Abstract
The successful translation of gene therapy for clinical application will require the assessment of transgene activity as a measure of the biological function of a therapeutic transgene. While current imaging permits the non-invasive detection of transgene expression, the critical need for quantitative imaging of the action of the expressed transgene has not been met. Magnetic resonance spectroscopic imaging (MRSI) was applied to quantitatively delineate both the concentration and activity of a cytosine deaminase-uracil phosphoribosyltransferase (CD-UPRT) fusion enzyme expressed from a transgene. MRSI enabled the generation of anatomically accurate maps of the intratumoral heterogeneity in fusion enzyme activity. We observed an excellent association between the CD-UPRT concentration and activity and the percentage of CD-UPRT+ cells. Moreover, the regional levels of UPRT activity, as measured by imaging, correlated well with the biological affect of this enzyme. This study presents a translational imaging strategy for precise, in vivo measurements of transgene activity with potential applications in both pre-clinical and clinical settings.
Introduction
The pharmacokinetic properties of a drug determine its therapeutic efficacy in vivo(1). This association holds true for all conventional drugs and suggests that the activity of novel therapeutic strategies, including gene therapy, must be thoroughly characterized in order to achieve therapeutic impact(2). In the excitement to introduce gene therapy for clinical trial, the characterization of transgene activity has been underemphasized(3),(4) and the inability to translate the successes of preclinical gene therapy studies into clinical treatments has underscored this deficiency in quantitative data1. We demonstrate quantitative in vivo magnetic resonance spectroscopic imaging (MRSI) of the regional activity and concentration of an exogenous enzyme expressed from a therapeutic transgene. We further establish that MRSI measurements of regional transgene activity can provide an effective index of the biological function of this enzyme. The ability of MRSI to facilitate the in vivo assessment of transgene activity suggests a potential role for this technique in optimizing the implementation of gene therapy.
To date, non-invasive imaging approaches used to monitor gene therapy strategies have primarily focused on semi-quantitative assessments of transgene expression(5-10). In contrast, the delineation of a transgene's regional activity is expected to enable the functional evaluation of gene therapy. Transgene activity can be assessed through serial, quantitative and noninvasive measurements of both the substrate specific for the transgene and its resultant metabolite(s). For example, the cytosine deaminase-uracil phosphoribosyltransferase (CD-UPRT) fusion enzyme metabolizes 5-fluorouracil (5FU) and 5-fluorocytosine (5FC) to discernible anabolites(11, 12) (Fig. 1). Magnetic resonance spectroscopic imaging (MRSI) is well-suited for this application as it allows the absolute quantitation of transgene specific substrates and metabolites with spatial encoding that provides the opportunity to detect local variations in the target tissue(13, 14). These substrates and metabolites may be distinguished in the acquired images based on their relative chemical shifts. In enabling the absolute quantitation of these resonances, MRSI provides the unique potential to characterize and to quantify transgene activity spatially and non-invasively in absolute terms.
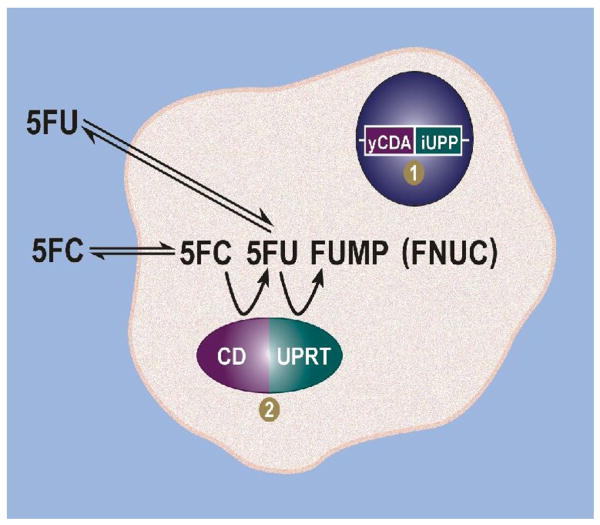
Figure 1.
The CD-UPRT fusion enzyme can be monitored based on its metabolism of MR-visible probes to intracellularly entrapped MR-visible anabolites. Cells transduced with the yCDA-iUPP fusion gene (1) express the CD-UPRT enzyme (2). CD converts 5FC to 5FU, while UPRT anabolizes 5FU to fluorouridine monophosphate (FUMP). FUMP and other fluorinated nucleotides are indistinguishable by MRS and apppear as a single resonance on MR spectra that is designated as FNuc. As anions, these fluoronucleotides are unable to cross the cell membrane and are therefore trapped within the cell.
Materials & Methods
Cytosine Deaminase-Uracil Phosphoribosyltransferase Expressing Tumors
Walker 256 cells were grown in minimal essential medium supplemented with 10% fetal calf serum. Cells transduced with a SFG retroviral vector(15) coding for CD-UPRT and neomycin resistance (Supplementary Figure 3) were selected in the presence of 500 μg/mL Geneticin (Invitrogen, Carlsbad, CA) and then a robustly CD-UPRT expressing clone (validated through cellular accumulation of [3H]-uracil) was propagated in antibiotic containing media. All tumors were inoculated by subcutaneous injection of 2×106 W256 cells (wild type or transduced) into the right flank of 5-6 week old male nude mice (Nu/Nu, NCI, Frederick, MD). CD-UPRT gene expression was detected in tumors by immunohistochemistry as long as seven weeks after inoculation. Transplanted tumors were implanted under general anesthesia (ketamine/xylazine) and wound closure was achieved using a topical tissue adhesive (Nexaband Liquid, Abbott Animal Health, Chicago, IL). Tumor volumes and doubling times were determined as described previously(16). To assay transgene expression in tumor tissue, Western blotting and immunohistochemical staining were performed as recommended by the manufacturer using a sheep anti-yCD polyclonal antibody (Biotrend Chemicals Inc, Cologne, Germany). The detection of apoptosis in tumor sections was achieved by immunohistochemical staining using the cleaved caspase-3 primary antibody according to the manufacturer's instructions (Cell Signaling Technology, Beverly, MA). Regional apoptosis was quantitated using MetaMorph software (Universal Imaging Corp., Downingtown, PA). For this purpose, regions of interest (ROI) were prescribed onto contiguous tumor sections (10 μm spacing) stained for cytosine deaminase gene expression. These ROI were overlaid onto sections stained for caspase-3. Apoptotic indices are reported as the average number of cells staining positively for caspase-3 per square millimeter.
