Abstract
Methylation at CpG sites is a critical epigenetic modification in mammals. Altered DNA methylation has been suggested to be a central mechanism in development, some disease processes and cellular senescence. Quantifying the extent and identity of epigenetic changes in the aging process is therefore potentially important for understanding longevity and age-related diseases. In the current study, we have examined DNA methylation at >27 000 CpG sites throughout the human genome, in frontal cortex, temporal cortex, pons and cerebellum from 387 human donors between the ages of 1 and 102 years. We identify CpG loci that show a highly significant, consistent correlation between DNA methylation and chronological age. The majority of these loci are within CpG islands and there is a positive correlation between age and DNA methylation level. Lastly, we show that the CpG sites where the DNA methylation level is significantly associated with age are physically close to genes involved in DNA binding and regulation of transcription. This suggests that specific age-related DNA methylation changes may have quite a broad impact on gene expression in the human brain.
INTRODUCTION
Genomic DNA methylation is an important, epigenetic modification in eukaryotes, essential for human life and playing a vital role in determining gene regulation. Alterations in DNA methylation are thought to be associated with diseases such as diabetes, schizophrenia, multiple sclerosis and cancer, as well as with processes such as cellular senescence (1–4).
Lately, there has been increasing interest in DNA methylation. This is not only because epigenetics is a plausible intermediary between the environment and the gene regulation but also due to the development of targeted arrays designed to comprehensively assay the epigenome (5). The application of targeted arrays affords the ability to accurately assess DNA methylation at thousands of individual CpG dinucleotides in large sample series. Using these arrays, we now have the tools to determine the pattern of locus-specific DNA methylation changes correlating with factors such as chronological age.
Recently, we mapped the landscape of DNA methylation status across a large series of brain tissues from neurologically normal donors (6). These data showed that DNA methylation was measurably different between distinct brain regions, and furthermore, that the DNA methylation levels at a substantive proportion of CpG sites are associated with genotype at proximal polymorphisms. Here, we extend our analyses of these data to test the effect of age on DNA methylation status in human brain. We find that there is a strong and significant correlation between CpG methylation and aging of the human brain.
RESULTS
We performed a series of experiments to map associations in DNA methylation with age in human brain tissue. Using Human Methylation27 BeadChips (Illumina Inc., CA, USA), we assayed DNA methylation at 27 578 CpG dinucleotides in a series of human brain samples. This work was performed in two stages. The first stage included tissue from frontal cortex, temporal cortex, pons and cerebellum from each of 150 human brains, collected from donors ranging in age from 16 to 101 years (6). The second stage included tissue from frontal cortex and cerebellum from 237 human brains, collected from donors ranging in age from 0.4 to 102 years (see Materials and Methods and Supplemental materials for details).
Association between CpG methylation levels and chronological age across brain regions
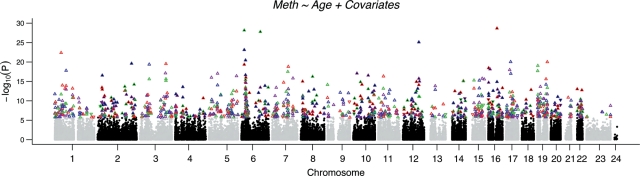
Analysis of the association between chronological age and DNA methylation levels at individual CpG sites in the stage I sample set revealed a large number of strongly associated loci (Fig. 1). After conservative correction for multiple testing, we identified 1141 associations between DNA methylation at CpG sites and age in stage I of the analysis. Of these, 589 loci were significant in one region only, 167 loci were significant in two regions, 86 loci were significant in three brain regions and DNA methylation levels at 10 CpG loci were significantly correlated with age in all four brain regions. Of all significant CpG sites detected, 932 were within our strict definition of CpG islands, 129 were not within islands and 80 were in regions that we could not unequivocally define as islands or non-islands.
Figure 1.
Manhattan plot showing association between methylation at individual CpG sites and chronological age. Plotted are P-values indicating strength of association between DNA methylation levels at >27 000 CpG sites and age in cerebellum (purple), frontal cortex (green), pons (blue) and temporal cortex (red). For each point, a positive association between DNA methylation and chronological age is indicated by upward pointing triangles; a negative association is indicated by downward pointing triangles.
