Abstract
Barrett's esophagus (BE) is a premalignant condition, where normal squamous epithelium is replaced by intestinal epithelium. BE is associated with an increased risk of developing esophageal adenocarcinoma (EAC). However, the BE cell of origin is not clear. We hypothesize that BE tissue originates from esophageal squamous cells, which can differentiate to columnar cells as a result of repeated exposure to gastric acid and bile acids, two components of refluxate implicated in BE pathology. To test this hypothesis, we repeatedly exposed squamous esophageal HET1A cells to 0.2 mM bile acid (BA) cocktail at pH 5.5 and developed an HET1AR-resistant cell line. These cells are able to survive and proliferate after repeated 2-h treatments with BA at pH 5.5. HET1AR cells are resistant to acidification and express markers of columnar differentiation, villin, CDX2, and cytokeratin 8/18. HET1AR cells have increased amounts of reactive oxygen species, concomitant with a decreased level and activity of manganese superoxide dismutase compared with parental cells. Furthermore, HET1AR cells express proteins and activate signaling pathways associated with inflammation, cell survival, and tumorigenesis that are thought to contribute to BE and EAC development. These include STAT3, NF-κB, epidermal growth factor receptor (EGFR), cyclooxygenase-2, interleukin-6, phosphorylated mammalian target of rapamycin (p-mTOR), and Mcl-1. The expression of prosurvival and inflammatory proteins and resistance to cell death could be partially modified by inhibition of STAT3 signaling. In summary, our study shows that long-term exposure of squamous cells to BA at acidic pH causes the cells to display the same characteristics and markers as BE.
Keywords: esophageal adenocarcinoma, reactive oxygen species, cytokeratin 8/18
barrett's esophagus (be) is a metaplastic lesion in the distal esophagus that commonly occurs in patients with gastroesophageal reflux disease (GERD). BE patients have a significantly increased risk for developing esophageal adenocarcinoma (EAC) (57). However, the exact mechanism by which GERD promotes BE is unclear.
Gastric acid and bile acids appear to be two major risk factors for the development of BE (40). Clinical studies have identified glycine-conjugated bile acids as the predominant bile acids appearing in the esophagus of patients with GERD (40). Bile acids are natural detergents synthesized in the liver and stored in the gall bladder, which are essential for the digestion of lipids. However, high levels of bile acids are known to promote gastrointestinal cancers (9). Bile acids were shown to induce oxidative stress, DNA damage, and mitochondrial damage (9). Furthermore, gastric acid and/or bile acids are known to induce oxidative stress and alter signaling pathways, such as MAPK, NF-κB, and STAT3 (2, 15, 56). These signaling pathways are associated with increased proliferation and decreased apoptosis of esophageal cells. Our recent study shows that gastric acid and bile acids in combination act synergistically to cause marked DNA damage (21). The DNA damage will lead in the majority of cells to the induction of cell death; however, few cells will attempt to adapt to these stressful conditions by the activation of stress response pathways. This may lead to a phenotypic switch and formation of metaplastic epithelium that is resistant to acid and bile acids and is able to survive repeated reflux episodes.
In the current study, we report the effects of repeated, long-term exposure of esophageal squamous HET1A cells to bile acids at pH 5.5. The goal was to develop and characterize squamous esophageal cells resistant to the acidic pH/bile acid combination. We hypothesized that long-term exposure of squamous cells to bile acids at pH 5.5 induces persistent stress that will lead to the clonal selection of cells that are resistant to cell death and exhibit activation of cell survival pathways, such as NF-κB or STAT3 signaling.
The identification of molecular changes within the normal esophageal tissue that contribute to BE, may assist in the development of new treatments for BE and biomarkers for early detection of esophageal adenocarcinoma.
METHODS
Cell lines and chemicals.
HET1A cells were provided by Dr. Curtis C. Harris (National Cancer Institute, Bethesda, MD). HET1A is a normal human esophageal epithelial cell line immortalized by transfection of the SV40 T antigen early region gene. These cells have epithelial morphology, stain positively for cytokeratin 13, and have remained nontumorigenic in athymic, nude mice for more than 12 mo (59). The cells were cultured in BRFF-EPM2 medium (Athena Environmental Sciences, Baltimore, MD) supplemented with 50 μg/ml gentamicin and 0.25 μg/ml fungizone.
Initially, HET1A cells were exposed for 10 min to medium acidified to pH 5.5 in the presence of 0.2 mM bile acid cocktail (BA). The cells were then split, and when they were 80–90% confluent, they were exposed again to 0.2 mM BA at pH 5.5 (on average once or twice a week). As the cells became adapted, the time of exposure was increased by 5 min. This procedure was repeated until the cells were able to survive and proliferate after exposure to 0.2 mM BA at pH 5.5 for 120 min. It took 90 wk to achieve this resistance. The cells were unexposed to bile acids at pH 5.5 for 21 days before being evaluated or the cells were exposed to 0.2 mM BA at pH 5.5 for 2 h and evaluated 24 h later as noted.
Bile acid cocktail consisted of an equimolar mixture of glycocholate, taurocholate, glycodeoxycholate, glycochenodeoxycholate, and deoxycholate at a final concentration of 0.2 mM. This cocktail reflects the mixture of bile acids to which the distal esophagus is ordinarily exposed during gastroesophageal reflux (40). AG490 was from Cayman Chemical (Ann Arbor, MI), Hydroethidine (HE), propidium iodide (PI), and Hoechst 3222 were from Molecular Probes, (Eugene, OR). All other chemicals were of the highest purity available.
Evaluation of cell death, cytotoxicity, and doubling time.
HET1A and HET1AR were treated for 2 h with either control medium or medium with 0.2 mM BA acidified to pH 5.5. Twenty-four hours after treatment, an aliquot of HET1A and HET1AR cells were cytospun onto slides using a Cytospin 2 (Shandon), fixed with 100% methanol for 6 min, air-dried, and stained with Giemsa stain for 4 h. Two hundred cells were evaluated, using bright field microscopy (×1,000), for apoptosis and necrosis, as described previously (12). Apoptotic cells were identified as cells with condensed chromatin, cell shrinkage, and nuclear fragmentation. In contrast, a pale cytoplasm, swelling, and disintegration of cytoplasmic and nuclear boundaries were the major characteristic features of necrotic cells.
Furthermore, cell viability/proliferation was analyzed using the CellTiter 96 AQueous nonradioactive cell proliferation assay {a[3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] (MTS) assay} (Promega, Madison, WI), according to the manufacturer's instructions, as described previously (20). This assay is a colorimetric assay for measuring the activity of mitochondrial enzymes that reduce MTS to purple formazan. Briefly, the cells were plated in 96-well plates, treated with indicated treatments for 2 h and evaluated 24 h later. Twenty microliters of MTS was added to each well, incubated at 37°C for 2 h, and the absorbance at 480 nm was read. Blank control values were subtracted from experimental and control samples, and the percentage of viable cells was calculated as follows: % viable cells = (A480 experiment − A480 blank control/A480 control − A480 blank control) × 100.
The increase in viable cell number compared with the initial number of cells was used to calculate the doubling time in the cells that were not exposed to any treatment for 21 days. HET1A and HET1AR (1 × 105) cells were plated in six-well plates, and after 24, 48, and 72 h, the cells were stained with trypan blue and counted.
Determination of oxidative stress.
Oxidative stress in esophageal cells was assessed by staining with HE, as described previously (18). Flow cytometry experiments were done in triplicate, and a minimum of 10,000 events were analyzed. Data are expressed as a histogram and as a graph indicating the percentage of HE-positive cells. In addition, fluorescent microscopy was employed to evaluate oxidative stress in live HET1A and HET1AR cells after staining with 2 μM HE for 30 min at 37°C (16).
Superoxide dismutase activities.