Radionuclide Uptake Assay
Accumulation assays using [6-3H]-fluorouracil (12 mCi/mmol) and [2-14C]-fluorocytosine (53 mCi/mmol) (Moravek Biochemicals, Brea, CA) were performed as described previously(17) and expressed as the cell to medium radiotracer accumulation ratio (mL/g)(18).
19FMRS
The mice were prepared and positioned for the experiments as described previously(11). Spectral parameters included 700 signal averages, 1024 data points, a 60° flip angle, a 1.7 s repetition time and a 12 kHz spectral width. A microsphere containing a 150 mM NaF (Sigma-Aldrich) aqueous solution doped with 15 mM Magnevist (Berlex Laboratories Inc., Montville, NJ) and positioned adjacent to the center of the coil was used as an external reference for quantitation. The NaF spectral parameters included 700 averages, 1024 data points, a 90° flip angle, a 600 ms repetition time and a 12 kHz spectral width.
Spins were quantitated using the AMARES algorithm(19). Cramer-Rao bounds (CRB) were used to estimate the achievable precision of this algorithm(20). In accordance with the literature, considerations relevant to data acquired using the external reference technique were included in the analysis(21, 22). Measured in vivo spin-lattice relaxation times of 5FU and fluoronucleotides (FNuc) were used to correct the metabolite signal areas for partial saturation effects(11).
19FMRSI
The MRSI sequence was utilized to acquire two-dimensional images in the axial plane with respect to the tumor at 4.7 Tesla with eight or thirteen phase encode steps at effective in-plane resolutions of 2.5 mm × 2.5 mm (FOV = 20 mm) or 3.0 mm × 3.0 mm (FOV = 24 mm). Each phase encode step was collected as the average of 76 FID signals in 1024 data points with a spectral width of 16 or 30 kHz, 60° flip angle and 1.7 s repetition time. The total imaging time was two hours and 17 minutes. Following 19FMRSI, proton images were acquired at 200.1 MHz for anatomic localization of the MRSI spectra. Imaging parameters included a 40 mm FOV, 256 × 256 matrix size, 16 signal averages, 100 ms repetition time and 16 ms echo time.
The SITOOLS(23) package was used to process the images and to generate the parametric maps. CD-UPRT enzyme concentrations and activity were determined by scaling the estimated Vmax due solely to UPRT by the ratio of the FNuc concentration in the relevant voxel to the FNuc concentration at the same time point in the modeled data. Corrections for intervoxel signal contamination were unnecessary for our voxel sizes (∼3.0 mm × 3.0 mm × 5.0 mm) as described previously(24) and confirmed empirically (data not shown).
Pharmacokinetic Modeling
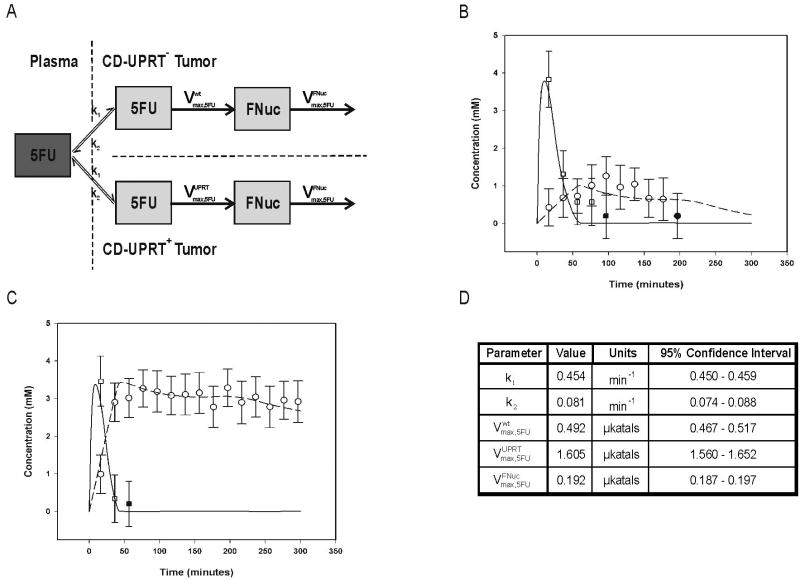
A five-compartment pharmacokinetic model was developed to simultaneously fit two, one-compartment and two, two-compartment models including 5FU in plasma, 5FU in both tumor types, as well as FNuc in each tumor type (Fig. 2a). The model is based on certain assumptions: (1) first order kinetics for the cellular transport of 5FU; (2) the enzyme mediated conversions of 5FU into FNuc and the further anabolism of FNuc were saturable and followed Michaelis-Menten constraints; (3) a single composite rate equation for the anabolism of FNuc. Certain kinetic parameters were assumed to be the same for both the CD-UPRT- and CD-UPRT+ tumors including (1) the rate constant describing the transport of 5FU into (k1) and out of (k2) the tissue; (2) the conversion of FNuc into anabolites not measured by MRS ( and Km,FNuc); (3) the half-saturation concentration for the 5FU to FNuc reaction (Km,5FU). The values for and were allowed to vary independently.
Figure 2.
Pharmacokinetic modeling of 5FU anabolism in CD-UPRT- and CD-UPRT+ tumors based on 19FMRS data. (a) Schematic representation of the 5-compartment model used in the pharmacokinetic analysis of 5FU metabolism in CD-UPRT- and CD-UPRT+ W256 tumor xenografts. The rate constants k1 and k2 describe the transport of 5FU into and out of the xenografts, while the Vmax parameters reflect the maximal rates of conversion of 5FU to FNuc and FNuc to higher molecular weight anabolites . The transport of 5FU in and out of the cell was assumed to follow first order kinetics, whereas enzyme mediated conversions of 5FU to FNuc and all subsequent anabolism of FNuc were assumed to be saturable and therefore to follow Michaelis-Menten constraints. Pharmacokinetic analysis of 5FU and its anabolites in CD-UPRT- (b) and CD-UPRT+ (c) tumors. 19FMRS-determined concentrations of 5FU (□, –––) and FNuc (○, – – –) were fit to the pharmacokinetic model (Fig. 2a). The initial time points at which 5FU (■) and FNuc (●) concentrations became subthreshold are indicated to represent the uncertainty that was associated with these measurements.