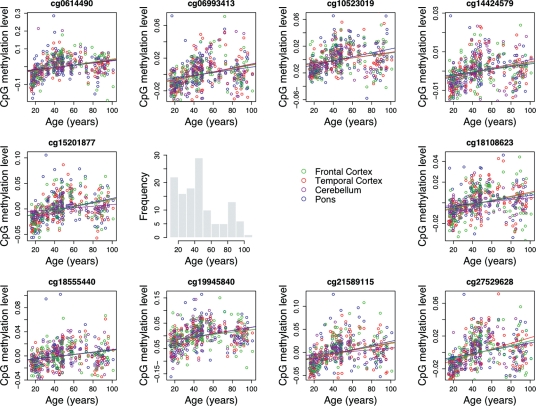
Next, we examined the 10 CpG sites that showed significant genome-wide association with chronological age across all four brain regions. The 10 loci were located within CpG islands and the DNA methylation levels at these sites were positively correlated with chronological age across each of the four tissues (Fig. 2). Analysis of independently ascertained stage II sample series confirmed that there are strong age associations at all of these loci (Table 1). Notably, the direction and magnitude of effect was consistent in both sample series. At these CpG sites, age accounted for 32–75% of the total variance in DNA methylation levels. This analysis was based on adjusted r2 values from the replication phase in order to avoid the possible impact of winner's curse on the discovery phase results and to achieve more accurate estimates.
Figure 2.
Covariate-adjusted methylation levels in cerebellum (purple), frontal cortex (green), pons (blue) and temporal cortex (red) for 10 CpG sites where methylation levels increase significantly with age in all four brain regions (based on a Bonferroni threshold for significance of P = 1.8 × 10−6). Notably for all 10 loci that met our conservative threshold for significance, methylation levels were positively associated with age.
Table 1.
Ten DNA methylation sites identified as significantly associated with chronological age in all tissues from stage I
| Name | Chr | Genomic position (bp) | Symbol | Distance to TSS | Stage I P-value | Stage II P-value | Beta coefficient range | Adjusted r2 estimates from stage I | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CRBLM | FCTX | PONS | TCTX | CRBLM | FCTX | CRBLM | FCTX | PONS | TCTX | ||||||
| cg06144905 | 17 | 24393906 | PIPOX | 138 | 7.3 × 10−7 | 6.2 × 10−13 | 3.4 × 10−7 | 1.1 × 10−18 | 5.8 × 10−8 | 3.2 × 10−18 | 1.5 × 10−3, 3.4 × 10−3 | 0.3085 | 0.4424 | 0.3524 | 0.627 |
| cg06993413 | 15 | 63597257 | DPP8 | 162 | 2.7 × 10−10 | 5.4 × 10−16 | 9.0 × 10−8 | 5.4 × 10−14 | 5.5 × 10−21 | 3.8 × 10−10 | 4.3 × 10−4, 9.8 × 10−4 | 0.5725 | 0.6796 | 0.3776 | 0.5975 |
| cg10523019 | 2 | 227408702 | RHBDD1 | 315 | 7.7 × 10−7 | 9.2 × 10−9 | 6.1 × 10−15 | 3.2 × 10−11 | 2.0 × 10−16 | 5.5 × 10−15 | 5.9 × 10−4, 1.2 × 10−3 | 0.5949 | 0.6909 | 0.6369 | 0.6641 |
| cg14424579 | 2 | 27127813 | FLJ21839 | 240 | 5.9 × 10−7 | 3.5 × 10−8 | 3.2 × 10−7 | 3.7 × 10−14 | 2.5 × 10−15 | 9.4 × 10−12 | 2.8 × 10−4, 4.0 × 10−4 | 0.6488 | 0.7015 | 0.5533 | 0.6195 |
| cg15201877 | 1 | 71285561 | PTGER3 | 518 | 5.4 × 10−10 | 1.3 × 10−14 | 1.1 × 10−13 | 1.6 × 10−18 | 5.1 × 10−8 | 8.5 × 10−26 | 3.4 × 10−4, 1.6 × 10−3 | 0.5928 | 0.7205 | 0.6902 | 0.8032 |
| cg18108623 | 17 | 30725434 | FLJ34922 | 701 | 1.7 × 10−6 | 9.3 × 10−12 | 1.8 × 10−7 | 9.6 × 10−18 | 3.1 × 10−8 | 1.2 × 10−22 | 3.0 × 10−4, 8.4 × 10−4 | 0.4937 | 0.717 | 0.6566 | 0.7531 |
| cg18555440 | 11 | 17698263 | MYOD1 | 528 | 1.8 × 10−6 | 2.2 × 10−8 | 3.6 × 10−7 | 2.6 × 10−10 | 1.0 × 10−26 | 6.0 × 10−11 | 5.7 × 10−4, 8.0 × 10−4 | 0.3153 | 0.5847 | 0.3713 | 0.6351 |
| cg19945840 | 1 | 1157899 | B3GALT6 | 391 | 4.2 × 10−10 | 1.7 × 10−6 | 1.8 × 10−12 | 5.0 × 10−10 | 6.9 × 10−20 | 2.3 × 10−17 | 1.9 × 10−3, 2.6 × 10−3 | 0.4704 | 0.3802 | 0.5503 | 0.5233 |
| cg21589115 | 19 | 54558926 | DKKL1 | 72 | 1.2 × 10−6 | 1.7 × 10−12 | 6.2 × 10−15 | 2.4 × 10−16 | 5.1 × 10−13 | 3.2 × 10−26 | 5.8 × 10−4, 1.8 × 10−3 | 0.5176 | 0.6528 | 0.588 | 0.7055 |
| cg27529628 | 12 | 99491350 | GAS2L3 | 270 | 8.1 × 10−15 | 1.8 × 10−16 | 8.2 × 10−12 | 7.8 × 10−26 | 2.3 × 10−27 | 3.8 × 10−26 | 7.1 × 10−4, 1.5 × 10−3 | 0.6565 | 0.7556 | 0.6507 | 0.812 |
Notably, the DNA methylation level at each of these CpG sites is significantly and consistently associated with chronological age in stage II. CRBLM, cerebellum; FCTX, frontal cortex; PONS, pons; TCTX, temporal cortex. Genomic position is based on hg18 of the human genome.