Cells (12 × 106) were sonicated in a buffer containing 100 mM diethanolamine; 100 mM triethanolamine at pH 7.45, centrifuged at 100,000 g for 1 h, and the supernatant was dialyzed for 16 h. CuZnSOD and MnSOD activities were determined using the method of Paoletti et al. (43), and the differential sensitivities of the enzymes to inactivation were determined by 50 mM diethylthiocarbamate (27). Activities were normalized to cellular protein measured using the BCA reagent following the manufacturer's instructions (Pierce Biotechnology, Rockford, IL).
Western blot analysis.
Western blot analysis was performed as previously described (18). The membranes were immunostained with antibodies against CuZnSOD (1:10,000; Abcam, Cambridge, MA), MnSOD (1:5,000; Upstate, Lake Placid, NY), Bag-3 (1:1,000; Imgenex, San Diego, CA), catalase (1:1,000; Rockland Immunochemicals, Gilbertsville, PA), cyclooxygenase-2 (1:250 COX-2; Cayman Chemical, Ann Arbor, MI), p53 (1:250; Santa Cruz Biotechnology, Santa Cruz, CA), EGFR (1:250; Santa Cruz Biotechnology), cytokeratin 8/18 (CK8/18, 1:250; Santa Cruz Biotechnology), phospho-STAT3tyr705 (1:300) and phospho-mTORser2448 (1:750; Cell Signaling, Boston, MA) β-actin (1:10,000; Calbiochem, Gibbstown, NJ), Mcl-1 (1:5,000;Thermo Scientific, Pittsburgh, PA), and then incubated with the appropriate secondary antibody conjugated to horseradish peroxidase (Pierce Biotechnology). The membranes were stripped using the Re-blot Western blot recycling kit (Chemicon International, Temecula, CA) and reprobed with a β-actin antibody or stained with Brilliant Blue G dye to confirm equal protein loading. The densities of individual bands were determined using QuantiScan software (Biosoft, Cambridge, UK).
Immunohistochemistry.
For fluorescence microscopy, the cells were grown on 4-chamber slides, fixed with formaldehyde, and permeabilized with methanol, as described previously (15). After blocking with 5% BSA, the cells were incubated overnight with antibodies against villin (1:100; BD Biosciences, San Jose, CA), CDX-2 (1:100; Biogenex, San Ramon, CA), STAT3 (1:100; Cell Signaling), pSTAT3 (1:100; Santa Cruz Biotechnology), or the p50 subunit of NF-κB (1:100; Santa Cruz Biotechnology). Alexa Fluor 488 secondary antibodies (1:100; Molecular Probes, Eugene, OR) were applied for 60 min. The slides were counterstained with Propidium iodide (PI) and coverslipped using VectaShield HardSet medium (Vector Laboratories, Burlingame, CA).
CK8/18 are markers of columnar epithelium that are not expressed by the squamous epithelium of the esophagus (10, 48). CK8/18 expression in HET1A and HET1AR cells was evaluated by immunohistochemistry according to protocols using the BenchMark XT IHC/ISH staining module (Ventana Medical Systems, Tucson, AZ). CK8/18 signal was detected using an iVIEW DAB detection kit (Ventana Medical Systems). The CK8/18 antibody was from Santa Cruz Biotechnology (1:100). Hematoxylin was used as a counterstain.
RNA preparation and real-time RT-PCR.
Total RNA was isolated from cells using the RNeasy Plus Mini Kit (Qiagen, Santa Clarita, CA), as described in the manufacturer's protocol. RNA concentration and purity were evaluated by NanoDrop (ThermoScientific, Wilmington, DE) at 260 nm/280 nm. Real-time RT-PCR assays were performed to quantify mRNA levels of IL-6, villin, and sodium hydrogen exchanger 1 (NHE1), as described previously (15).
IL-6 secretion.
A Quantikine HS ELISA kit was used to determine the concentration of IL-6 in conditioned media (R&D Systems, Minneapolis, MN). The assay was performed according to the manufacturer's instructions.
Intracellular pH measurement.
Intracellular pH was measured in cells by epifluorescence microscopy using the pH-sensitive dye 2′-7′-bis(carboxyethyl)-5(6)-carboxyfluorescein (BCECF; Invitrogen, Carlsbad, CA). Dye loading and calculation of the rate of recovery after acid load were performed as described previously (51). Cells were perfused with serum-free and phenol red-free RPMI at pH 7.4 to obtain a stable baseline intracellular pH (pHi). The solution was then changed to medium with Hcl to pH 5.5 or medium supplemented with 20 mM NH4Cl for 10 min. Minimal pHi (pHimin) was determined as an average of the 4 lowest pHi values. Data are from at least three independent experiments. In each experiment, data from a minimum 30 individual cells were evaluated.
Statistical evaluation.
The data are expressed as the means ± SE. The statistical difference between two cell lines was determined by a Student's t-test using a 95% confidence level.
RESULTS
Characterization of HET1AR resistance to acid and bile acids.
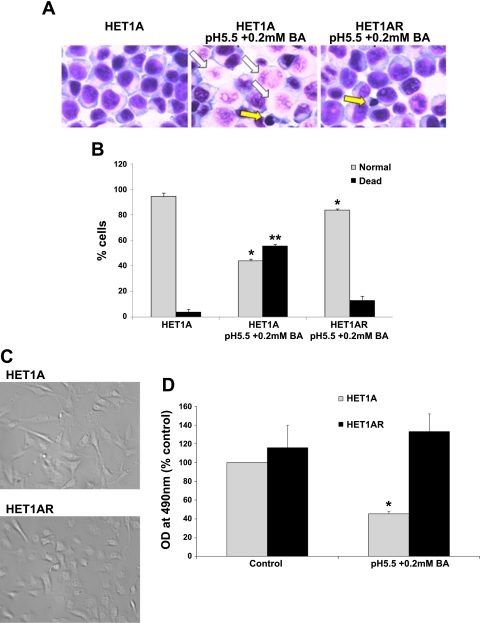
To mimic chronic reflux disease, squamous esophageal HET1A cells were exposed to medium acidified to pH 5.5 with 0.2 mM BA for increasing time intervals. An untreated long-passage HET1A cell line was concurrently maintained, as a control. These HET1AR new cells are able to survive and proliferate after exposure to 0.2 mM BA at pH 5.5 for 120 min. When parental HET1A cells were exposed for 120 min to pH 5.5 in combination with 0.2 mM BA and evaluated 24 h later, a majority of the cells died (Fig. 1, A and B). We found that only 13% of the resistant cells died after the exposure to 0.2 mM BA at pH 5.5 (3% by apoptosis and 10% by necrosis). In contrast, 56% of the parental cells died after a similar treatment, primarily by necrosis (43%); however, apoptotic cells (12%) were also found (Fig. 1A). Evaluation at later time points indicated that all HET1A cells were unable to proliferate and died; however, the HET1AR cells remained viable. In addition, HET1AR cells were morphologically different compared with parental cells (Fig. 1C). HET1AR cells appeared small and round, while the parental cells appeared spindle-shaped and more spread. The doubling time of HET1AR cells not exposed to any treatment was not significantly different from control HET1A cells (27.6 ± 4.6 h compared with 27.4 ± 3.6 h, n = 3).
Fig. 1.
HET1AR cells are resistant to the cytotoxic effects of bile acids in combination with acid. HET1A and HET1AR cells were treated with control medium or medium at pH 5.5 with 0.2 mM bile acid cocktail (BA) for 2 h, medium was replaced, and the cells were evaluated 24 h later by morphological criteria after staining with Giemsa stain (A). A: representative images indicating dead cells (necrotic and apoptotic) and alive cells after staining with Giemsa stain (magnification ×600). Several necrotic and apoptotic cells are indicated by white and yellow arrows, respectively. B: graph of % normal and dead HET1A and HET1AR cells treated as above. *Significant difference compared with normal HET1A cells (P < 0.05). **Significant difference compared with dead HET1A cells (P < 0.05). C: morphology of the parental HET1A and HET1AR cells at ×400 magnification. D: data from the MTS assay of HET1A and HET1AR (not exposed to acid and bile acids for 21 days) treated for 2 h with 0.2 mM BA at pH 5.5 and evaluated 24 h later. Data are expressed as means ± SE. *Significant difference compared with control cells (P < 0.05).