Each 19FMRS tissue metabolite measurement was converted into a weighted average over five tumors using the inverse of the uncertainty as the weight. The uncertainty in each measurement was estimated to be at its CRB. The inverse of these uncertainties was later used during the fitting process to weight each data point. Metabolite concentrations that were below the detection limit were assumed to be at a median concentration of 200 μM and were assigned an uncertainty of 600 μM based on empirically determined thresholds (400 μM) and consistent with measured CRBs.
LC/MS was used to measure plasma 5FU concentrations drawn from non-tumor bearing mice (n = 3 per time point) administered 150 mg/kg 5FU. The resulting 5FU plasma curve was used as a forcing-function for the plasma 5FU compartment. Each of the five compartments was associated with data from the 19FMRS derived time-series. A nonlinear fitting procedure was used to achieve the best weighted least-squares fit of each of the four tissue compartments to each of the four measured time series, simultaneously. The half-saturation concentration for the UPRT mediated reaction (Km,5FU) was fixed at 25 μM(25). Five of the remaining parameters (k1, k2, , and ) were allowed to vary during the iterative fitting procedure. Because of the potential that Km,FNuc would lack a clearly defined minimum, the fitting procedure was run repeatedly with Km,FNuc values ranging from 0.1 μM to 1680 mM. 25 fits were evaluated in total and the target function and parameter values noted (Supplementary Fig. 4a, b). Solutions to the rate equations and the nonlinear fitting were performed using MATLAB v5.3 (Mathworks, Natick, MA) routines.
Statistical Analysis
Mean values are reported as mean ± SEM. Radiotracer accumulation was analyzed using the paired two-tailed Student t-test. Apoptosis indices were analyzed using a one-way analysis of variance.
Results
The CD-UPRT fusion enzyme was selected as a model transgene system for the imaging of transgene activity because of the versatility it offers with respect to magnetic resonance visible probes (Fig.1). We initially compared 5FC and 5FU as potential probes for the assessment of CD-UPRT activity. Radionuclide uptake studies using [14C]-5FC or [3H]-5FU were performed on wild type Walker 256 (W256) carcinosarcoma cells (CD-UPRT-) as well as W256 cells stably expressing the CD-UPRT fusion gene (CD-UPRT+) (Supplementary Fig. 1). These data, which demonstrate the accumulation of radiotracer at an appreciably higher rate and to substantially higher levels for incubations of CD-UPRT+ cells with [3H]-5FU as compared to [14C]-5FC, suggest that 5FU should provide better sensitivity and, relatedly, superior temporal and spatial resolution than 5FC as an in vivo MR probe for CD-UPRT levels and activity.
In addition to an effective reporter-probe system, imaging transgene activity requires a pharmacokinetic model to characterize the function of the transgene in living tissue. Toward this end, we next fit absolute metabolite concentrations derived from 19FMRS time series measurements (Supplementary Figure 2) to a pharmacokinetic model describing the kinetics of 5FU in CD-UPRT+ and CD-UPRT- tumors (Fig. 2a). Fits of the 19FMRS-derived metabolite concentrations to this model (Fig. 2b, c), enabled the parameters controlling the transport and metabolite conversions between the various compartments to be determined for wild type and transduced tumors (Fig. 2d). The measured substrate concentrations (5FU) greatly exceeded the reported UPRT Km (25 μM)(25) such that the UPRT was producing FNuc at or near to its maximal rate. As shown in Fig. 2d, the maximum rate of 5FU anabolism in CD-UPRT+ tumors was more than three fold larger than that associated with the activity of endogenous enzymes in CD-UPRT- tumors . The UPRT specific was determined to be 1.113 μkatals; this represents the maximum enzymatic rate specific to UPRT and, as such, is also a direct measure of the CD-UPRT protein concentration. Moreover, because the pharmacokinetic model follows Michaelis-Menten (non-linear) kinetics, this UPRT-specific rate can be used to determine the activity of the CD-UPRT fusion enzyme for any single-time-point measurement of FNuc concentration.
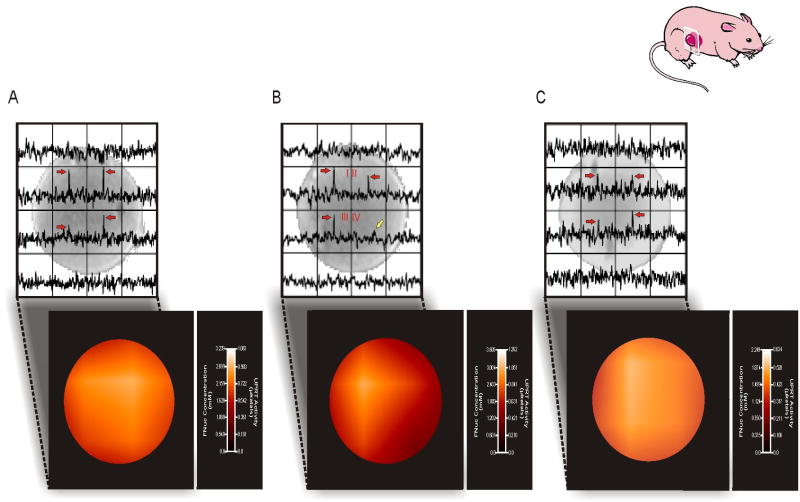
This relationship allowed the application of the pharmacokinetic model to achieve quantitative images of transgene activity measuring regional CD-UPRT concentration and activity. Following the i.v. injection of 150 mg/kg (450 mg/m2) 5FU into mice bearing CD-UPRT+ tumors, two-dimensional 19FMRS images were acquired. These images were overlaid onto corresponding two-dimensional proton images in order to co-register the 19FMRSI spectra with tumor anatomy (Fig. 3). Each voxel of the 19FMRS image shows the level of FNuc within a 0.045 cc tumor volume with an in-plane resolution of 3.0 mm × 3.0 mm. The homogeneous delivery of 5FU was indicated by the uniform distribution of fluorinated anesthetic observed throughout the tumor by MRSI (resonance located 75 ppm from 5FU, data not shown). The measured FNuc concentration within these voxels enabled the generation of parametric maps of regional UPRT activity (Fig. 3a). In all but one of these tumors (n=8), UPRT activity appeared to be homogeneous. The image shown in Figure 3b voxels I, II and III demonstrate similar levels of UPRT activity (∼0.841 μkatals) while UPRT activity in voxel IV is undetectable despite the presence of significant amounts of substrate 5FU. In addition to the detection of heterogeneous enzyme activity within CD-UPRT+ tumors, Figure 3c demonstrates that 19FMRSI enabled the assessment of CD-UPRT concentrations and UPRT activity in mixed tumors constituted from equal parts wild type and CD-UPRT transduced W256 cells. As expected, the FNuc concentrations and the range of enzyme activity in mixed tumors is approximately half that of fully transduced tumors. This difference is further reflected in the decreased signal-to-noise ratio associated with the spectra in the mixed tumors as compared to that in the fully transduced tumors.