Substantive enrichment of CpG methylation sites positively correlated with chronological age
Initial inspection of the 10 loci where the DNA methylation level was associated with the chronological age revealed that each of the associations represented a positive correlation. Further, the majority of all significantly associated loci in each tissue showed that this positive association was the trend. This is nicely illustrated in Figure 1, where positive correlations between chronological age and DNA methylation appear to be in extreme excess (positively associated loci are indicated by upward pointing triangles). 95.4% of significant results passing the Bonferroni correction of 1.8E−6 in stage I showed a positive association with age, whereas only 56.0% of non-significant results had positive regression coefficients, illustrating that this consistent direction of effect far exceeds chance (Z-statistic = 26.7, P < 0.0001). This enrichment of positive associations was also seen in the replication data set with 78.6% of significant associations having a positive direction of effect and 55.1% of non-significant results having a positive direction of effect (Z-statistic = 20.4, P < 0.0001).
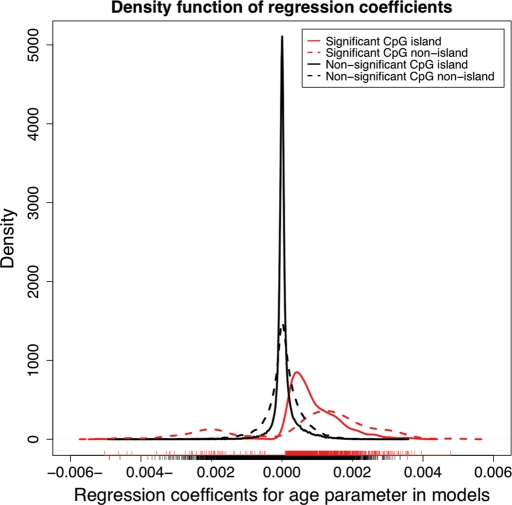
Previous data suggest that the direction of association between age and methylation differ upon whether the CpG dinucleotide is located within or outside of a CpG island (7). Analysis of the regression coefficients from our stage I data showed an excess of CpG sites where DNA methylation positively correlated with age within islands compared with those sites outside of CpG islands. Of the age-associated sites within CpG islands, the correlation between DNA methylation and chronological age was positive in more than 98% of sites. In contrast, a substantially lower proportion of associated sites outside of CpG islands showed a positive correlation between DNA methylation levels and age (76%; Z-statistic = 12.1, P < 0.0001 in discovery phase and P < 0.0001 in replication phase; Fig. 3).
Figure 3.
An excess of CpG sites where DNA methylation is positively associated with age exists in significant results.
We were concerned about the possible confounding effect of unreliable designation into island/non-island status and therefore repeated this analysis using a more restrictive definition of sites inside and outside of islands. This more restrictive definition required a CpG site to meet the criteria for being located within an island in all three resources used for annotation: EPI score (8), UCSC genome browser sequence-based annotation of CpG sites (9) and Illumina documentation. Restricting the definition did not change the excess of positive correlations within islands versus non-islands (Supplementary Material, Fig. S1). These data are supported by previous work performed in human blood (10). Our analysis illustrates that those sites where DNA methylation was negatively correlated with age are 16 times more likely to be located outside of a CpG island versus within a CpG island.