To confirm the morphological data, HET1A and HET1AR cells were exposed to a 120-min treatment with 0.2 mM BA at pH 5.5 and evaluated 24 h later by the MTS assay. More than 50% of the parental HET1A cells did not survive the treatment compared with untreated cells (Fig. 1D). In contrast, HET1AR cells did not show any decrease in the number of surviving cells after treatment with 0.2 mM BA at pH 5.5 compared with the untreated cells (Fig. 1D).
HET1AR cells display markers associated with metaplastic and columnar epithelium.
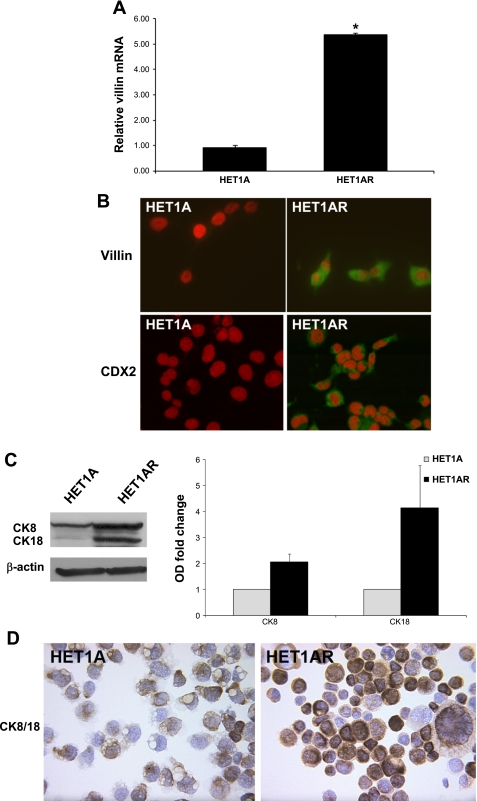
Villin is a marker of intestinal differentiation, which is commonly used to detect the presence of intestinal metaplasia. Villin mRNA levels were elevated more than five-fold in HET1AR cells compared with the HET1A cells (P < 0.05, Fig. 2A). Furthermore, increased villin immunostaining was found in the cytoplasm of HET1AR cells (Fig. 2B). We also evaluated, in both cell lines, the expression of CDX2, a transcription factor that regulates early stages of intestinal differentiation. We did not find increased mRNA levels, but immunostaining, in conjunction with fluorescent microscopy, revealed an increased signal of CDX2 in the cytoplasm of the HET1AR cells (Fig. 2B). Moreover, immunohistochemistry and immunoblotting show increased expression of columnar epithelial markers, cytokeratins 8/18 (CK8/18), in the HET1AR-resistant cells (Fig. 2, C and D).
Fig. 2.
Columnar marker expression in HET1AR cells. Control HET1A and resistant HET1AR cells were evaluated by RT-PCR, immunoblotting, or immunohistochemistry for the presence of columnar differentiation markers. A: relative villin mRNA levels, data are expressed as means ± SE from three independent experiments. *Significant difference (P < 0.05). Villin and CDX2 (green signal) were detected in the cytoplasm of the HET1AR cells, while no villin and CDX2 were detected in the control HET1A cells (B). The red signal indicates nuclei (propidium iodide). Immunoblots of CK8/18 and β-actin and a graph representing the relative densities of the immunoblot signal in HET1A and HET1AR cells are shown in C. D: representative images of HET1A and HETAR cells immunostained for columnar markers CK8 and CK18 (brown color, magnification ×400).
HET1AR display increased production of reactive oxygen species.
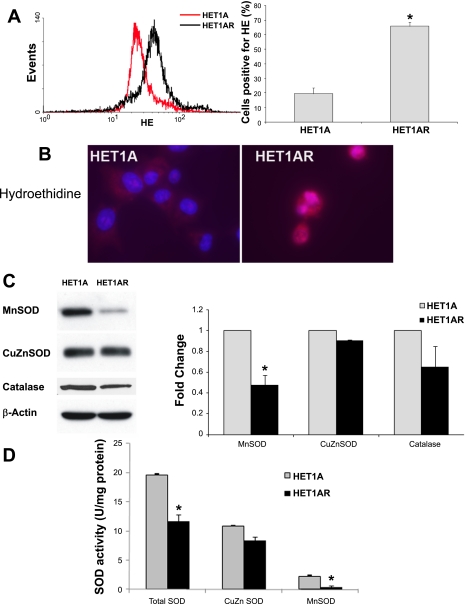
Since previous studies suggest that oxidative stress plays a role in BE pathogenesis, we wanted to determine whether exposure to 0.2 mM BA at pH 5.5 increased the baseline levels of reactive oxygen species (ROS). HET1AR cells were exposed to 0.2 mM BA at pH 5.5 for 2 h and evaluated 24 h later. Using flow cytometry to measure the fluorescence of HE, a probe that forms a fluorescent product in the presence of ROS, we found that HET1AR cells had significantly elevated levels of ROS (Fig. 3A). We found that 65.8 ± 2.8% of the HET1AR cells stained positively with HE compared with only 19.5 ± 3.8% of the parental HET1A cells (P < 0.05; Fig. 3A). Fluorescent microscopy confirmed these results, illustrating a high intensity of HE staining in the HET1AR cells, while the parental cells did not stain with HE (Fig. 3B).
Fig. 3.
Reactive oxygen species (ROS) and expression and activities of antioxidant proteins in HET1A cells and HET1AR cells. A: overlay of flow cytometry histograms of control HET1A cells and resistant HET1AR cell lines. The graph in A shows the flow cytometry analysis, indicating the percentage of cells staining positive with HE. B: representative images of live cells stained with HE (red color, magnification ×600). Immunoblots of MnSOD, CuZnSOD, catalase and β-actin and a graph showing the relative densities of catalase and MnSOD immunoblot signals in HET1A and HET1AR cells are shown in C. D: total superoxide dismutase (SOD), CuZnSOD, and MnSOD activities in HET1A and HET1AR cells. Data are expressed as a means ± SE. *Significant difference (P < 0.05).
HET1AR have decreased MnSOD protein and activity.
Next, we evaluated the antioxidant defense enzymes in the parental and resistant cells. We found that the HET1AR cells have significantly decreased protein expression of MnSOD, (Fig. 3C). Enzymatic activity of MnSOD was also significantly decreased in the HET1AR cells compared with parental cells (P < 0.05, Fig. 3D). Although, CuZnSOD protein and activity were not significantly decreased (Fig. 3, C and D), total SOD activity was significantly lower in the HET1AR cells (P < 0.05; Fig. 3D). The protein levels of another antioxidant enzyme, catalase, was also slightly, but not significantly, decreased in HET1AR cells compared with HET1A (Fig. 3C).
Activation of IL-6/STAT3 and NF-κB in HET1AR cells.
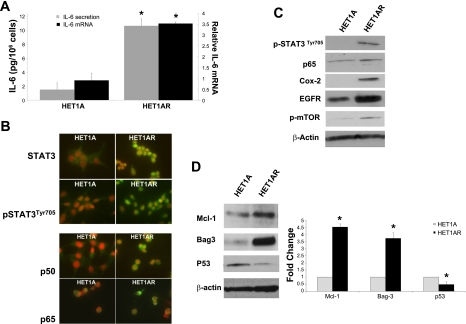
IL-6/STAT3 signaling plays a key role in many cellular processes, such as cell growth and apoptosis resistance, and is increased in BE and EAC (17). Consistent with these findings, the expression of IL-6 mRNA and secretion of IL-6 was significantly increased in HET1AR compared with the parental HET1A cells (Fig. 4A).
