Abstract
Clinical data that may be used in a secondary capacity to support research activities are regularly stored in three significantly different formats: (1) structured, codified data elements; (2) semi-structured or unstructured narrative text; and (3) multi-modal images. In this manuscript, we will describe the design of a computational system that is intended to support the ontology-anchored query and integration of such data types from multiple source systems. Additional features of the described system include (1) the use of Grid services-based electronic data interchange models to enable the use of our system in multi-site settings and (2) the use of a software framework intended to address both potential security and patient confidentiality concerns that arise when transmitting or otherwise manipulating potentially privileged personal health information. We will frame our discussion within the specific experimental context of the concept-oriented query and integration of correlated structured data, narrative text, and images for cancer research.
Key words: Image retrieval, information retrieval, ontologies, text mining, Grid computing
INTRODUCTION
Within academic medical centers, large amounts of multi-dimensional, heterogeneous data are collected electronically on an ongoing basis. This data includes clinical parameters derived during the patient care process, as well as financial and operational information. Clinical data can take many forms, including but not limited to (1) structured, codified data elements; (2) semi-structured or unstructured narrative text; and (3) multi-modal images.1 While such data is readily available to clinical providers and administrators, accessing the same data for research or business intelligence purposes is often a challenge, usually because of regulatory compliance requirements and concerns over patient privacy and confidentiality. Furthermore, data stored in operational systems is not necessarily structured in a manner that lends itself to integrative longitudinal or class-based query and analysis—a requirement that often exists in the research context.2 Therefore, even when regulatory and patient privacy and confidentiality concerns are adequately addressed, it often remains difficult to query and access such data for research and business intelligence. One of the ways institutions have tried to address this problem is by extracting clinical, operational, and financial data from source systems and subsequently storing it in a centralized data warehouse that uses a data model optimized for longitudinal and/or class-based queries.3,4 However, imaging data sets are not usually physically stored in such warehouses because of concerns over data storage capacity and are instead commonly stored and managed using a separate Picture Archiving and Communication System (PACS). However, for purposes of clinical,5 translational, and educational research,6–8 the integration and retrieval of image data along with structured data and narrative text is highly desirable.8,9
There are several examples in the current literature of integrative and ontology-anchored image search or query tools;2,10–12 however, to the best of our knowledge, none of these tools have been shown to also support the simultaneous query and subsequent integration with image data sets of structured data and narrative text. We believe that ability to execute such truly integrative, ontology-anchored queries across multiple data types is critical to the ability to execute highly effective clinical and translational research. Therefore, to address the preceding gap in knowledge, we have formulated a model computational system that is intended to support the integrative query of structured data, narrative text, and image data sets in support of research activities. This system has also been designed to address the challenges posed by regulatory compliance, patient privacy/confidentiality concerns, and the need to facilitate multi-center research paradigms. The model we will describe is motivated by two types of research-oriented end users, specifically (1) clinical researchers who need to perform queries such as “find all patients with brain CT and MRI images and who have a prior medical history of congestive heart failure, high blood pressure, and diabetes;” and (2) imaging informatics researchers who need to obtain image sets defined by both a specific anatomic location and phenotypic parameters to evaluate techniques such as the use of computer-aided diagnosis algorithms.13 In either scenario, such a query requires the ability to locate patients with the specified phenotypic parameters, utilizing some combination of structured data and narrative text, and then subsequently, query a PACS system to identify and obtain image sets for such patients that are potentially generated by one or more modalities (e.g., computed tomography, CT; magnetic resonance imaging, MRI; etc.) and that correspond to the desired anatomic location(s).
Given the preceding motivation and use cases, in the following sections of this manuscript, we will (1) provide pertinent and contributing background material concerning information needs in the clinical and translational research domains, Grid-computing electronic data interchange platforms, ontology-anchored information retrieval techniques, image retrieval tools, applicable regulatory compliance, and patient privacy/confidentiality concerns that must be addressed when using clinical data for research purposes, and the specific experimental context for our model formulation—The Ohio State University Medical Center (OSUMC) Information Warehouse (IW); (2) describe the methods and system design approaches used for our model formulation process; (3) report upon initial feasibility evaluation results relating to the described model; and (4) describe the implications, limitations, and next steps for our work.