Comparison of age-related CpG methylation changes across brain regions
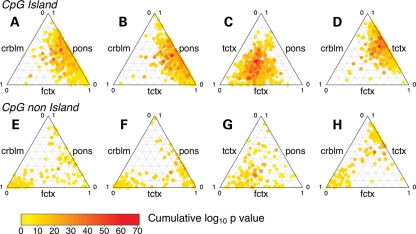
We were interested in whether the associations found between DNA methylation at individual CpG sites and chronological age were consistent across brain regions. To test this idea, we compared association P-values across cerebellum, frontal cortex, pons and temporal cortex data sets at individual CpG sites where the DNA methylation level showed a significant association with chronological age in at least one of the four tissues. This analysis revealed that age-associated CpG sites are most similar in frontal cortex and temporal cortex and that these two tissues are in turn quite similar to pons (Fig. 4). In contrast, the pattern of age-associated CpG sites observed in the cerebellum was by far the most distinct of the four regions tested.
Figure 4.
Ternary plots showing concordance of combined P-values from phase I analyses across all four brain regions stratified by CpG island or non-island status. (A–H) The combined magnitude of association per CpG site for all possible combinations of brain regions. The cumulative log10 P-value is the additive combination of P-values (transformed to a −log10 scale) across three brain regions spatially indicated in the ternary plots. For each side of the ternary plots, the 0–1 scale is related to the untransformed P-values, with each side labeled per brain region analyzed in the discovery phase of analyses. These plots show that most robust associations, with the largest combined P-values across multiple brain regions (on the −log10 scale) occur at CpG islands.
The number and identity of samples in stage I was marginally different between the four tissue regions tested due to occasional sample or assay failure. To ensure consistency across data sets, we compared age-associated CpG sites across tissues on a subset of donors from stage I for whom data on each of the four tissues were available (n = 84). These analyses revealed that uniqueness of associations in cerebellar tissue was not due to the sampling bias. The relative similarity between the frontal cortex and the temporal cortex tissues remained (Supplementary Material, Fig. S2).
Next, we expanded this analysis to include data derived from the additional frontal cortex and cerebellar samples typed in stage II (Fig. 5). We also saw significant associations occurring in both the frontal and the cerebellar datasets (as well as in all four regions). As before, the most associated methylation sites display relatively concordant methylation levels in both tissues. These findings are consistent with previous reports from our group and others showing that the patterns of both DNA methylation and expression are quite different in cerebellum compared with other brain tissues (6,11).
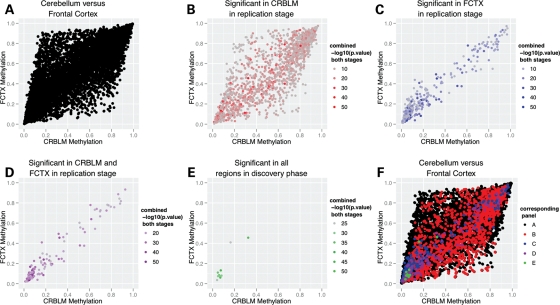
Figure 5.
Data from stage II comparing results from cerebellum and frontal cortex, showing that significant results across two or more tissues often occur at similar methylation levels across various tissues, seemingly robust to the magnitude of methylation. (A) The dispersion of all results in stage II across varying levels of methylation in both tissue types regardless of statistical significance, whereas (F) presents the same data but also overlays the subsets of the data shown in (B)–(E). (B)–(E) describe subsets of results significant across a number of brain regions evident in the panel labels within the figure. The cumulative log10 P-value is the additive combination of P-values (transformed to a −log10 scale) across brain regions for the same CpG site.
Gene ontology/functional annotation analysis
Functional relationships were investigated using the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) by investigating age-related CpG sites for enrichment of gene ontology (GO) terms. Six hundred eighty-three unique EntrezGene identifiers were cross-referenced in the DAVID database, using Illumina gene annotation for significantly associated CpG sites from our initial stage I analysis. These 683 were considered our experimental pool in the clustering analysis.
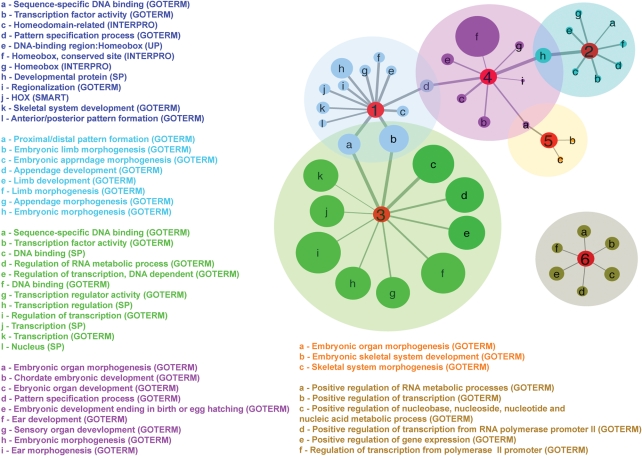
A total of 228 clusters were generated. Six clusters with the highest degrees of enrichment are shown in Figure 6 and described in Table 2. These clusters illustrate a strong enrichment for genes related to DNA binding, morphogenesis and regulation of transcription.