Fig. 4.
The expression of prosurvival factors and proapoptotic and antiapoptotic proteins. A: IL-6 mRNA and IL-6 secreted to medium as determined by ELISA assay in HET1A and HET1AR cells. Data represent the means ± SE from three independent experiments. B: representative images of STAT3, activated (phosphorylated) STAT3 (pSTAT3 ) the p50 and p65 subunits of NF-κB in control HET1A, and resistant HET1AR cells (STAT3, pSTAT3, p50, and p65 are indicated by a green color; note that the majority of signal in the resistant cells is present in the nuclei, magnification ×600). Nuclei are counterstained with propidium iodide (red). C: immunoblots of pSTAT3(Tyr705), p65 subunit of NF-κB, COX-2, EGFR, and p-mTOR. Mcl-1, Bag-3, and p53 are shown in D. Graph shows the fold change in relative densities of Mcl-1, Bag-3, and p53 on the immunoblots. The data were obtained from three independent experiments. *Significant difference (P < 0.05).
Expression, cellular localization, and phosphorylation (activation) of STAT3 were next evaluated in HET1AR and HET1A. Immunostaining, in conjunction with fluorescent microscopy, showed that STAT3 protein expression is upregulated, and STAT3 is translocated into the nuclei of HET1AR cells (Fig. 4B). The expression of activated (phosphorylated) STAT3 (Tyr705) was also increased in the nuclei of the HET1AR cells (Fig. 4B). Western blots confirmed the presence of phosphorylated STAT3 in the HET1AR cells (Fig. 4C).
The transcription factor NF-κB, which regulates the expression of genes associated with inflammation and tumorigenicity, is often increased in BE and EAC (62). The immunofluorescence staining showed that the expression of the p50 and p65 subunits of NF-κB was increased in the HET1AR cells compared with the HET1A cells. The signal was primarily detected in the nuclei, suggesting that NF-κB was translocated into the nucleus, where it can activate the transcription of target genes (Fig. 4B). In addition, we confirmed the increase in p65 subunit expression in HET1AR cells by Western blot (Fig. 4C); however, this increase was not substantial.
HET1AR cells express proteins that are up-regulated in BE and EAC.
In our next experiments, we evaluated the expression of prosurvival and proinflammatory proteins that are commonly elevated in biopsies obtained from patients with BE and/or EAC. Overexpression of COX-2 and EGFR, which are involved in cell survival and proliferation, is highly associated with BE and EAC development (36). By analyzing Western blots for these proteins, we observed that COX-2 is absent in HET1A cells but robustly expressed in HET1AR cells (Fig. 4C). Immunoblotting revealed that EGFR was increased by more than twofold in HET1AR cells compared with the HET1A cells (Fig. 4C). Mammalian target of rapamycin (mTOR) is another important regulator of cell survival, proliferation, and angiogenesis. The phosphorylated (activated) form of this protein was overexpressed in the HET1AR compared with the HET1A cells (Fig. 4C).
On the basis of the increased activated STAT3 in the HET1AR cells, we examined Mcl-1 levels. Mcl-1 is an antiapoptotic protein regulated by STAT3 that is increased in BE (17). As expected, Mcl-1 was increased in the HET1AR cells (Fig. 4D). Bag-3, another protein that is linked to resistance to cell death and is induced in response to oxidative stress (45, 46), was also markedly elevated in the HET1AR cells (Fig. 4D). In contrast, the tumor suppressor p53 was decreased by more than 50% in HET1AR compared with HET1A cells (P < 0.05, Fig. 4D).
Inhibition of STAT3 signaling downregulated the expression of prosurvival proteins.
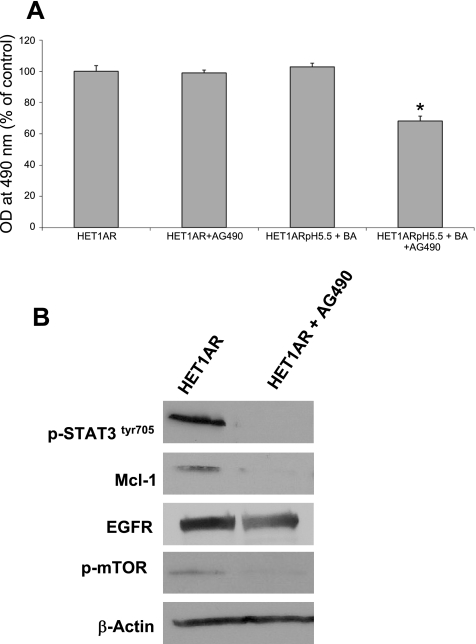
To determine whether the elevated STAT3 signaling contributes to the resistance phenotype, we tested whether inhibition of STAT3 signaling can affect protein expression/activation and cell survival. For these experiments, we used AG490, an inhibitor of JAK2/STAT3 signaling (41). First, we tested whether AG490 can reverse resistance to the treatment with 0.2 mM BA at pH 5.5 in the HET1AR cells. We found, by MTS assay, that pretreatment with AG490 for 30 min followed by incubation with 0.2 mM BA at pH 5.5 for 2 h and 24 h in medium containing AG490, reduced cell survival (Fig. 5A). AG490 alone or 0.2 mM BA at pH 5.5 alone had no significant effect on survival.
Fig. 5.
AG490 reverses resistance to acid and bile acids and decreases the expression of proteins associated with cell survival in HET1AR cells. HET1AR cells were pretreated with AG490 and exposed to acid and bile acids for 2 h and evaluated 24 h later. A: data from the MTS assay of HET1AR. Data are expressed as means ± SE. *Significant difference (P < 0.05). B: immunoblots show the expression of p-STAT3, Mcl-1, EGFR, and p-mTOR after 24 h incubation with AG490.
Furthermore, treatment with 10 μM AG490 for 24 h completely inhibited STAT3 activation (Fig. 5B). Mcl-1 expression was eliminated in HET1AR cells following incubation with AG490 (Fig. 5B). AG490 treatment also caused a decrease in EGFR expression and mTOR activation (Fig. 5B).These data suggest that increased STAT3 signaling partially contributes to the resistance phenotype of the HET1AR cells.
HET1AR cells are resistant to acidification.
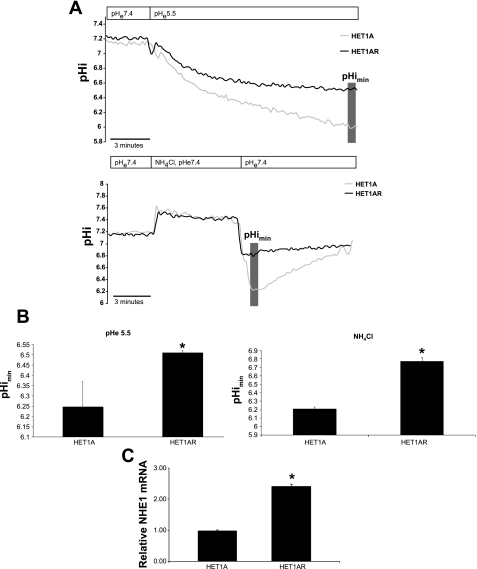
Our recent studies show that the NHE1 is highly expressed in clinical samples of BE (21). NHE1 is an important regulator of intracellular pH (64). Because of the acid and bile acid resistance of the HET1AR cells, we analyzed the ability of these cells to regulate intracellular pH and the expression of NHE1. We analyzed the minimal intracellular pHi (pHimin) in both cell lines following a 10-min exposure to pH 5.5 or acidification by NH4Cl. Serum-free, phenol red-free RPMI supplemented with sodium bicarbonate was used as the bathing solution during pHi readings. The baseline pHi was not significantly different; 7.28 ± 0.28 and 7.23 ± 0.35 in HET1A and HET1AR cells, respectively. However, following 10 min of an acid pulse (pH 5.5), pHimin in HET1AR cells was 6.51 ± 0.01, while in HET1A cells, pHimin was 6.24 ± 0.12 (P < 0.05, Fig. 6, A and B). Acidification induced by treatment with 20 mM NH4Cl also showed a statistically significant difference between HET1A and HET1AR cells; minimal intracellular pHi was 6.21 ± 0.02 and 6.77 ± 0.05, respectively (P < 0.05, Fig. 6, A and B). When we examined the expression of NHE1, we found a significant increase in NHE1 mRNA in HET1AR cells compared with the HET1A cells (Fig. 6C).