Figure 3.
In vivo imaging of transgene activity. 19FMRS images of FNuc (red arrows) and associated parametric maps of regional CD-UPRT levels and activity within CD-UPRT+ xenografts are shown. The inset cartoon indicates the slice orientation of the acquired images. 19FMRS enabled images demonstrating homogeneous enzyme activity (a) to be distinguished from xenografts exhibiting regional heterogeneities in CD-UPRT activity (b). An area of deficient CD-UPRT activity is evident within voxel IV and is underscored by the presence of unmetaboilized 5FU (yellow arrow). 19FMRSI was also able to discern CD-UPRT enzyme levels and activity in mixed xenografts grown from equal parts CD-UPRT+ and CD-UPRT- cells (c). Homogeneous CD-UPRT activity was seen; however, this activity is reduced in comparison to CD-UPRT+ tumors (a).
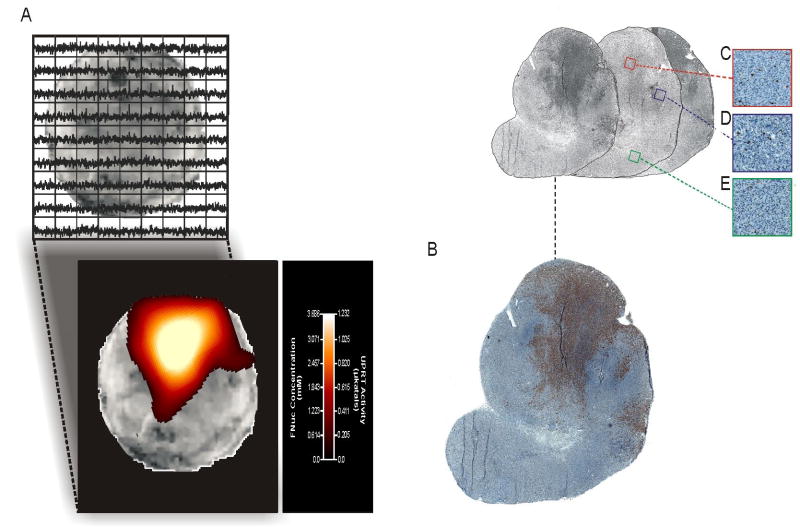
Finally, we undertook experiments to investigate the relationship between MRSI measured UPRT activity and the biological activity of this enzyme. For this purpose we developed a mouse model in which sections of CD-UPRT+ and CD-UPRT- tumors were transplanted side-by-side into the flanks of athymic nu/nu mice (n=6). A representative example is shown in Figure 4. Quantitative 19FMRS images of these co-transplanted tumors demonstrated a discreet pattern of regional UPRT activity (Fig. 4a). In order to correlate transgene action, gene expression and biological function, the acquired spectroscopic images were co-registered with tumor sections stained for CD-UPRT gene expression as well as caspase activity. As expected, tumor regions demonstrating UPRT activity by imaging correlated to those regions staining for CD-UPRT gene expression (Fig. 4b). Moreover, as early as six hours following 5FU therapy, a significant difference in FNuc-induced apoptosis was observed in tumor regions exhibiting UPRT activity and expression as compared to regions showing neither (Fig. 4c, d, e). Taken together, these data suggest that the imaging of transgene activity provides a relevant index of gene therapy.
Figure 4.
In vivo imaging of transgene activity provides an index of biological enzyme function. (a) 19FMRS image and associated parametric map demonstrating circumscribed regions of CD-UPRT levels and activity within a CD-UPRT+/CD-UPRT- co-transplanted tumor xenograft at a digital resolution of 1.8 mm × 1.8 mm. (b) The regional distribution of UPRT activity closely parallels the expression pattern of the CD-UPRT transgene as determined by immunohistochemical staining. Apoptotic indices measured in a contiguous slice of tumor tissue through staining for cleaved capase-3 demonstrated that regions with low, medium and high apoptotic indices (c, d, e) correlated well with transgene activity, (13.2 apoptotic cells/mm2 ± 2.9, 23.8 cells/mm2 ± 1.8 and 61.2 apoptotic cells/mm2 ± 2.4 respectively, P<0.0001).
Discussion
The studies presented here introduce a paradigm for the in vivo assessment of transgene activity through quantitative MRS imaging. The quantitation of in vivo metabolites in absolute units of concentration allowed the modeling of expressed transgene activity in absolute, SI coherent, units of enzyme activity. In developing the pharmacokinetic model, we determined that a Michaelis-Menten analysis is required for single time point spectroscopic imaging of transgene activity(26). Using this analysis, we achieved the first reported quantitative images of transgene action in absolute terms and were able to discern regional heterogeneities in the activity of the CD-UPRT fusion enzyme. MRSI enabled unique insights into regional heterogeneity that would not be discernible by other non-invasive imaging modalities. This point is underscored by the presence of a 5FU resonance and the noticeable absence of an FNuc resonance in voxel IV of Fig. 3b. Assessments of this tumor using positron emission tomography would be infeasible because signals resulting from 5FU and FNuc would be indistinguishable leading to the false impression of homogenous enzyme activity. In addition, we found that CD-UPRT concentration and activity were proportional to the percentage of CD-UPRT+ cells. Furthermore, regional levels of UPRT activity as measured by imaging correlated with metabolite-induced apoptosis evincing that the non-invasive assessment of transgene activity can perhaps provide predictive insights into the therapeutic competency of a gene therapy system.