Figure 6.
A map of enriched functional clusters for genes proximal to age-associated CpG sites based on DAVID functional annotation clustering. This figure shows the inferred functional relatedness of clustered genes at a greater than 4-fold level of enrichment among significant results in phase I of this study, as described in Table 2. Line thickness connecting nodes indicates relative P-value of the term within the cluster, with the thickest line representing the most significant term (P = 1.6 × 10−19) and the thinnest lines representing the least significant term (P = 2.5 × 10−5). The numbers in the red ‘hub’ nodes denote the cluster number described in Table 2, with relative size of each node representing the count of each particular node from DAVID analyses, and nodes connected across clusters showing functional overlaps.
Table 2.
Functional annotation clusters generated from DAVID (http://david.abcc.ncifcrf.gov/) showing at least 4-fold enrichment
| Count | P-value | FDR-adjusted P-value | Pass FDR | |
|---|---|---|---|---|
| Annotation cluster 1, enrichment score: 13.97 | ||||
| Sequence-specific DNA binding | 80 | 9.40E − 19 | 1.41E − 15 | True |
| Transcription factor activity | 105 | 5.60E − 18 | 8.42E − 15 | True |
| Homeodomain related | 42 | 1.60E − 15 | 2.63E − 12 | True |
| Pattern specification process | 48 | 1.70E − 15 | 2.95E − 12 | True |
| DNA-binding region: homeobox | 37 | 2.20E − 15 | 3.80E − 12 | True |
| Homeobox, conserved site | 40 | 5.00E − 15 | 7.90E − 12 | True |
| Homeobox | 40 | 5.00E − 15 | 7.29E − 12 | True |
| Developmental protein | 78 | 5.50E − 14 | 7.89E − 11 | True |
| Regionalization | 39 | 7.70E − 14 | 1.37E − 10 | True |
| HOX | 40 | 4.90E − 13 | 6.25E − 10 | True |
| Skeletal system development | 46 | 5.30E − 11 | 9.43E − 08 | True |
| Anterior/posterior pattern formation | 29 | 6.00E − 11 | 1.06E − 07 | True |
| Annotation cluster 2, enrichment score: 11.2 | ||||
| Embryonic morphogenesis | 57 | 1.60E − 19 | 2.82E − 16 | True |
| Embryonic limb morphogenesis | 23 | 1.70E − 11 | 3.00E − 08 | True |
| Embryonic appendage morphogenesis | 23 | 1.70E − 11 | 3.00E − 08 | True |
| Appendage development | 25 | 1.80E − 11 | 3.23E − 08 | True |
| Limb development | 25 | 1.80E − 11 | 3.23E − 08 | True |
| Limb morphogenesis | 24 | 4.60E − 11 | 8.10E − 08 | True |
| Appendage morphogenesis | 24 | 4.60E − 11 | 8.10E − 08 | True |
| Proximal/distal pattern formation | 11 | 7.70E − 08 | 1.37E − 04 | True |
| Annotation cluster 3, enrichment score: 10.62 | ||||
| DNA binding | 146 | 3.10E − 19 | 4.49E − 16 | True |
| Sequence-specific DNA binding | 80 | 9.40E − 19 | 1.41E − 15 | True |
| Transcription factor activity | 105 | 5.60E − 18 | 8.42E − 15 | True |
| Regulation of RNA metabolic process | 135 | 1.20E − 12 | 2.06E − 09 | True |
| Regulation of transcription, DNA-dependent | 129 | 1.90E − 11 | 3.40E − 08 | True |
| DNA binding | 155 | 2.50E − 11 | 3.70E − 08 | True |
| Transcription regulator activity | 119 | 2.90E − 10 | 4.37E − 07 | True |
| Transcription regulation | 125 | 1.10E − 08 | 1.58E − 05 | True |
| Regulation of transcription | 158 | 5.10E − 08 | 9.00E − 05 | True |
| Transcription | 121 | 4.00E − 07 | 5.69E − 04 | True |
| Transcription | 123 | 1.20E − 05 | 2.20E − 02 | True |
| Nucleus | 209 | 4.60E − 05 | 6.59E − 02 | False |
| Annotation cluster 4, enrichment score: 8.28 | ||||
| Embryonic morphogenesis | 57 | 1.60E − 19 | 2.82E − 16 | True |
| Pattern specification process | 48 | 1.70E − 15 | 2.95E − 12 | True |
| Embryonic organ development | 34 | 1.10E − 12 | 1.98E − 09 | True |
| Embryonic organ morphogenesis | 26 | 9.20E − 10 | 1.63E − 06 | True |
| Embryonic development ending in birth or egg hatching | 44 | 1.50E − 09 | 2.61E − 06 | True |
| Chordate embryonic development | 43 | 3.50E − 09 | 6.19E − 06 | True |