BACKGROUND AND SIGNIFICANCE
In this section, we will summarize several areas that contribute to or otherwise inform the model formulation work reported in later sections of the manuscript.
Information Needs in the Clinical and Translational Research Domains
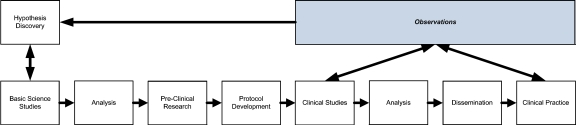
The relationship between the information needs of clinical and translational researchers and currently available information technology (IT) can be understood by conceptualizing the translational research process (which subsumes clinical research) as a sequential information-flow model as illustrated in Figure 1.
Fig 1.
Illustration of translational research information flow model (adapted from Payne, et al.69).
At each stage in this model, a combination of dual-purpose and research-specific IT systems may be utilized. Examples of such systems that can support translational research include (1) literature search tools such as PubMed and OVID,14 (2) protocol authoring15 and data mining tools,16 (3) simulation and visualization tools,17 (4) research-specific web portals,18 (5) electronic data collection or capture tools,19 (6) participant screening tools,20 (7) electronic health records (EHRs),21 (8) computerized physician order entry (CPOE) systems,22 (9) decision support systems,23 and (10) picture archiving and communications systems (PACS).24,25
Numerous reports have described increased translational capacity, data quality, and decreased clinical trial protocol deviations resulting from the use of such IT systems.17,19 In addition, the use of IT26 in studies involving multiple, geographically distributed research sites has been shown to have demonstrable benefits in terms of increased efficiency and decreased resource requirements.22,27 However, despite the promise that the integration of research-specific and dual-purpose IT systems with the translational research enterprise holds, institutional informatics infrastructures providing such integration are generally absent, an outcome largely attributed to socio-technical factors.28 Given such problematic adoption of IT in the translational research domain, it is critical for new systems and technology frameworks to provide significant value to investigators and research staff to overcome potential barriers to acceptance.
A particular concern in the context of translational research and the utilization of information technology to support such activities is the need to maintain proper security and confidentiality of privileged or protected health information (PHI). In many cases, providing for such security, confidentiality, and ethical conduct of research requires the provision of partially or completely deidentified data sets (including imaging data) to researchers. In general, two overriding frameworks exist that dictate such needs, Institutional Review Boards (IRBs) and the Health Insurance Portability and Accountability Act (HIPAA), as summarized briefly below:
Institutional Review Boards (IRBs) are federally mandated oversight bodies who have the responsibility to monitor, approve, and ensure the regulatory compliance of human-subjects research (of note, IRBs also exist to oversee animal research).
The HIPAA, which includes compulsory security standards pertaining to the protection and use of a well-defined collection of privileged data elements known as PHI.29,30 HIPAA guidelines mandate the removal of over 18 specified identifiers from PHI before its research use. Significant challenges exist when attempting to transact in such PHI to support research operations, especially when employing technologies such as the Grid-based electronic data interchange models described earlier.31–35 These challenges include ensuring that appropriate access controls are maintained throughout a distributed architecture, the certification that consumers of such data have a valid and documented purpose for accessing PHI, and maintaining the confidentiality of such data while in-transit or being stored throughout a potentially heterogeneous computing environment. An example of a HIPAA compliant research data repository that contains image data and phenotypic data is the Reference Image Database to Evaluate Response (RIDER) archive of CT scans for lung cancer patients.7
Grid-Computing Electronic Data Interchange Platforms
We define a computational Grid per the convention used by Foster and Kesslman as “…a hardware and software infrastructure that provides dependable, consistent, pervasive, and inexpensive access to high-end computational capabilities… concerned, above all, with large-scale pooling of resources, whether compute cycles, data, sensors, or people.”36 Grid computing has become the new means for creating distributed infrastructures and virtual organizations for multi-institutional research and enterprise applications. The use of Grid computing has evolved from being a platform targeted at large-scale computing applications to a new architecture paradigm for sharing information, data, and software, as well as computational and storage resources. A vast array of middleware systems and toolkits has been developed to support the implementation of Grid computing infrastructures. These include middleware and tools for service deployment and remote service invocation, security, resource monitoring and scheduling, high-speed data transfer, metadata and replica management, component-based application composition, and workflow management. Central to the ability to develop such Grid computing middleware and toolkits is the existence of widely accepted standards. The Grid services framework builds on and extends web services for scientific applications. It defines mechanisms for such additional features as stateful services, service notification, and management of service/resource lifetime. A primary example of a Grid computing initiative focusing upon the biomedical domain is the NCI’s Cancer Biomedical Informatics Grid (caBIG, https://www.cabig.nci.nih.gov/) program, which uses a Grid computing infrastructure, named caGrid, to provide a common, extensible, collaborative platform for data interchange and analysis between cancer researchers and institutions.