Fig. 6.
Intracellular pH in HET1A and HET1AR cells. Intracellular pH was determined by microfluorometry in cells perfused with acidic medium (pH 5.5) or 20 mM NH4Cl. A: pHi traces, pH of medium used (pHe) is indicated in the top boxes. B: minimal intracellular pH. C: mRNA expression of NHE1 in HET1A and HET1AR cells. The data are expressed as a means ± SE (n > 3). In each experiment, ∼15–30 cells were evaluated. *Statistically significant difference (P < 0.05).
DISCUSSION
BE is characterized as a metaplastic lesion, where normal esophageal epithelium is replaced by intestinal epithelium. BE appears to result from chronic, repeated irritation of the esophageal mucosa by gastric acids, proteases, and bile acids that lasts several years (8, 44). Several different theories exist on the cell of BE origin (57). Originally, it was proposed that the cells from gastoresophageal junction migrate to the distal part of esophagus as a consequence of damage caused by refluxate. Other theories propose abnormal differentiation of pluripotent stem cells in the squamous esophagus or submucosal glands. Finally, BE may evolve from native squamous epithelium that undergoes transdifferentiation as a result of chronic exposure to low pH and bile acids (28). To test this hypothesis and mimic chronic exposure to gastric acid and bile acids, we developed resistant cells from squamous esophageal cells that are able to survive and proliferate after repeated exposures to bile acids at acidic pH. Importantly, these new resistant cells activate signaling pathways and express inflammatory, oncogenic, and metaplastic markers similar to those seen in BE.
Currently, two squamous esophageal cell models are used in in vitro experiments to establish the role of acid and/or bile acids in the development of BE from squamous epithelium. These two models are 1) human keratinocytes (EPC-hTERT, NES) that are immortalized with human telomerase (hTERT) transfection; and 2) HET1A cells that are immortalized by SV40 T antigen. Both of these models have limitations. Telomerase maintains telomere integrity; however, recent studies show that telomerase is involved in many different processes, independent of the ability to maintain telomeres, such as apoptosis resistance, altered gene expression, and DNA damage. (11, 58) For example, the expression of hTERT is linked to increased resistance to apoptosis, an upregulation of growth promoting genes, and a downregulation of growth inhibitory genes (55). Furthermore, recent studies showed that ectopic expression of hTERT in epithelial cells resulted in reduced basal levels of active p53-, and p53-dependent signaling (7). We used HET1A cells since these cells are nontumorigenic; however, we recognize that the SV40T antigen can also cause p53 and pRB inactivation and changes in DNA methylation (32). In this study, we compared parental long-passage control HET1A to resistant HET1AR cells to determine the major changes induced by long-term treatment with bile acids at acidic pH. The cells were repeatedly exposed to 0.2 mM bile acid cocktail at pH 5.5 for up to 120 min. These concentrations and time exposures are relevant to BE patients, since these patients experience simultaneous reflux of acid and bile acids for long time periods compared with normal individuals (40, 61). The total percentage of time when bilirubin absorbance was ≥0.14 in the esophagus (an indication of bile reflux) was 46% (∼11 h/24 h) in BE patients but only 1.5% in normal controls (61). Similarly, the total percentage of time when the esophageal pH was <4 was 22.8% (∼5.5 h/24 h) in BE patients with dysplasia but only 0.4% in normal controls (61).
Development of the resistance phenotype in the HET1AR cells was accompanied by morphological changes and increases in several markers of columnar differentiation, villin, CDX2, and CK8/18. These markers are also increased in BE metaplasia (19, 38). Furthermore, these results are in agreement with recent studies, indicating that bile acids at low pH may induce an increase in the expression of CDX2, CK8/18, and villin in other model systems (5, 6, 13, 24, 25, 66). At this point, it is not clear why we did not observe an increase in CDX2 mRNA, while villin mRNA was significantly increased. It is possible that turnover or stability of CDX2 mRNA in these new cells derived from squamous cells is different compared with true columnar cells.
Epidemiological and clinical studies indicate that malignant transformation of BE is associated with oxidative stress, decreased antioxidant capacity, and low glutathione content (26, 53, 54) . Consistent with these studies, we have shown that the cells resistant to bile acids at pH 5.5 have increased levels of ROS, while the activities and expression of antioxidant enzymes are decreased. The major enzymes responsible for the attenuation of ROS are the family of superoxide dismutases, which convert superoxide radicals into hydrogen peroxide, and catalase, which converts hydrogen peroxide to water and oxygen. The loss of SOD and catalase increases levels of ROS, thus creating conditions favorable for increased proliferation and mutation (22). We found a significant decrease in the total superoxide dismutase activity. The decrease in total SOD activity is likely due to decreased MnSOD; CuZnSOD protein and activity were unchanged. The loss of MnSOD function was previously shown to be associated with increased incidence of BE (37). Furthermore, a recent study by Li et al. (35) suggests that exposure to hydrophobic bile acids decreases MnSOD activity. Our data demonstrate that MnSOD activity can be significantly decreased by chronic treatment with bile acids at low pH.
Many tumor types have decreased MnSOD compared with normal tissue (31); however, repression of MnSOD during transformation is not well understood. One possibility is increased methylation of the promoter (23). A second possibility is decreased FOXO3a-induced MnSOD transcription due to increased activation of Akt (52). MnSOD is also regulated by a combination of Sp1, p53, nucleophosmin, and NF-κB (14). Recent evidence indicates that the relative abundance of the different NF-κB subunits contributes to the positive or negative regulation of MnSOD (14). Increased p50/p50 homodimers result in repression, while an increase in p50/p65 heterodimers positively regulates MnSOD (14, 60). The relative increase in nuclear p50 in the HET1AR cells could contribute to the observed decrease in MnSOD.
The expression of catalase was also decreased in resistant HET1AR cells; however, the difference did not reach statistical significance. Furthermore, increased levels of Bag-3, protein that has a central role in the protection of cancer cells against cell death induced by oxidative stress, were found in HET1AR cells (47). These results indicate HET1AR cells have increased basal levels of ROS; thus, Bag-3 may be one of the proteins responsible for the cells' ability to survive.
The transcription factor NF-κB regulates cell proliferation, inflammation, and survival; its activity is upregulated in dysplastic BE and EAC (3, 42). Many studies showed that exposure of esophageal cancer cells to bile acids at low pH induces NF-κB activation (30, 63). We observed increased expression of the p50 subunit of NF-κB primarily in the nuclei of HET1AR cells, suggesting constitutive activation of NF-κB. The p65 subunit of NF-κB was also elevated in HET1AR cells; however, this increase was not robust. Furthermore, COX-2, the NF-κB-regulated protein implicated in BE pathogenesis (1), was also upregulated in HET1AR cells.