| Sensory organ development | 31 | 1.20E − 07 | 2.11E − 04 | True |
| Ear development | 17 | 3.00E − 06 | 5.36E − 03 | True |
| Ear morphogenesis | 13 | 1.50E − 05 | 2.64E − 02 | True |
| Inner ear morphogenesis | 11 | 6.30E − 05 | 1.11E − 01 | False |
| Inner ear development | 13 | 1.00E − 04 | 1.84E − 01 | False |
| In utero embryonic development | 17 | 1.00E− 02 | 1.64E + 01 | False |
| Annotation cluster 5, enrichment score: 5.45 | ||||
| Embryonic organ morphogenesis | 26 | 9.20E − 10 | 1.63E − 06 | True |
| Skeletal system morphogenesis | 19 | 7.10E − 06 | 1.26E − 02 | True |
| Embryonic skeletal system development | 15 | 1.50E − 05 | 2.68E − 02 | True |
| Embryonic skeletal system morphogenesis | 10 | 1.60E − 03 | 2.83E + 00 | False |
| Annotation cluster 6, enrichment score: 4.39 | ||||
| Positive regulation of gene expression | 53 | 6.40E − 06 | 1.14E − 02 | True |
| Regulation of transcription from RNA polymerase II promoter | 61 | 1.60E − 05 | 2.89E − 02 | True |
| Positive regulation of transcription from RNA polymerase II promoter | 38 | 1.70E − 05 | 3.07E − 02 | True |
| Positive regulation of RNA metabolic process | 45 | 2.10E − 05 | 3.75E − 02 | True |
| Positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 54 | 2.40E − 05 | 4.24E − 02 | True |
| Positive regulation of transcription | 50 | 2.50E − 05 | 4.39E − 02 | True |
| Positive regulation of transcription, DNA-dependent | 44 | 3.70E − 05 | 6.48E − 02 | False |
| Positive regulation of nitrogen compound metabolic process | 54 | 6.00E − 05 | 1.06E − 01 | False |
| Positive regulation of cellular biosynthetic process | 56 | 8.50E − 05 | 1.51E − 01 | False |
| Positive regulation of macromolecule biosynthetic process | 54 | 8.60E − 05 | 1.53E − 01 | False |
| Positive regulation of biosynthetic process | 56 | 1.30E − 04 | 2.30E − 01 | False |
| Positive regulation of macromolecule metabolic process | 63 | 4.90E − 04 | 8.65E − 01 | False |
DISCUSSION
A large number of individual CpG sites were shown to be strongly associated with chronological age and DNA methylation levels. A potential confound of note is the dynamic cellular composition of human brain, particularly that the proportion of neurons to glia may change with chronological aging. Because we have identified consistent results across multiple brain regions, this is not likely to be a confound among the 10 CpG sites that showed significant genome-wide association with chronological age across all four brain regions. Of the 10 loci identified, one locus within the MYOD1 gene has previously been reported to be associated with age-related methylation changes in the brain and the pleura (7). In addition, 3 of the 10 loci identified were also shown to be associated with age in human blood (10). Collectively, these data support the notion that the age-associated CpG sites identified in our data are not an artifact of age-associated alterations in cellularity, but rather reflect an underlying biological change in DNA methylation status.
The consistent findings from our group and others showing that the patterns of both DNA methylation and gene expression are quite different in cerebellum compared with other brain tissues (6,11) may be attributed to the unique nuclei of cerebellar purkinje neurons, which are large, euchromatic structures that exhibit a greater proportion of 5-hydroxymethylcystosine modifications compared with other neuronal populations (12). Although these factors alone may not account for the substantive divergence observed in our current analyses, they do illustrate that the genetic component of this tissue exhibits tangible differences from that within other brain regions. Thus, it is not surprising that age-related DNA methylation sites would be most divergent in cerebellar tissue compared with the other brain regions tested here.