Knowledge-Anchored Information Retrieval
Ontologies and terminologies (also described as vocabularies) are a type of conceptual knowledge collection comprised of definitions of both atomic units of knowledge (e.g., facts) and the network of hierarchical and/or semantic relationships between those atoms.37 Ontologies can be either formal or semi-formal, which corresponds to their level of ontological commitment that can be described as the degree to which the output of computational agents that reason based upon the ontology is consistent with the ontologies’ definition and structure. Controlled terminologies or vocabularies that do not satisfy the preceding definition of what constitutes an ontology but that do contain definitions of concepts and hierarchical relationships between such definitions are another example of a conceptual knowledge collection. Instances of ontologies in the biomedical domain include SNOMED-CT and the NCI thesaurus, whereas commonly used controlled terminologies include ICD9-CM and Current Procedural Terminology (CPT). A frequently employed resource when reasoning over or upon the content of such ontologies and terminologies and their potential interrelationships is the Unified Medical Language System (UMLS), a meta-thesaurus of over 100 biomedical ontologies and vocabularies incorporating in excess of 1 million concepts and 4 million synonyms, which is maintained by the National Institutes of Health (NIH)–National Library of Medicine (NLM).38–40 The primary benefit of using the UMLS is the ability to identify a concept of interest and, subsequently, discover synonymous definitions for that concept in multiple source ontologies or terminologies. In addition, it is also possible to reason upon all possible hierarchical and semantic relationships between concepts in the UMLS based upon the relational structures subsumed from the included source ontologies and terminologies. Such conceptual knowledge collections are highly useful when performing information retrieval tasks, as they allow for object-oriented, semantic, or class-based definitions or expansions of queries. For example, if one were to pose a query of the following type (represented in pseudo-code): “SELECT ALL WHERE PROCEDURE = ‘MRI’ and ANATOMIC LOCATION = ‘lower extremity,’” it would be difficult to identify and return such patients, as it is highly unlikely that such imaging procedures would be codified and stored as having an anatomic location of “lower extremity.” Instead, it is likely that such procedures would be codified as having an anatomic location such as “foot,” “ankle,” “lower leg,” or “knee.” However, if the concept “lower extremity” is contained in an ontology or terminology and is related (hierarchically and/or semantically) to concepts including the preceding specific anatomic locations (e.g., foot, etc.), then, by reasoning upon the contents of that knowledge collection, the earlier query could be expanded to “SELECT ALL WHERE PROCEDURE = ‘MRI’ and ANATOMIC LOCATION = (‘foot’, ‘ankle’, ‘lower leg’, ‘knee’).”