In this application, MRSI offers the potential for significant versatility as spectroscopic imaging of several MR-visible nuclei including hydrogen, fluorine, phosphorus, and carbon(27) have been described. Transgene products that modify substrates containing these nuclei can potentially be monitored by MRSI reporter gene imaging. Several MRS reporter strategies have been developed including creatine kinase, arginine kinase and β-galactosidase(28-30). These strategies are not limited to intracellular proteins; MRSI has also been used to demonstrate extracellular enzyme function in vivo(31). This breadth of applications not withstanding, several technical issues for the quantitative assessment of expressed transgenes by MRSI must be considered. The sensitivity of MRSI is inherently limited and sufficient metabolite concentrations must be sustained during image acquisition in order to achieve adequate spatial resolution. For example, the rapid anabolism of 5FU by UPRT resulted in high concentrations of FNuc that were sustained for more than five hours in vivo. In contrast, the metabolism of 5FC by CD-UPRT does not result in persistently elevated levels of fluorinated anabolites(12) suggesting that 5FC would be a less effective probe for MRSI of CD-UPRT (data not shown). The anabolite levels required to image CD-UPRT activity in vivo are intrinsically related to the administered dose of 5FU as well as the level of transgene expression. The described experiments involved a bolus i.v. injection of 150 mg/kg 5FU which represents the maximum tolerated dose (MTD) of this agent in mice. Longitudinal studies of a single animal using this dosing regimen would be tolerated with a ten day interval between 5FU injections. The monitoring interval can be shortened by using lower doses of 5FU. Our data indicate that the administration of as little as half of the MTD should be sufficient to achieve quantitative images of CD-UPRT activity. In many cases, even when metabolites are readily MR-visible in vivo, poor estimation of spin-lattice relaxation times in the tissue of interest may contribute to uncertainties in quantitation of tissue metabolite concentrations. We have previously reported the in vivo spin-lattice relaxation times of both 5FU and FNuc in subcutaneous W256 tumors(11). In the setting of adequate substrate delivery, the sensitivity of our approach would be limited by the number of CD-UPRT+ cells. If we consider that a 1 cc tumor contains 109 clonogenic cells(32) with an empiric 19FMRSI detection limit of 650μM for a 0.045 cc tumor volume(33), a conservative analysis of our data indicates that our approach should enable the detection of as few as 8×106 CD-UPRT+ cells at 4.7 Tesla.
The delineation of transgene activity holds significant implications for the optimization of gene therapy in oncology as well as other disciplines. The correlation of expressed transgene levels and activity with dosing schedules and/or routes of administration could be used to evaluate the efficacy of vector delivery. Similarly, the ability of MRSI to demonstrate differences in the activity of the expressed transgene could be applied to assess regional differences in vector distribution. Further, the accurate quantitation of tissue metabolites for the delineation of pharmacokinetic parameters in absolute units could enable comparisons with conventional biochemical assays and facilitate the translation of in vitro data for in vivo application. For example, the in vitro 5FU IC50 of 1.04×10-8 M, in combination with the in vitro [3H]-FNuc accumulation rate of 0.35 mL·g-1·min-1 for CD-UPRT+ cells, indicates that UPRT enzyme activity producing an FNuc concentration of ∼1.58×10-5 mM·g-1 will inhibit cell growth by 50%. The average FNuc concentration achieved in a single voxel of the imaged CD-UPRT+ tumor shown in Fig. 3a was 5.36×10-3 ± 3.10×10-4 mM·g-1. This intratumoral FNuc concentration was more than 300-fold higher than FNuc concentrations corresponding to the in vitro IC50 of 5FU. The in vivo efficacy of this regimen is indicated by an associated CD-UPRT+ tumor doubling time of 13.9 ± 0.55 days as compared with 1.8 ± 0.06 days for untreated CD-UPRT+ tumors. Indeed, the presented data suggest that the enzyme concentration and activity observed by in vivo MRSI are therapeutically meaningful.
Importantly, our approach could be applied to acquire this data in a clinical setting. The described magnetic resonance (MR) techniques are feasible using existing technology. Clinical MRSI is routinely applied(34, 35) and quantitative MRSI of 5FU pharmacokinetics in patients has been described previously(36) on available 1.5T MR scanners. The current availability of higher field strength magnets for clinical studies (3T-9T) affords the opportunity for improved sensitivity in MRSI. On-going clinical trials of 5FC-CD cancer gene therapy(37) suggest that MRSI monitoring of gene therapy could have immediate implications forBhujwalla the investigation of gene therapy pharmacokinetics. Thus, MRSI offers the potential to gain new and essential insights into transgene activity in both preclinical and clinical settings.
Supplementary Material
Supplementary Figure 1. Probe efficiency was compared in radiouptake studies using (a) [14C]-5FC (with enlarged inset) and (b) [3H]-5FU. The amount of accumulated radioactivity in CD-UPRT+ ( ) and CD-UPRT- (
) and CD-UPRT- ( ) was measured relative to the amount of radioactivity remaining in the medium following the incubations (n=3 per time point). Uptake was linear with time and the net accumulation rate (Ki) was determined from the slope of the relevant regression equation. The data are summarized below each plot.
) was measured relative to the amount of radioactivity remaining in the medium following the incubations (n=3 per time point). Uptake was linear with time and the net accumulation rate (Ki) was determined from the slope of the relevant regression equation. The data are summarized below each plot.
Expression of the CD-UPRT fusion gene enabled cells to accumulate radiotracer at an increased rate as compared to CD-UPRT- cells. CD-UPRT+ cells incubated with [3H]-5FU accumulated radiotracer at a rate that was an order of magnitude faster than that of CD-UPRT- cells. This accumulation rate was also substantially faster than that observed for CD-UPRT+ cells incubated with [14C]-5FC. Moreover, the increase in accumulation for CD-UPRT+ cells incubated with [3H]-5FU resulted in intracellular tracer levels that were a full log order higher than in CD-UPRT- cells (P=0.0043). CD-UPRT+ cells incubated with [14C]-5FC demonstrated significant, while less dramatic, increases in radiotracer levels relative to CD-UPRT- cells (P=0.0014).
Supplementary Figure 2. Representative serial 19FMR spectra of 5FU and anabolite FNuc in wild type (CD-UPRT-) (a) and transduced (CD-UPRT+) (b) subcutaneous xenografts. Spectral acquisition was initiated 7 minutes after i.v. injection of 5FU. The median time point relative to the time of injection is indicated for each spectrum. Western blots of 19FMRS studied tumors demonstrating expression of the 41kD CD-UPRT protein corresponding to the spectral data are shown (c). Lane 1, CD-UPRT- tumor tissue, Lane 2, CD-UPRT+ tumor tissue.