The classes of genes identified at age-associated sites included DNA-binding factors and transcription factors, illustrating a strong enrichment for genes related to DNA binding, morphogenesis and regulation of transcription. Given the functional nature of these clusters, it is conceivable that altered epigenetic regulation at these loci may give rise to quite broad changes in transcriptional potential during the aging process. The clustering of age-associated CpG methylation sites that are proximal to genes associated with DNA binding could have several biological implications. Given that the genes in the DAVID clusters (Table 2) are not associated with DNA damage argues against these associations being a response to pathological events such as reactive oxygen species-mediated damage of nucleic acids. Rather, these genes are responsible for transcription with the homeobox proteins being especially prominent. This suggests that age-related alterations in methylation might be important for the maintenance of transcriptional programs in aging tissues. In this context, it is of interest that there are relatively few mRNA changes that show linear association with aging, although there is a tendency for gene expression to show higher variance as organisms age (13). An interesting possibility therefore, is that the accumulation of DNA methylation may be important in maintenance of consistent gene expression patterns with age.
Summary
Here, we describe CpG sites that exhibit strong age-associated changes in DNA methylation. We see a large number of statistically significant age-associated changes in DNA methylation, despite a conservative correction for multiple testing. Many of these CpG sites were significant in multiple tissues and occur at higher frequencies than one would expect by chance. We saw a highly significant enrichment of age-associated methylation changes at CpG islands of functionally related transcripts. Finally, the majority of such associations were positive, showing that methylation tends to increase with age.
We observed an excess of shared age-associated CpG sites across more than one of the four selected brain tissues, suggesting that altered cellular composition was not the underlying cause of changing the DNA methylation profile, but rather that there was a common regulatory mechanism across the brain regions. The classes of genes identified at the age-associated sites included DNA-binding factors and transcription factors; therefore, one might surmise that the age-associated changes in methylation are likely to be associated with the maintenance of transcriptional programs.
In conclusion, we present a comprehensive analysis of DNA methylation across the four distinct human brain tissues. Our data suggest that there are specific loci where the DNA methylation level changes with the chronological age in the human brain and underscores the necessity to study DNA methylation in aging research in order to understand the underlying mechanism and its functional effects.
MATERIALS AND METHODS
Human brain tissue samples
For stage I analysis, fresh, frozen tissue samples of the frontal and temporal cortices, caudal pons and cerebellum regions were obtained from 150 neurologically normal Caucasian subjects, resulting in 600 tissue samples (6). For stage II analysis, fresh, frozen tissue samples of the frontal cortex and cerebellum regions were obtained from an additional 237 neurologically normal Caucasian subjects, resulting in 474 tissue samples across all samples in both stages I and II. These numbers are prior to quality control. Genomic DNA was phenol–chloroform extracted from brain tissues and quantified on the Nanodrop1000 spectrophotomer prior to bisulfite conversion.
CpG methylation
Bisulfite conversion of 1 µg of genomic DNA was performed using Zymo EZ-96 DNA Methylation Kit as per the manufacturer's protocol. CpG methylation status of >27 000 sites was determined using Illumina Infinium HumanMethylation27 BeadChip, as per the manufacturer's protocol. Data were analyzed in BeadStudio software (Illumina Beadstudio v.3.0). The threshold call rate for inclusion of samples in analysis was 95%. Quality control of sample handling included comparison of genders reported by the brain banks with the gender of the same samples determined by analyzing methylation levels of CpG sites on the X chromosome. Beta values were extracted for sites on chromosome X and loaded into the TM 4 MeV tool. These data were then clustered by sample. Based on the methylation levels for chromosome X loci, these data split into two primary groups based on gender. Calls generated by this method were then compared with sample information reported by the brain bank. Samples where genders did not match between brain bank and methylation data were excluded from our analyses. Forty-seven tissue samples from subjects were excluded due to the low methylation call rate or gender discrepancies, and seven additional subjects were excluded due to the low call rate or gender discrepancies from genotyping data utilized for a separate project.
CpG methylation analysis
For all available samples, stratified by brain region, multivariate linear regression was performed to test the effect of age on CpG methylation at each CpG site in the publicly available data. Regression models were adjusted for the following covariates: hybridization and amplification batch, study center responsible for sample collection, post-mortem interval and gender. The Bonferroni correction of 1.8E − 6 was used to account for the effects of multiple testing phenomenon after testing the associations of >27 000 CpG sites per brain region in the stratified analyses (27 476 in pons, 27 310 in cerebellum, 27 532 in frontal cortex and 27 538 in temporal cortex).
Any CpG site passing the Bonferroni thresholds for significance (1.8E − 6) in all four brain regions was carried forward from the discovery phase of the project. Ten CpG sites that met these criteria and were analyzed using the same statistical models as implemented in the discovery phase, in an independent set of frontal cortex and cerebellum samples.