In the context of our model system formulation, there is a particular focus on the use of controlled terminologies, specifically ICD9-CM (CM, clinical modification to the International Classification of Diseases, ninth revision). Within the USA, ICD9-CM is the most commonly used medical coding system for procedures and diagnoses. Such codes are manually assigned to patient records by medical information management and billing experts based on numerous information sources, including (1) clinician-provided progress and/or diagnostic reports (usually consisting of free text) and lab results, (2) quantitative findings such as laboratory data, and (3) prior medical history (in both coded and non-coded forms).41,42 A patient encounter (which could encompass either an outpatient visit or a multi-day inpatient admission) can be characterized by multiple diagnosis and procedure codes usually summarized by what is known as a primary code that represents the motivation for that encounters (e.g., the diagnosis or procedure that incurred the encounter). ICD9-CM codes are hierarchically organized. For example, the codes between 160 and 165 correspond to malignant neoplasms of respiratory and intra-thoracic organs, with associated subdiagnoses of such neoplasms being indicated using decimalized variants of the codes, such as code 162.2 that specifically defines “primary disease in the upper lung lobes.”
In addition to the use of ICD9-CM, our model system formulation also employs an advanced type of knowledge-anchored information retrieval known text mining. At the most basic level, text mining involves the use of a computational agent, usually informed by one or more conceptual knowledge collections to parse (i.e., decompose text into constituent components at one or more levels of granularity ranging from paragraphs to words) and tag (i.e., apply a codified concept identifier to a parsed component that is representative of its lexical and/or semantic meaning) narrative, free text. Such parsed and tagged text can then be queried as structured data. An example of the current state-of-the-art in biomedical text mining tools includes the open-source MetaMap Transfer (MMTx) application provided by the NLM, which uses the contents of the UMLS as a knowledge source to inform the parsing and tagging of biomedical narrative text (codifying such text in terms of UMLS concepts).43–46 In addition to open-source applications, numerous commercial database vendors such as IBM, Microsoft, and Oracle provide free-text indexing and mining capabilities within the scope of their database management systems.47–49 However, these capabilities are usually limited to keyword-based searches. One exception to the previous statement is the free text search functions provided by Oracle, which do have limited thesaurus-based capabilities. Yet, in the case of the thesaurus-based text mining functionality provided by Oracle, there is a noticeable lack of available biomedical thesauri that can be utilized for such purposes. Additional examples of commercial text mining tools include (1) IBM’s UIMA,50 which employs an ontology-anchored approach to concept tagging, and (2) Vivisimo,51 which supports concept-oriented search of tagged narrative text where such tagging can be informed by the use of commonly available ontologies or terminologies.
Image Retrieval Tools
In the case of medical images, the most commonly used storage repositories are Picture Archival and Communication Systems (PACS). Most PACS systems support the Digital Imaging and Communications in Medicine (DICOM) standard52 (currently version 3). In PACS that are compliant with the DICOM standard, medical images are stored and retrieved using patient metadata (e.g., descriptors such as medical record number, MRN). The primary focus of image retrieval functionality within modern PACS is to support conventional clinical operations and not necessarily research requirements. An example use case in terms of clinical operations would be a radiologist querying the system for a patient’s latest MRI or CT scan using the patient’s MRN, and then reviewing the imagery for the related visit. Recently, there have been efforts to improve the image retrieval process to support image-related research efforts,34,35,53–55 as well as to enable better integration of imaging data with Electronic Health Records (EHRs).56–58 However, the preceding efforts are still relatively immature, and wide-scale adoption of such tools and processes is limited.
Research uses of most clinical data, including imaging data, presents additional challenges beyond those associated with clinical uses and, in particular, are due to the frequent need to deidentify such data sets. To address the deidentification of imaging data sets for research purposes, the commonly employed practice is to generate a deidentified duplicate of the desired DICOM image as found in the source, production PACS, and then store the duplicate in a research-specific PACS instance.7,55,59 While this approach is feasible for the retrieval and use of well-defined cases for a given experimental context, it does not readily support the integration with and subsequent retrieval and deidentification of related imaging and phenotypic data.