Our data are consistent with elevated IL-6/STAT3 signaling playing a role in the development of BE (15, 17). Our previous studies showed that, compared with duodenum and squamous epithelium, BE cells secrete large amounts of IL-6 and express activated STAT3, which leads to increased levels of antiapoptotic proteins (17). A more recent study demonstrated that BE cell lines that lack functional p53, and express active H-Ras, secrete IL-6 and activate the STAT3 signaling pathway (65). In the current study, we found increased IL-6 levels and STAT3 activation in HET1AR cells. Mcl-1, the anti-apoptotic Bcl-2 family protein regulated by the STAT3 pathway, was also significantly increased in HET1AR cells. The inhibition of STAT3 signaling with AG490 decreased Mcl-1 expression and partially sensitized the HET1AR cells to cell death induced by bile acids at low pH, indicating that increased STAT3 signaling may contribute to the observed resistance in the HET1AR cells.
Mutations or amplifications of EGFR or EGFR family members are implicated in BE and EAC development (33, 36). Bile acids alone or in combination with acid may activate EGFR (5). In agreement with these studies, EGFR expression was significantly increased in HET1AR cells compared with the HET1A cells. The activity of mTOR, another protein that plays a significant role in cell proliferation, survival, and migration was elevated in HET1AR cells. Interestingly, EGFR expression and mTOR activation could be downregulated by a AG490 treatment, indicating again the importance of STAT3 signaling in esophageal tumorigenesis. Previous reports showed that p53 mutation and loss of heterozygosity is one of the best defined markers of BE oncogenesis (29). The most common response to stress induced by the tumor suppressor gene p53 is cell cycle arrest or DNA repair. If the DNA damage cannot be repaired, p53 induces cell death. Our data show that the protein levels of p53 are significantly decreased in HET1AR cells compared with the HET1A cells. Interestingly, p53 can repress IL-6 transcription (4). Therefore, it is possible that a decrease in p53 expression in combination with IL-6/STAT3 activation may play an important role in BE pathogenesis.
Repeated exposures to acid may also lead to changes in intracellular pH (pHi) and the increased ability of cells to adapt to acid exposure. Our data demonstrate that cell acidification is diminished in esophageal cells resistant to bile acids at low pH compared with control cells. This finding suggests that HET1AR cells have evolved an ability to adapt to an acidic environment and perhaps extrude intracellular protons faster. However, we did not observe a significant difference in baseline pH, suggesting that the acid extrusion modulation is likely mediated by enzymes that are activated only in the presence of acid. We hypothesized that this acid extrusion may be mediated by increased expression of integral H+ transporters, such as the NHE1. Indeed, NHE1 mRNA levels were significantly increased in HET1AR cells compared with parental HET1A cells. However, selective inhibition of NHE-1 using Zoniporide (39) did not affect the pHimin during treatment with either acidified medium or during NH4Cl prepulse (data not shown). This suggests that other mechanisms, such as Cl−/HCO3− exchange (34, 49), different isoforms of NHE, or the difference in cell volume, may contribute to the altered acidification in the HET1AR cells.
Our recent studies show NHE1 is highly expressed in BE and EAC (21). Previous studies showed that NHE1 plays an important role in regulation of cell survival, migration, tumor growth, and invasion (50). Thus, it is possible that in addition to its function as a proton antiporter, NHE1 may be linked to increased risk of BE development.
Metaplasia, such as BE, arises in tissues undergoing continuous regeneration after chronic trauma. Transdifferentiation will occur if the combination of transcription factors that are normally expressed is altered in the course of regeneration because of mutation or environmental effects. It can only occur in a few cells, and if the new tissue type has a growth advantage, it will expand to become a macroscopic lesion. We have shown that repeated long-term exposure to bile acids at low pH in cells that originate from squamous epithelium activate transcription factors and expression of proteins that are associated with columnar phenotype. While our data support the transdifferentiation theory, other theories cannot be excluded, especially the abnormal differentiation of stem cells from squamous epithelium into columnar epithelium as a result of the exposure to refluxate.
In summary, the exposure of esophageal epithelium to bile acids at acidic pH is an important factor in BE pathogenesis. HET1AR cells resistant to 0.2 mM BA at pH 5.5 serve as an in vitro model showing that squamous esophageal cells, repeatedly exposed to these two major components of refluxate, express hallmarks typical of BE. This model cell line may contribute to a better understanding of the molecular mechanisms underlying BE pathogenesis and provide important insights into the development of new therapeutic strategies for BE. On the basis of the data from this study, we propose that not only should the reflux of gastric acid be controlled but control of duodenal reflux may also be critical in patients with GERD.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
ACKNOWLEDGMENTS
The work was supported by GI SPORE Grant P50 CA95060 from NCI (K. Dvorak) and HD039657 (B. Dvorak) from National Institute of Child Health and Human Development.
REFERENCES
- 1. Abdel-Latif MM, Kelleher D, Reynolds JV. Potential role of NF-κB in esophageal adenocarcinoma: as an emerging molecular target. J Surg Res 153: 172–180, 2009 [DOI] [PubMed] [Google Scholar]
- 2. Abdel-Latif MM, O'Riordan J, Windle HJ, Carton E, Ravi N, Kelleher D, Reynolds JV. NF-κB activation in esophageal adenocarcinoma: relationship to Barrett's metaplasia, survival, and response to neoadjuvant chemoradiotherapy. Ann Surg 239: 491–500, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Abdel-Latif MM, O'Riordan JM, Ravi N, Kelleher D, Reynolds JV. Activated nuclear factor-κB and cytokine profiles in the esophagus parallel tumor regression following neoadjuvant chemoradiotherapy. Dis Esophagus 18: 246–252, 2005 [DOI] [PubMed] [Google Scholar]
- 4. Angelo LS, Talpaz M, Kurzrock R. Autocrine interleukin-6 production in renal cell carcinoma: evidence for the involvement of p53. Cancer Res 62: 932–940, 2002 [PubMed] [Google Scholar]
- 5. Avissar NE, Toia L, Hu Y, Watson TJ, Jones C, Raymond DP, Matousek A, Peters JH. Bile acid alone, or in combination with acid, induces CDX2 expression through activation of the epidermal growth factor receptor (EGFR). J Gastrointest Surg 13: 212–222, 2009 [DOI] [PubMed] [Google Scholar]
- 6. Bajpai M, Liu J, Geng X, Souza RF, Amenta PS, Das KM. Repeated exposure to acid and bile selectively induces colonic phenotype expression in a heterogeneous Barrett's epithelial cell line. Lab Invest 88: 643–651, 2008 [DOI] [PubMed] [Google Scholar]