We implanted mice subcutaneously with either CD-UPRT+ or CD-UPRT- W256 cells and selected 175-250 mm3 tumors for experiment (n = 5 per cohort). Following the intravenous (i.v.) injection of 150 mg/kg 5FU, its anabolism to FNuc was monitored by serial 19F spectra in tumors (Supplementary Fig 2 a and b). The identification of 5FU and its FNuc metabolites was made based on documented chemical shifts(38, 39). The intratumoral concentration of 5FU peaked in the initial spectrum and subsequently decreased rapidly for both tumor types. CD-UPRT- tumors produced modest FNuc signals reaching an average peak concentration of only 1.24 ± 0.31 mM. In contrast, the intratumoral FNuc concentration in CD-UPRT+ rapidly reached a plateau, achieving an average concentration of 3.34 ± 0.27 mM, and decreased only slightly thereafter. These data demonstrate that 5FU is an effective probe for detecting the rapid accumulation of a marker metabolite (FNuc) specific for UPRT activity by 19FMRS. The 19FMRS data were validated through Western blot analysis performed following each experiment which confirmed that the expression of the CD-UPRT fusion gene was limited to the CD-UPRT+ tumors.
Supplementary Figure 3. Schematic representation of the bicistronic retroviral vector. The viral long terminal repeats (LTR) control expression of the Saccharomyces cerevisiae cytosine deaminase (yCDA)-Haemophilus influenzae uracil phosphoribosyltransferase (iUPP) fusion gene, while expression of the neomycin phosphotransferase (NEO) from the single transcript is mediated by an encephalomyocarditis virus derived internal ribosome entry site (IRES)(40). The coding sequences for yCDA (Erbs P et al. Curr Genet. 1997 Jan;31(1):1-6) and iUPP (Fleischmann RD et al. Science. 1995 Jul 28;269(5223):496-512) have been described previously and were linked by a glycine adaptor. Unique restriction sites used for cloning are indicated. Plasmid containing yCDA cDNA was kindly provided by Professor Brian Ross, University of Michigan. An iUPP encoding cosmid was obtained from ATCC (Rockville, MD).
Supplementary Figure 4. Plots showing impact of the different fixed values for the half-saturation concentration of the FNuc to anabolites reaction, Km,FNuc used in the pharmacokinetic. (a) Plot of the final weighted residuals versus Km,FNuc. The model produced virtually identical results for values of Km,FNuc less than about 100 μM, below which the rate equation describing the anabolism of FNuc is effectively zero order and therefore completely determined by . The fits became significantly worse for values of Km,FNuc above 100 μM only stabilizing when the rate equation effectively became linear at a Km,FNuc above about 420 mM. (b) Plot of the fit determined values for (–––) and (– – –) versus Km,FNuc. Both of these Vmax values were largely independent of the value for Km,FNuc thus, any uncertainty in the precise value for Km,FNuc is of little consequence.
Acknowledgments
The authors thank Dr. Andrew Maudsley for the use of the SITOOLs software package. In addition, the authors would like to acknowledge the excellent work of the Molecular Cytology Core Facility of Sloan-Kettering Institute with special thanks to Betul Altayozer, Eric Suh and Katia Manova-Todorova.
Funding: This research was supported by grants from the National Institutes of Health (R24CA83084, P50CA86438).
Footnotes
Orkin, S. H. and Motulsky, A. G. Report and Recommendations of the Panel to Assess the NIH Investment in Research on Gene Therapy. pp. Available at http://www.nih.gov/news/panelrep.html. National Institutes of Health, 1995.
References
- 1.Benet LZ, Kroetz DL, Sheiner LB. Goodman & Gilman's Pharmacokinetics: The Dynamics of Drug Absorption, Distribution, and Elimination. 9th. New York: McGraw-Hill; 1996. [Google Scholar]
- 2.Jain RK. The next frontier of molecular medicine: delivery of therapeutics. Nat Med. 1998;4:655–7. doi: 10.1038/nm0698-655. [DOI] [PubMed] [Google Scholar]
- 3.Pislaru S, Janssens SP, Gersh BJ, Simari RD. Defining gene transfer before expecting gene therapy: putting the horse before the cart. Circulation. 2002;106:631–6. doi: 10.1161/01.cir.0000019621.18368.b7. [DOI] [PubMed] [Google Scholar]
- 4.Kamiya H, Akita H, Harashima H. Pharmacokinetic and pharmacodynamic considerations in gene therapy. Drug Discov Today. 2003;8:990–6. doi: 10.1016/s1359-6446(03)02889-7. [DOI] [PubMed] [Google Scholar]
- 5.Contag PR, Olomu IN, Stevenson DK, Contag CH. Bioluminescent indicators in living mammals. Nat Med. 1998;4:245–7. doi: 10.1038/nm0298-245. [DOI] [PubMed] [Google Scholar]
- 6.Yang M, Baranov E, Moossa AR, Penman S, Hoffman RM. Visualizing gene expression by whole-body fluorescence imaging. Proc Natl Acad Sci U S A. 2000;97:12278–82. doi: 10.1073/pnas.97.22.12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tjuvajev JG, Finn R, Watanabe K, et al. Noninvasive imaging of herpes virus thymidine kinase gene transfer and expression: a potential method for monitoring clinical gene therapy. Cancer Res. 1996;56:4087–95. [PubMed] [Google Scholar]
- 8.Louie AY, Huber MM, Ahrens ET, et al. In vivo visualization of gene expression using magnetic resonance imaging. Nat Biotechnol. 2000;18:321–5. doi: 10.1038/73780. [DOI] [PubMed] [Google Scholar]
- 9.Weissleder R, Tung CH, Mahmood U, Bogdanov A., Jr In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nat Biotechnol. 1999;17:375–8. doi: 10.1038/7933. [DOI] [PubMed] [Google Scholar]
- 10.Weissleder R, Moore A, Mahmood U, et al. In vivo magnetic resonance imaging of transgene expression. Nat Med. 2000;6:351–5. doi: 10.1038/73219. [DOI] [PubMed] [Google Scholar]
- 11.Gade TP, Spees WM, Le HC, et al. In vivo 5-fluorouracil and fluoronucleotide T1 relaxation time measurements using the variable nutation angle method. Magn Reson Med. 2004;52:169–73. doi: 10.1002/mrm.20131. [DOI] [PubMed] [Google Scholar]