OSUMC Information Warehouse
The Information Warehouse (IW) at the Ohio State University Medical Center (OSUMC) is a comprehensive repository integrating data from over 80 clinical, operational, and research systems throughout the institution. The IW serves a broad variety of customers in all mission areas at OSUMC, including (1) clinical operations, (2) administration, (3) education, and (4) research. Content in the IW includes structured medical and financial data, clinical free-text reports, tissue and genomic data, and limited numbers of medical images. Data stored in the IW can be queried from and presented to end users in identifiable, partially deidentified, or completely deidentified forms, depending on applicable IRB and institutional policies and requirements. As described earlier, the IW does include medical free text from sources such as radiology reports, pathology reports, as well as structured and codified data elements such as age, sex, and diagnosis. However, imaging (e.g., PET, CT, MRI) data are stored in a separate picture archival systems (PACS), with only limited physically duplicated image data within the IW. This physical separation of most image data makes the integrative query of multiple data types throughout OSUMC, including image data, extremely challenging. It should be noted that this problem is not unique to this IW but quite common in other IWs.
METHODS
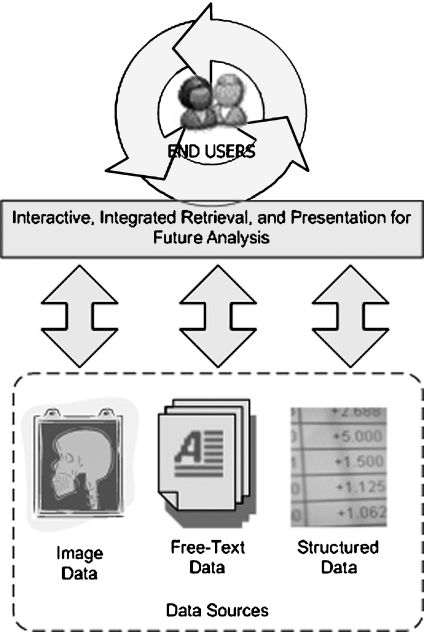
As stated at the outset of this manuscript, we are reporting upon the development and implementation of a model system intended to enable the integrative, knowledge-anchored query of multiple information types, including structured, narrative text, and image data, in support of research requirements. Our model system is based upon a framework60–62 that is intended to provide a convenient interactive environment for such integrative data retrieval tasks (Fig. 2). The resulting system that was implemented based upon this framework allows end users to retrieve images based on characteristics defined in correlative text and structured data elements stored within the OSUMC IW. Such retrieval and presentation of image datasets using phenotypic context derived from narrative text and structured data involves the handling of all data types related to the image, including the image itself, as well as associated heterogeneous, multi-dimensional textual and structured data. To enable such a data handling process, our framework leverages knowledge-anchored information retrieval techniques, specifically combining the UMLS Meta-Thesaurus (UMLS MT)63,64 knowledge collection with text mining platforms incumbent to the Oracle database management system employed by the IW. The use of the UMLS MT allows the expansion of simple keyword-based queries into concept-oriented queries that can then be applied to the contents of clinical narrative text and structured data elements such as diagnosis codes. Because of the research orientation of our framework, we have also incorporated mechanisms to partially or completely deidentify the PHI contained in any returned dataset. This provides compliance with the applicable regulatory requirements, including HIPAA. Finally, to allow for the elegant evolution of our framework in light of constantly emerging technology platforms and standards, as well as the need to enable research activities that span institutional and geographic boundaries, our framework incorporates a multi-tiered service-oriented architecture (SOA). A primary benefit of this multi-tiered SOA is the ability to utilize emergent, research-oriented electronic data interchange platforms, such as the previously introduced caGrid middleware.65
Fig 2.
Conceptual model for the our software framework and model system implementation.
Motivating Use Case
For the remainder of this manuscript, we will frame our discussion using the following motivating use case: An investigator is interested in lung cancer patients. In addition to images, the investigator would like to assess findings in both radiology and pathology reports, as well as diagnosis codes, for each patient from whom images are obtained. The specific findings of interest in the radiology reports should mention lung nodules, whereas the pathology reports should mention adenocarcinoma.
The interaction between our system (Fig. 3) and the researcher (i.e., end-user) would incorporate the following components:
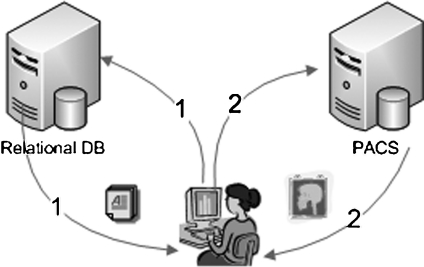
Fig 3.