- 7. Beliveau A, Bassett E, Lo AT, Garbe J, Rubio MA, Bissell MJ, Campisi J, Yaswen P. p53-dependent integration of telomere and growth factor deprivation signals. Proc Natl Acad Sci USA 104: 4431–4436, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bernstein C, Bernstein H, Payne CM, Garewal H. Field defects in progression to adenocarcinoma of the colon and esophagus. Electronic J Biotechnol 3: 167–183, 2000 [Google Scholar]
- 9. Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res 589: 47–65, 2005 [DOI] [PubMed] [Google Scholar]
- 10. Chang CL, Lao-Sirieix P, Save V, De La Cueva Mendez G, Laskey R, Fitzgerald RC. Retinoic acid-induced glandular differentiation of the oesophagus. Gut 56: 906–917, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cong Y, Shay JW. Actions of human telomerase beyond telomeres. Cell Res 18: 725–732, 2008 [DOI] [PubMed] [Google Scholar]
- 12. Crowley-Weber CL, Dvorakova K, Crowley C, Bernstein H, Bernstein C, Garewal H, Payne CM. Nicotine increases oxidative stress, activates NF-κB and GRP78, induces apoptosis and sensitizes cells to genotoxic/xenobiotic stresses by a multiple stress inducer, deoxycholate: relevance to colon carcinogenesis. Chem Biol Interact 145: 53–66, 2003 [DOI] [PubMed] [Google Scholar]
- 13. Das KM, Kong Y, Bajpai M, Kulkarni D, Geng X, Mishra P, Banerjee D, Hirshfield K. Transformation of benign Barrett's epithelium by repeated acid and bile exposure over 65 weeks: A novel in-vitro model. Int J Cancer 128: 274–282, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dhar SK, Xu Y, Chen Y, St. Clair DK. Specificity protein 1-dependent p53-mediated suppression of human manganese superoxide dismutase gene expression. J Biol Chem 281: 21698–21709, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dvorak K, Chavarria M, Payne CM, Ramsey L, Crowley-Weber C, Dvorakova B, Dvorak B, Bernstein H, Holubec H, Sampliner RE, Bernstein C, Prasad A, Green SB, Garewal H. Activation of the interleukin-6/STAT3 antiapoptotic pathway in esophageal cells by bile acids and low pH: relevance to Barrett's esophagus. Clin Cancer Res 13: 5305–5313, 2007 [DOI] [PubMed] [Google Scholar]
- 16. Dvorak K, Payne CM, Chavarria M, Ramsey L, Dvorakova B, Bernstein H, Holubec H, Sampliner RE, Guy N, Condon A, Bernstein C, Green SB, Prasad A, Garewal HS. Bile acids in combination with low pH induce oxidative stress and oxidative DNA damage: relevance to the pathogenesis of Barrett's oesophagus. Gut 56: 763–771, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dvorakova K, Payne CM, Ramsey L, Holubec H, Sampliner R, Dominguez J, Dvorak B, Bernstein H, Bernstein C, Prasad A, Fass R, Cui H, Garewal H. Increased expression and secretion of interleukin-6 in patients with Barrett's esophagus. Clin Cancer Res 10: 2020–2028, 2004 [DOI] [PubMed] [Google Scholar]
- 18. Dvorakova K, Waltmire CN, Payne CM, Tome ME, Briehl MM, Dorr RT. Induction of mitochondrial changes in myeloma cells by imexon. Blood 97: 3544–3551, 2001 [DOI] [PubMed] [Google Scholar]
- 19. Glickman JN, Chen YY, Wang HH, Antonioli DA, Odze RD. Phenotypic characteristics of a distinctive multilayered epithelium suggests that it is a precursor in the development of Barrett's esophagus. Am J Surg Pathol 25: 569–578, 2001 [DOI] [PubMed] [Google Scholar]
- 20. Goldman A, Condon A, Adler E, Minnella M, Bernstein C, Bernstein H, Dvorak K. Protective effects of glycoursodeoxycholic acid in Barrett's esophagus cells. Dis Esophagus 23: 83–93, 2010 [DOI] [PubMed] [Google Scholar]
- 21. Goldman A, Shahidullah M, Goldman D, Khailova L, Watts G, Delamere N, Dvorak K. A novel mechanism of acid and bile acid-induced DNA damage involving Na+/H+ exchanger: implication for Barrett's oesophagus. Gut 59: 1606–1616, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hodge DR, Peng B, Pompeia C, Thomas S, Cho E, Clausen PA, Marquez VE, Farrar WL. Epigenetic silencing of manganese superoxide dismutase (SOD-2) in KAS 6/1 human multiple myeloma cells increases cell proliferation. Cancer Biol Ther 4: 585–592, 2005 [DOI] [PubMed] [Google Scholar]
- 23. Hodge DR, Xiao W, Peng B, Cherry JC, Munroe DJ, Farrar WL. Enforced expression of superoxide dismutase 2/manganese superoxide dismutase disrupts autocrine interleukin-6 stimulation in human multiple myeloma cells and enhances dexamethasone-induced apoptosis. Cancer Res 65: 6255–6263, 2005 [DOI] [PubMed] [Google Scholar]
- 24. Hu Y, Williams VA, Gellersen O, Jones C, Watson TJ, Peters JH. The pathogenesis of Barrett's esophagus: secondary bile acids upregulate intestinal differentiation factor CDX2 expression in esophageal cells. J Gastrointest Surg 11: 827–834, 2007 [DOI] [PubMed] [Google Scholar]
- 25. Huo X, Zhang HY, Zhang XI, Lynch JP, Strauch ED, Wang JY, Melton SD, Genta RM, Wang DH, Spechler SJ, Souza RF. Acid and bile salt-induced CDX2 expression differs in esophageal squamous cells from patients with and without Barrett's esophagus. Gastroenterology 139: 194–203 e191, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Inayama M, Hashimoto N, Tokoro T, Shiozaki H. Involvement of oxidative stress in experimentally induced reflux esophagitis and esophageal cancer. Hepatogastroenterology 54: 761–765, 2007 [PubMed] [Google Scholar]
- 27. Iqbal J, Whitney P. Use of cyanide and diethyldithiocarbamate in the assay of superoxide dismutases. Free Radic Biol Med 10: 69–77, 1991 [DOI] [PubMed] [Google Scholar]
- 28. Jankowski JA, Harrison RF, Perry I, Balkwill F, Tselepis C. Barrett's metaplasia. Lancet 356: 2079–2085, 2000 [DOI] [PubMed] [Google Scholar]
- 29. Jenkins GJ, Doak SH, Parry JM, D'Souza FR, Griffiths AP, Baxter JN. Genetic pathways involved in the progression of Barrett's metaplasia to adenocarcinoma. Br J Surg 89: 824–837, 2002 [DOI] [PubMed] [Google Scholar]
- 30. Jenkins GJ, Harries K, Doak SH, Wilmes A, Griffiths AP, Baxter JN, Parry JM. The bile acid deoxycholic acid (DCA) at neutral pH activates NF-κB and induces IL-8 expression in oesophageal cells in vitro. Carcinogenesis 25: 317–323, 2004 [DOI] [PubMed] [Google Scholar]
- 31. Kinnula VL, Crapo JD. Superoxide dismutases in malignant cells and human tumors. Free Radic Biol Med 36: 718–744, 2004 [DOI] [PubMed] [Google Scholar]
- 32. Kong J, Nakagawa H, Isariyawongse BK, Funakoshi S, Silberg DG, Rustgi AK, Lynch JP. Induction of intestinalization in human esophageal keratinocytes is a multistep process. Carcinogenesis 30: 122–130, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kwak EL, Jankowski J, Thayer SP, Lauwers GY, Brannigan BW, Harris PL, Okimoto RA, Haserlat SM, Driscoll DR, Ferry D, Muir B, Settleman J, Fuchs CS, Kulke MH, Ryan DP, Clark JW, Sgroi DC, Haber DA, Bell DW. Epidermal growth factor receptor kinase domain mutations in esophageal and pancreatic adenocarcinomas. Clin Cancer Res 12: 4283–4287, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lao-Sirieix P, Corovic A, Jankowski J, Lowe A, Triadafilopoulos G, Fitzgerald RC. Physiological and molecular analysis of acid loading mechanisms in squamous and columnar-lined esophagus. Dis Esophagus 21: 529–538, 2008 [DOI] [PubMed] [Google Scholar]
- 35. Li Y, Reuter NP, Li X, Liu Q, Zhang J, Martin RC. Colocalization of MnSOD expression in response to oxidative stress. Mol Carcinog 49: 44–53, 2010 [DOI] [PubMed] [Google Scholar]