Post hoc, we categorized CpG sites as within or outside of CpG islands. This categorization was based on annotation as a CpG island if the CpG site was described as an island in at least two resources out of three used for annotation: EPI score (10), UCSC genome browser sequence based annotation of CpG sites (9) or Illumina documentation. Non-island CpG sites were defined as sites not annotated as within an island in any of the three resources used for annotation.
DAVID analysis
Functional relationships were investigated using DAVID (http://david.abcc.ncifcrf.gov/). Enrichment of selected GO terms among age-associated CpG sites was examined using the functional annotation clustering module. Six hundred eighty-three unique EntrezGene identifiers in the David database were cross-referenced from the Illumina gene annotation for significantly associated CpG sites from our discovery analyses, where a CpG site passed the Bonferroni correction in any brain region specific analysis. These 683 genes were considered our experimental pool in the clustering analysis.
To account for possible bias in the Illumina array design (i.e. bias introduced by the array being enriched for CpG sites nearby a certain functional class of gene), 14 495 unique EntezGene identifiers were cross referenced between the entire Illumina CpG array annotation and the DAVID database, with this second gene set serving as the background level of enrichment for genes on the array. Default settings were used for the derivation of clusters and false-discovery rates were used to correct for multiple testing. A total of 228 clusters were generated, with six clusters with enrichment scores showing a greater than 4-fold enrichment of clustered terms.
Additional analyses
Replication was deemed successful if the association between age and methylation passed the Bonferroni threshold for significance of 1.8E − 6 in analyses of both the frontal cortex and the cerebellum data sets. Since the replication data set included a significant number of individuals in the lower age ranges compared with the data used in the discovery phase, two additional iterations of the replication model were utilized to further scrutinize results, first by excluding all samples with age at sampling under 16 years, then excluding all samples under 18 years. Neither of these secondary models caused any marked attenuation of the P-values in the replication results. In addition, a fourth set of models using additional covariates of component vectors 1 and 2 from multidimensional scaling of genotype data for these samples did not significantly alter the results of the regression models.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services; project Z01 AG000932-02.
ACKNOWLEDGEMENTS
We would like to thank the tissue donors and brain banks for their support of this project. Brain tissue was obtained from the Baltimore Longitudinal Study on Aging at the Johns Hopkins School of Medicine, the Miami Brain Bank at the University of Miami, the Sun Health Research Institute Tissue Bank and from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD, USA. This study used the high-performance computational capabilities of the Biowulf Linux cluster (http://biowulf.nih.gov). Statistical analyses were conducted using R (R Development Core Team, 2005).
REFERENCES
- 1.Feng J., Fan G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int. Rev. Neurobiol. 2009;89:67–84. doi: 10.1016/S0074-7742(09)89004-1. [DOI] [PubMed] [Google Scholar]
- 2.Ling C., Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58:2718–2725. doi: 10.2337/db09-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Urdinguio R.G., Sanchez-Mut J.V., Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol. 2009;8:1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- 4.Wilson A.S., Power B.E., Molloy P.L. DNA hypomethylation and human diseases. Biochem. Biophys. Acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Schones D.E., Zhao K. Genome-wide approaches to studying chromatin modifications. Nat. Rev. Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibbs J.R., van der Brug M.P., Hernandez D.G., Traynor B.J., Nalls M.A., Lai S.L., Arepalli S., Dillman A., Rafferty I.P., Troncoso J., Johnson R., Zielke H.R., et al. Abundant quantitative trait Loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen B.C., Houseman E.A., Marsit C.J., Zheng S., Wrensch M.R., Wiemels J.L., Nelson H.H., Karagas M.R., Padbury J.F., Bueno R., et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bock C., Walter J., Paulsen M., Lengauer T. CpG island mapping by epigenome prediction. PLoS Comput. Biol. 2007;3:e110. doi: 10.1371/journal.pcbi.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 10.Rakyan V.K., Down T.A., Maslau S., Andrew T., Yang T.P., Beyan H., Whittaker P., McCann O.T., Finer S., Valdes A.M., et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010;20:434–439. doi: 10.1101/gr.103101.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ladd-Acosta C., Pevsner J., Sabunciyan S., Yolken R.H., Webster M.J., Dinkins T., Callinan P.A., Fan J.B., Potash J.B., Feinberg A.P. DNA methylation signatures within the human brain. Am. J. Hum. Genet. 2007;81:1304–1315. doi: 10.1086/524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kriaucionis S., Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Southworth L.K., Owen A.B., Kim S.K. Aging mice show a decreasing correlation of gene expression within genetic modules. PLoS Genet. 2009;5:e1000776. doi: 10.1371/journal.pgen.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.