- 12.Hamstra DA, Lee KC, Tychewicz JM, et al. The use of 19F spectroscopy and diffusion-weighted MRI to evaluate differences in gene-dependent enzyme prodrug therapies. Mol Ther. 2004;10:916–28. doi: 10.1016/j.ymthe.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 13.Rajanayagam V, Fabry ME, Gore JC. In vivo quantitation of water content in muscle tissues by NMR imaging. Magn Reson Imaging. 1991;9:621–5. doi: 10.1016/0730-725x(91)90050-v. [DOI] [PubMed] [Google Scholar]
- 14.Brown TR, Kincaid BM, Ugurbil K. NMR chemical shift imaging in three dimensions. Proc Natl Acad Sci U S A. 1982;79:3523–6. doi: 10.1073/pnas.79.11.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riviere I, Brose K, Mulligan RC. Effects of retroviral vector design on expression of human adenosine deaminase in murine bone marrow transplant recipients engrafted with genetically modified cells. Proc Natl Acad Sci U S A. 1995;92:6733–7. doi: 10.1073/pnas.92.15.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rygaard K, Spang-Thomsen M. Quantitation and gompertzian analysis of tumor growth. Breast Cancer Res Treat. 1997;46:303–12. doi: 10.1023/a:1005906900231. [DOI] [PubMed] [Google Scholar]
- 17.Doubrovin M, Ponomarev V, Beresten T, et al. Imaging transcriptional regulation of p53-dependent genes with positron emission tomography in vivo. Proc Natl Acad Sci U S A. 2001;98:9300–5. doi: 10.1073/pnas.161091198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tjuvajev JG, Joshi A, Callegari J, et al. A general approach to the non-invasive imaging of transgenes using cis-linked herpes simplex virus thymidine kinase. Neoplasia. 1999;1:315–20. doi: 10.1038/sj.neo.7900053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naressi A, Couturier C, Devos JM, et al. Java-based graphical user interface for the MRUI quantitation package. Magma. 2001;12:141–52. doi: 10.1007/BF02668096. [DOI] [PubMed] [Google Scholar]
- 20.Cavassila S, Deval S, Huegen C, van Ormondt D, Graveron-Demilly D. Cramer-Rao bounds: an evaluation tool for quantitation. NMR Biomed. 2001;14:278–83. doi: 10.1002/nbm.701. [DOI] [PubMed] [Google Scholar]
- 21.Bottomley PA, Charles HC, Roemer PB, et al. Human in vivo phosphate metabolite imaging with 31P NMR. Magn Reson Med. 1988;7:319–36. doi: 10.1002/mrm.1910070309. [DOI] [PubMed] [Google Scholar]
- 22.Bottomley PA, Hardy CJ. Rapid, reliable in vivo assays of human phosphate metabolites by nuclear magnetic resonance. Clin Chem. 1989;35:392–5. [PubMed] [Google Scholar]
- 23.Maudsley AA, Lin E, Weiner MW. Spectroscopic imaging display and analysis. Magn Reson Imaging. 1992;10:471–85. doi: 10.1016/0730-725x(92)90520-a. [DOI] [PubMed] [Google Scholar]
- 24.Li CW, Negendank WG, Murphy-Boesch J, Padavic-Shaller K, Brown TR. Molar quantitation of hepatic metabolites in vivo in proton-decoupled, nuclear Overhauser effect enhanced 31P NMR spectra localized by three-dimensional chemical shift imaging. NMR Biomed. 1996;9:141–55. doi: 10.1002/(SICI)1099-1492(199606)9:4<141::AID-NBM403>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 25.Rasmussen UB, Mygind B, Nygaard P. Purification and some properties of uracil phosphoribosyltransferase from Escherichia coli K12. Biochim Biophys Acta. 1986;881:268–75. doi: 10.1016/0304-4165(86)90013-9. [DOI] [PubMed] [Google Scholar]
- 26.Stegman LD, Rehemtulla A, Beattie B, et al. Noninvasive quantitation of cytosine deaminase transgene expression in human tumor xenografts with in vivo magnetic resonance spectroscopy. Proc Natl Acad Sci U S A. 1999;96:9821–6. doi: 10.1073/pnas.96.17.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Graaf RA. In vivo NMR Spectroscopy: Principles and Techniques. New York: John Wiley & Sons; 1998. [Google Scholar]
- 28.Koretsky AP, Brosnan MJ, Chen LH, Chen JD, Van Dyke T. NMR detection of creatine kinase expressed in liver of transgenic mice: determination of free ADP levels. Proc Natl Acad Sci U S A. 1990;87:3112–6. doi: 10.1073/pnas.87.8.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui W, Otten P, Li Y, Koeneman KS, Yu J, Mason RP. Novel NMR approach to assessing gene transfection: 4-fluoro-2-nitrophenyl-beta-D-galactopyranoside as a prototype reporter molecule for beta-galactosidase. Magn Reson Med. 2004;51:616–20. doi: 10.1002/mrm.10719. [DOI] [PubMed] [Google Scholar]
- 30.Walter G, Barton ER, Sweeney HL. Noninvasive measurement of gene expression in skeletal muscle. Proc Natl Acad Sci U S A. 2000;97:5151–5. doi: 10.1073/pnas.97.10.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aboagye EO, Artemov D, Senter PD, Bhujwalla ZM. Intratumoral conversion of 5-fluorocytosine to 5-fluorouracil by monoclonal antibody-cytosine deaminase conjugates: noninvasive detection of prodrug activation by magnetic resonance spectroscopy and spectroscopic imaging. Cancer Res. 1998;58:4075–8. [PubMed] [Google Scholar]
- 32.Hall E. Solid tumor system and reoxygenation. 2nd. Hagerstown, MD: Harper & Row; 1978. [Google Scholar]
- 33.Procissi D, Claus F, Burgman P, et al. In vivo 19F magnetic resonance spectroscopy and chemical shift imaging of tri-fluoro-nitroimidazole as a potential hypoxia reporter in solid tumors. Clin Cancer Res. 2007;13:3738–47. doi: 10.1158/1078-0432.CCR-06-1563. [DOI] [PubMed] [Google Scholar]
- 34.Kurhanewicz J, Vigneron DB, Hricak H, Narayan P, Carroll P, Nelson SJ. Three-dimensional H-1 MR spectroscopic imaging of the in situ human prostate with high (0.24-0.7-cm3) spatial resolution. Radiology. 1996;198:795–805. doi: 10.1148/radiology.198.3.8628874. [DOI] [PubMed] [Google Scholar]
- 35.Zakian KL, Eberhardt S, Hricak H, et al. Transition zone prostate cancer: metabolic characteristics at 1H MR spectroscopic imaging--initial results. Radiology. 2003;229:241–7. doi: 10.1148/radiol.2291021383. [DOI] [PubMed] [Google Scholar]
- 36.Li CW, Negendank WG, Padavic-Shaller KA, O'Dwyer PJ, Murphy-Boesch J, Brown TR. Quantitation of 5-fluorouracil catabolism in human liver in vivo by three-dimensional localized 19F magnetic resonance spectroscopy. Clin Cancer Res. 1996;2:339–45. [PubMed] [Google Scholar]