- 36. Li Y, Wo JM, Ray MB, Jones W, Su RR, Ellis S, Martin RC. Cyclooxygenase-2 and epithelial growth factor receptor up-regulation during progression of Barrett's esophagus to adenocarcinoma. World J Gastroenterol 12: 928–934, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Y, Wo JM, Su RR, Ray MB, Martin RC. Loss of manganese superoxide dismutase expression and activity in rat esophagus with external esophageal perfusion. Surgery 141: 359–367, 2007 [DOI] [PubMed] [Google Scholar]
- 38. MacLennan AJ, Orringer MB, Beer DG. Identification of intestinal-type Barrett's metaplasia by using the intestine-specific protein villin and esophageal brush cytology. Mol Carcinog 24: 137–143, 1999 [DOI] [PubMed] [Google Scholar]
- 39. Masereel B, Pochet L, Laeckmann D. An overview of inhibitors of Na+/H+ exchanger. Eur J Med Chem 38: 547–554, 2003 [DOI] [PubMed] [Google Scholar]
- 40. Nehra D, Howell P, Williams CP, Pye JK, Beynon J. Toxic bile acids in gastro-oesophageal reflux disease: influence of gastric acidity. Gut 44: 598–602, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nielsen M, Kaltoft K, Nordahl M, Ropke C, Geisler C, Mustelin T, Dobson P, Svejgaard A, Odum N. Constitutive activation of a slowly migrating isoform of Stat3 in mycosis fungoides: tyrphostin AG490 inhibits Stat3 activation and growth of mycosis fungoides tumor cell lines. Proc Natl Acad Sci USA 94: 6764–6769, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O'Riordan JM, Abdel-latif MM, Ravi N, McNamara D, Byrne PJ, McDonald GS, Keeling PW, Kelleher D, Reynolds JV. Proinflammatory cytokine and nuclear factor-κB expression along the inflammation-metaplasia-dysplasia-adenocarcinoma sequence in the esophagus. Am J Gastroenterol 100: 1257–1264, 2005 [DOI] [PubMed] [Google Scholar]
- 43. Paoletti F, Aldinucci D, Mocali A, Caparrini A. A sensitive spectrophotometric method for the determination of superoxide dismutase activity in tissue extracts. Anal Biochem 154: 536–541, 1986 [DOI] [PubMed] [Google Scholar]
- 44. Richter JE. Importance of bile reflux in Barrett's esophagus. Dig Dis 18: 208–216, 2000 [DOI] [PubMed] [Google Scholar]
- 45. Rosati A, Ammirante M, Gentilella A, Basile A, Festa M, Pascale M, Marzullo L, Belisario MA, Tosco A, Franceschelli S, Moltedo O, Pagliuca G, Lerose R, Turco MC. Apoptosis inhibition in cancer cells: a novel molecular pathway that involves BAG3 protein. Int J Biochem Cell Biol 39: 1337–1342, 2007 [DOI] [PubMed] [Google Scholar]
- 46. Rosati A, Di Salle E, Luberto L, Quinto I, Scala G, Turco MC, Pascale M. Identification of a Btk-BAG3 complex induced by oxidative stress. Leukemia 23: 823–824, 2009 [DOI] [PubMed] [Google Scholar]
- 47. Rosati A, Leone A, Del Valle L, Amini S, Khalili K, Turco MC. Evidence for BAG3 modulation of HIV-1 gene transcription. J Cell Physiol 210: 676–683, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Salo JA, Kivilaakso EO, Kiviluoto TA, Virtanen IO. Cytokeratin profile suggests metaplastic epithelial transformation in Barrett's oesophagus. Ann Med 28: 305–309, 1996 [DOI] [PubMed] [Google Scholar]
- 49. Sarosi GA, Jr, Jaiswal K, Herndon E, Lopez-Guzman C, Spechler SJ, Souza RF. Acid increases MAPK-mediated proliferation in Barrett's esophageal adenocarcinoma cells via intracellular acidification through a Cl−/HCO3− exchanger. Am J Physiol Gastrointest Liver Physiol 289: G991–G997, 2005 [DOI] [PubMed] [Google Scholar]
- 50. Schelling JR, Abu Jawdeh BG. Regulation of cell survival by Na+/H+ exchanger-1. Am J Physiol Renal Physiol 295: F625–F632, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shahidullah M, Mandal A, Delamere NA. Responses of sodium-hydrogen exchange to nitric oxide in porcine cultured nonpigmented ciliary epithelium. Invest Ophthalmol Vis Sci 50: 5851–5858, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shukla S, Shukla M, Maclennan GT, Fu P, Gupta S. Deregulation of FOXO3A during prostate cancer progression. Int J Oncol 34: 1613–1620, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sihvo EI, Ruohtula T, Auvinen MI, Koivistoinen A, Harjula AL, Salo JA. Simultaneous progression of oxidative stress and angiogenesis in malignant transformation of Barrett esophagus. J Thorac Cardiovasc Surg 126: 1952–1957, 2003 [DOI] [PubMed] [Google Scholar]
- 54. Sihvo EI, Salminen JT, Rantanen TK, Ramo OJ, Ahotupa M, Farkkila M, Auvinen MI, Salo JA. Oxidative stress has a role in malignant transformation in Barrett's oesophagus. Int J Cancer 102: 551–555, 2002 [DOI] [PubMed] [Google Scholar]
- 55. Smith LL, Coller HA, Roberts JM. Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat Cell Biol 5: 474–479, 2003 [DOI] [PubMed] [Google Scholar]
- 56. Souza RF, Shewmake K, Terada LS, Spechler SJ. Acid exposure activates the mitogen-activated protein kinase pathways in Barrett's esophagus. Gastroenterology 122: 299–307, 2002 [DOI] [PubMed] [Google Scholar]
- 57. Spechler SJ, Fitzgerald RC, Prasad GA, Wang KK. History, molecular mechanisms, and endoscopic treatment of Barrett's esophagus. Gastroenterology 138: 854–869, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stewart SA, Hahn WC, O'Connor BF, Banner EN, Lundberg AS, Modha P, Mizuno H, Brooks MW, Fleming M, Zimonjic DB, Popescu NC, Weinberg RA. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc Natl Acad Sci USA 99: 12606–12611, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stoner GD, Kaighn ME, Reddel RR, Resau JH, Bowman D, Naito Z, Matsukura N, You M, Galati AJ, Harris CC. Establishment and characterization of SV40 T-antigen immortalized human esophageal epithelial cells. Cancer Res 51: 365–371, 1991 [PubMed] [Google Scholar]
- 60. Tong X, Yin L, Washington R, Rosenberg DW, Giardina C. The p50-p50 NF-κB complex as a stimulus-specific repressor of gene activation. Mol Cell Biochem 265: 171–183, 2004 [DOI] [PubMed] [Google Scholar]
- 61. Vaezi MF, Richter JE. Role of acid and duodenogastroesophageal reflux in gastroesophageal reflux disease. Gastroenterology 111: 1192–1199, 1996 [DOI] [PubMed] [Google Scholar]
- 62. Wilson KT, Fu S, Ramanujam KS, Meltzer SJ. Increased expression of inducible nitric oxide synthase and cyclooxygenase-2 in Barrett's esophagus and associated adenocarcinomas. Cancer Res 58: 2929–2934, 1998 [PubMed] [Google Scholar]
- 63. Wu J, Gong J, Geng J, Song Y. Deoxycholic acid induces the overexpression of intestinal mucin, MUC2, via NF-κB signaling pathway in human esophageal adenocarcinoma cells. BMC Cancer 8: 333, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zachos NC, Tse M, Donowitz M. Molecular physiology of intestinal Na+/H+ exchange. Annu Rev Physiol 67: 411–443, 2005 [DOI] [PubMed] [Google Scholar]
- 65. Zhang HY, Zhang X, Huo X, Hormi-Carver K, Spechler SJ, Souza RF. Increased expression, secretion and activation of IL-6 during the neoplastic progression of Barrett's esophagus (Abstract). Gastroenterology 136: A-92, 2009 [Google Scholar]
- 66. Zhou G, Sun YG, Wang HB, Wang WQ, Wang XW, Fang DC. Acid and bile salt up-regulate BMP4 expression in human esophageal epithelium cells. Scand J Gastroenterol 44: 926–932, 2009 [DOI] [PubMed] [Google Scholar]