By first querying for available meta-data in one or more relational databases, users are able to identify patients of interest; later, corresponding images can be retrieved from a PACS.
Query Construction
The construction of a query intended to identify and retrieve patients that meet the given criteria could be decomposed into two parts: (1) a structured data search query and (2) a free-text search query:
Structured data search query. In this case, the user is looking for ICD9-CM codes that correspond to different types of lung cancer. At this stage, if the user already knows the codes for lung cancer (e.g., 162, 162.2, etc.), he or she can provide those codes. For those users not familiar with the ICD9-CM coding system, a code lookup facility is provided, informed by the UMLS knowledge collection. This lookup system supports keyword-based searches. Once ICD9-CM codes are entered or selected, construction of the structured query is complete.
Free-text search query. The free-text query in this instance requires the assessment of two different report types: radiology and pathology, using the initial keywords “lung nodules” and “carcinoma,” respectively. The system’s end user is provided with the ability to expand these keywords using the contents of the UMLS knowledge collection. Once the end user has identified the report types, entered the search keywords, and expanded the search scope per the contents of the UMLS, the construction of the free-text search query is complete.
During the construction of both query types (free text and structured), the presentation tier solicits end-user data entry as appropriate and passes that data to a Grid service in an application tier. This Grid service then interacts with a local UMLS instance that is available via a data sources tier. Asynchronous calls that take place in the presentation tier (e.g., the user interface) allow such transactions to occur in parallel, so that the user can construct both queries simultaneously.
Query Execution
Once the user is satisfied with the queries as constructed previously, he or she can execute the query and identify matching patients or cases. At this point in the system workflow, the presentation tier passes the query to the application tier that, in turn, interacts with the data sources tier. All queries are executed in parallel; queries on diagnosis codes for lung cancer, free text queries for lung nodules (radiology reports), and queries for adenocarcinoma (pathology reports) all return results simultaneously. Once all available results have been returned, the presentation tier joins them and presents them to the user.
Data Browsing and Retrieval
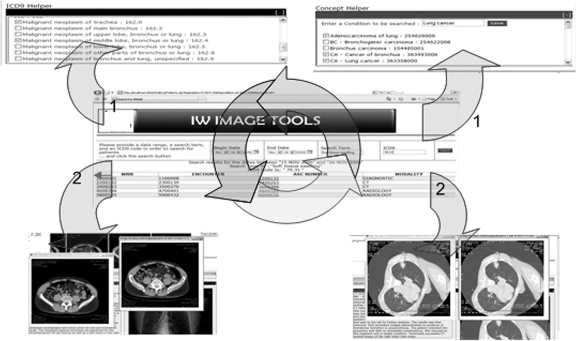
Upon completion of the preceding phase of the systems workflow, end users may browse the returned patients or cases. Once a patient or case is selected, the presentation tier calls the Grid services in application tier to retrieve the patient or case-specific data. Instead of presenting abstract results for such a query, the radiology report containing the concept of “lung nodule,” the pathology report containing the concept of “adenocarcinoma,” any structured codified data pertaining to the “diagnosed with lung cancer” characteristic, and finally, the corresponding images are presented collectively for further evaluation and review (Fig. 4).
Fig 4.
The interface provides interactive assistance to users in order to map keywords used during query formulation to appropriate diagnosis codes (ICD9-CM). Once a query is executed, users may browse the result sets using a hierarchical “drill down” model.
