMed] [Google Scholar]
- Perrella G, Brusini P, Spelat R, Hossain P, Hopkinson A, Dua HS. Expression of haematopoietic stem cell markers, CD133 and CD34 on human corneal keratocytes. Br J Ophthalmol. 2007;91:94–99. doi: 10.1136/bjo.2006.097352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatigorsky J. Gene sharing in lens and cornea: facts and implications. Prog Retin Eye Res. 1998;17:145–174. doi: 10.1016/s1350-9462(97)00004-9. [DOI] [PubMed] [Google Scholar]
- Portellinha W, Belfort R., Jr Central and peripheral corneal thickness in newborns. Acta Ophthalmol (Copenh) 1991;69:247–250. doi: 10.1111/j.1755-3768.1991.tb02719.x. [DOI] [PubMed] [Google Scholar]
- Rice DS, Huang W, Jones HA, Hansen G, Ye GL, Xu N, Wilson EA, Troughton K, Vaddi K, Newton RC, Zambrowicz BP, Sands AT. Severe retinal degeneration associated with disruption of semaphorin 4A. Invest Ophthalmol Vis Sci. 2004;45:2767–2777. doi: 10.1167/iovs.04-0020. [DOI] [PubMed] [Google Scholar]
- Sivak B, Sivak J. Results Probl Cell Differ. Vol. 31. M. E. Fini, Springer; 2000. Vertebrate eye development; pp. 1–14. [DOI] [PubMed] [Google Scholar]
- Snider P, Hinton RB, Moreno-Rodriguez RA, Wang J, Rogers R, Lindsley A, Li F, Ingram DA, Menick D, Field L, Firulli AB, Molkentin JD, Markwald R, Conway SJ. Periostin Is Required for Maturation and Extracellular Matrix Stabilization of Noncardiomyocyte Lineages of the Heart. Circ Res. 2008 doi: 10.1161/CIRCRESAHA.107.159517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Lee YG, Houston J, Petroll WM, Chakravarti S, Cavanagh HD, Jester JV. Neonatal corneal stromal development in the normal and lumican-deficient mouse. Invest Ophthalmol Vis Sci. 2003;44:548–557. doi: 10.1167/iovs.02-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thill M, Schlagner K, Altenahr S, Ergun S, Faragher RG, Kilic N, Bednarz J, Vohwinkel G, Rogiers X, Hossfeld DK, Richard G, Gehling UM. A novel population of repair cells identified in the stroma of the human cornea. Stem Cells Dev. 2007;16:733–745. doi: 10.1089/scd.2006.0084. [DOI] [PubMed] [Google Scholar]
- Varela JC, Goldstein MH, Baker HV, Schultz GS. Microarray analysis of gene expression patterns during healing of rat corneas after excimer laser photorefractive keratectomy. Invest Ophthalmol Vis Sci. 2002;43:1772–1782. [PubMed] [Google Scholar]
- Wadehra M, Sulur GG, Braun J, Gordon LK, Goodglick L. Epithelial membrane protein-2 is expressed in discrete anatomical regions of the eye. Exp Mol Pathol. 2003;74:106–112. doi: 10.1016/s0014-4800(03)00009-1. [DOI] [PubMed] [Google Scholar]
- You L, Kruse FE. Differential effect of activin A and BMP-7 on myofibroblast differentiation and the role of the Smad signaling pathway. Invest Ophthalmol Vis Sci. 2002;43:72–81. [PubMed] [Google Scholar]
- Zhou L, Beuerman RW, Huang L, Barathi A, Foo YH, Li SF, Chew FT, Tan D. Proteomic analysis of rabbit tear fluid: Defensin levels after an experimental corneal wound are correlated to wound closure. Proteomics. 2007;7:3194–3206. doi: 10.1002/pmic.200700137. [DOI] [PubMed] [Google Scholar]
- Zieske JD, Francesconi CM, Guo X. Cell cycle regulators at the ocular surface. Exp Eye Res. 2004;78:447–456. doi: 10.1016/s0014-4835(03)00205-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene expression in arbitrary units of 1087 expressed at ≥ 500 arbitrary units in any one sample type.
Gene-specific primer information for semi-quantitative RT-PCR