- 37.Pandha HS, Martin LA, Rigg A, et al. Genetic prodrug activation therapy for breast cancer: A phase I clinical trial of erbB-2-directed suicide gene expression. J Clin Oncol. 1999;17:2180–9. doi: 10.1200/JCO.1999.17.7.2180. [DOI] [PubMed] [Google Scholar]
- 38.Wolf W, Presant CA, Servis KL, et al. Tumor trapping of 5-fluorouracil: in vivo 19F NMR spectroscopic pharmacokinetics in tumor-bearing humans and rabbits. Proc Natl Acad Sci U S A. 1990;87:492–6. doi: 10.1073/pnas.87.1.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koutcher JA, Sawyer RC, Kornblith AB, et al. In vivo monitoring of changes in 5-fluorouracil metabolism induced by methotrexate measured by 19F NMR spectroscopy. Magn Reson Med. 1991;19:113–23. doi: 10.1002/mrm.1910190111. [DOI] [PubMed] [Google Scholar]
- 40.Gallardo HF, Tan C, Sadelain M. The internal ribosomal entry site of the encephalomyocarditis virus enables reliable coexpression of two transgenes in human primary T lymphocytes. Gene Ther. 1997;4:1115–9. doi: 10.1038/sj.gt.3300506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Probe efficiency was compared in radiouptake studies using (a) [14C]-5FC (with enlarged inset) and (b) [3H]-5FU. The amount of accumulated radioactivity in CD-UPRT+ ( ) and CD-UPRT- (
) and CD-UPRT- ( ) was measured relative to the amount of radioactivity remaining in the medium following the incubations (n=3 per time point). Uptake was linear with time and the net accumulation rate (Ki) was determined from the slope of the relevant regression equation. The data are summarized below each plot.
) was measured relative to the amount of radioactivity remaining in the medium following the incubations (n=3 per time point). Uptake was linear with time and the net accumulation rate (Ki) was determined from the slope of the relevant regression equation. The data are summarized below each plot.
Expression of the CD-UPRT fusion gene enabled cells to accumulate radiotracer at an increased rate as compared to CD-UPRT- cells. CD-UPRT+ cells incubated with [3H]-5FU accumulated radiotracer at a rate that was an order of magnitude faster than that of CD-UPRT- cells. This accumulation rate was also substantially faster than that observed for CD-UPRT+ cells incubated with [14C]-5FC. Moreover, the increase in accumulation for CD-UPRT+ cells incubated with [3H]-5FU resulted in intracellular tracer levels that were a full log order higher than in CD-UPRT- cells (P=0.0043). CD-UPRT+ cells incubated with [14C]-5FC demonstrated significant, while less dramatic, increases in radiotracer levels relative to CD-UPRT- cells (P=0.0014).
Supplementary Figure 2. Representative serial 19FMR spectra of 5FU and anabolite FNuc in wild type (CD-UPRT-) (a) and transduced (CD-UPRT+) (b) subcutaneous xenografts. Spectral acquisition was initiated 7 minutes after i.v. injection of 5FU. The median time point relative to the time of injection is indicated for each spectrum. Western blots of 19FMRS studied tumors demonstrating expression of the 41kD CD-UPRT protein corresponding to the spectral data are shown (c). Lane 1, CD-UPRT- tumor tissue, Lane 2, CD-UPRT+ tumor tissue.
We implanted mice subcutaneously with either CD-UPRT+ or CD-UPRT- W256 cells and selected 175-250 mm3 tumors for experiment (n = 5 per cohort). Following the intravenous (i.v.) injection of 150 mg/kg 5FU, its anabolism to FNuc was monitored by serial 19F spectra in tumors (Supplementary Fig 2 a and b). The identification of 5FU and its FNuc metabolites was made based on documented chemical shifts(38, 39). The intratumoral concentration of 5FU peaked in the initial spectrum and subsequently decreased rapidly for both tumor types. CD-UPRT- tumors produced modest FNuc signals reaching an average peak concentration of only 1.24 ± 0.31 mM. In contrast, the intratumoral FNuc concentration in CD-UPRT+ rapidly reached a plateau, achieving an average concentration of 3.34 ± 0.27 mM, and decreased only slightly thereafter. These data demonstrate that 5FU is an effective probe for detecting the rapid accumulation of a marker metabolite (FNuc) specific for UPRT activity by 19FMRS. The 19FMRS data were validated through Western blot analysis performed following each experiment which confirmed that the expression of the CD-UPRT fusion gene was limited to the CD-UPRT+ tumors.
Supplementary Figure 3. Schematic representation of the bicistronic retroviral vector. The viral long terminal repeats (LTR) control expression of the Saccharomyces cerevisiae cytosine deaminase (yCDA)-Haemophilus influenzae uracil phosphoribosyltransferase (iUPP) fusion gene, while expression of the neomycin phosphotransferase (NEO) from the single transcript is mediated by an encephalomyocarditis virus derived internal ribosome entry site (IRES)(40). The coding sequences for yCDA (Erbs P et al. Curr Genet. 1997 Jan;31(1):1-6) and iUPP (Fleischmann RD et al. Science. 1995 Jul 28;269(5223):496-512) have been described previously and were linked by a glycine adaptor. Unique restriction sites used for cloning are indicated. Plasmid containing yCDA cDNA was kindly provided by Professor Brian Ross, University of Michigan. An iUPP encoding cosmid was obtained from ATCC (Rockville, MD).
Supplementary Figure 4. Plots showing impact of the different fixed values for the half-saturation concentration of the FNuc to anabolites reaction, Km,FNuc used in the pharmacokinetic. (a) Plot of the final weighted residuals versus Km,FNuc. The model produced virtually identical results for values of Km,FNuc less than about 100 μM, below which the rate equation describing the anabolism of FNuc is effectively zero order and therefore completely determined by . The fits became significantly worse for values of Km,FNuc above 100 μM only stabilizing when the rate equation effectively became linear at a Km,FNuc above about 420 mM. (b) Plot of the fit determined values for (–––) and (– – –) versus Km,FNuc. Both of these Vmax values were largely independent of the value for Km,FNuc thus, any uncertainty in the precise value for Km,FNuc is of little consequence.