Three-Tiered Software Framework
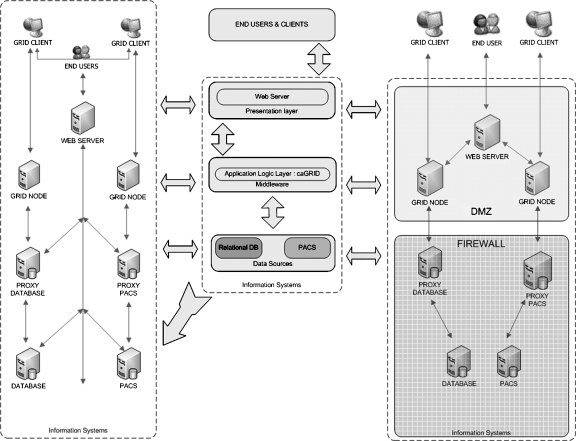
There are several complementary goals within this framework. The realization of all these goals requires the use of components that satisfy both infrastructure constraints (e.g., available platforms and prevailing data interchange standards) and institutional or regulatory requirements as they pertain to the use of PHI for research purposes. The tiered architecture is one of the common techniques used today for the separation of presentation, application, and data66,67 for web-based applications. We utilize this approach within our framework to achieve flexibility and efficiency in terms of development, deployment, and management. The framework used in our model system (Fig. 5) consists specifically of the following three tiers:
Presentation tier provides end-user interaction capabilities based upon the features of the framework via web interfaces.
Application tier utilizes standards-compliant Grid services to interact with and apply logic to multiple, heterogeneous data sources.
Data sources tier support access to data sources such as relational database management systems and PACS.
Fig 5.
The multi-tier implementation of our framework provides a means to implement Grid based or web based electronic data interchange platform within a service-oriented architecture. End-user access to privileged data is managed in one or more ways: (1) a fully privileged user within the institutional firewall (left-hand side) can access the data in anyway they prefer; (2) an external user (located outside the firewall, right-hand side) is subject to additional restriction to access data; and (3) the multi-tier service-oriented approach allows for the deployment of custom services easily.
In the following subsection, detailed descriptions of the design approaches and functionality that pertains to each such tier will be provided.
Presentation Tier. As stated previously, the primary goal of this framework and the resulting system is to provide simple yet flexible ways of identifying and retrieving images from a PACS system that are characterized by one or more heterogeneous sources of phenotypic data. Simplicity is delivered through a unified user interface that deals with all available data types, whereas flexibility is demonstrated by the expandability of queries. Our user interface gives the end user the flexibility to execute such queries by first allowing searches on existing sources of phenotypic data such as free-text reports and diagnosis codes (or any other patient-related data). Once the patients are identified according to all available meta-data, the PACS system is then queried, and associated images are retrieved (Fig. 4). Because the number of retrieved images depends on the number of identified patients, proper utilization of existing free text (radiology and pathology reports) and structured data (diagnosis codes) is crucial in the search criteria. This framework brings together knowledge-anchored free-text capabilities provided by a combination of the UMLS knowledge collection and the text indexing and keyword search capabilities of Oracle. The utilization of the UMLS and Oracle’s text indexing allow our framework to provide interactive query expansion capabilities on free-text documents, along with assistance on diagnosis code selection through an interactive web-based user interface. Selected images are retrieved using PixelMed, an open source Java-based toolkit.68 The web interface component of the presentation tier enables users to construct and execute queries based upon both free-text (e.g., radiology and pathology) and structured data elements (e.g., diagnosis codes) in an interactive fashion. Once the patients are identified based upon specified query parameters, their images along with radiology reports, pathology reports, and diagnosis codes (ICD9-CM) are presented to the users. During the preceding query construction and result set presentation process, end users interact with and are assisted by the presentation layer in the following ways.
UMLS-Based Text Query Expansion. Within the IW, free-text reports are stored in relational databases (Oracle, version 10gR2), where they are indexed for fast text searches. However, those indexes only allow keyword-based searches. As introduced earlier, we have adopted a combination of the UMLS Meta-Thesaurus (MT) and MetaMap (MMTx) to convert users’ keyword queries into conceptual queries. UMLS MT allows us to present alternatives to or expansions of a given query criteria. For example, the user might enter the search term “melanocarcinoma” when searching the contents of pathology reports but would receive few results because of the infrequent use of that specific term in such text (in the test dataset to be described later in the evaluation section of this manuscript, such a search returns no text reports). However, if this keyword were expanded to include the synonym “melanoma” per the contents of the UMLS MT, the user will likely retrieve many more reports (again, in our test dataset, such a search returns 2,706 text reports with the specified term). The MMTx API allows us to parse clinically relevant terms and phrases that can then be expanded and used for query operations when the initial user-entered search criteria consists of larger text constructs (e.g., a sentence or paragraph). For example, the concept of “chronic obstructive lung disease” can be extracted from the sentence “Patient has chronic obstructive lung disease” and can subsequently be mapped to as a corresponding UMLS concept.
UMLS-Based Diagnosis Code Selection. When users are unsure which ICD9-CM code to choose when querying structured data elements that are encoded using that terminology, context-specific assistance that enabled those users to select appropriate code(s) is provided. Specifically, during our UMLS MT installation, controlled vocabularies and dictionaries related to ICD9-CM codes were included. This allows us to provide hierarchical searches on ICD9-CM codes initiated by user supplied text. For example, during the construction of a query, a user may not know the proper codes for a targeted type of lung cancer. In this case, the user may use the assistance mechanism to navigate the ICD9-CM hierarchy, traversing through the concepts “neoplasm” and “lung cancer” to visualize and select the appropriate specific lung cancer codes.
Image Handling and Display. After constructing and executing queries, users may browse the result set using a hierarchical “drill-down” model. For example, the interface allows end users to drill down through multiple layers of granularity in a CT study, beginning with a series and ending at a single image. In addition, the presentation layer allows the user to compare images within a series. Corresponding radiology or pathology reports are also displayed when retrieving and presenting image data (Fig. 4).
The systems presentation layer is implemented using Java Server Pages (JSP) running on Apache Tomcat. In addition, asynchronous calls to the application tier are used to provide a more dynamic human–computer interaction experience.
Application Tier
To support relational database connectivity sufficient to access both textual and structured data elements and to support the use of the UMLS knowledge collection as described earlier, an Ontology Tools Package (OTP) was created within the application tier. In addition, to enable the query, retrieval, and manipulation of images from our PACS system, we utilized the functionality provided by the PixelMed open source Java toolkit. These two packages form the core of our application tier and are designed and implemented to manage interaction between the presentation and data source tiers. Furthermore, these packages are wrapped as caGrid services as described in the following sections.
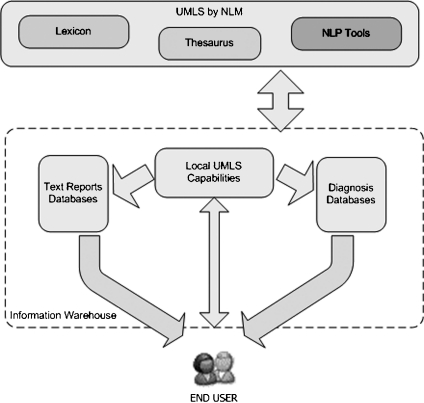
Ontology Tools Package. The OTP is a collection of functions from several existing API’s, including both the MMTx API from the National Library of Medicine and Oracle’s JDBC drivers. Additionally, OTP allows for queries on diagnostic data tables to be executed and linked with UMLS MT, and expanded conceptual search queries on radiology and pathology reports. For performance considerations, local copies of controlled vocabularies and dictionaries from the UMLS MT are maintained and utilized by the OTP. When a user interacts with the relational databases targeted by our system, OTP’s functions (Fig. 6) are utilized during the query construction, execution, and retrieval of the resulting datasets, as described below:
Query construction with OTP. During the construction of text queries, OTP takes free text entered by end users and expands that free text and returns synonyms and other semantically relevant concepts through a process of reasoning upon the previously described local UMLS tables. However, the final selection of such search terms to define a conceptual text query is based upon end-user evaluation of such suggested expansions. Similarly, for the construction of diagnosis code-based queries, user-supplied free-text entries are expanded with the help of UMLS. Instead of keywords, ICD9-CM codes and their descriptions are returned to the user. Again, end users decide which of the returned diagnosis codes are to be included with their query.
Query execution with OTP. During the query execution process, OTP executes the generated query against text and structured data tables, links the datasets, and returns identifiers for further retrieval of other correlated data such as radiology images contained in a PACS. This data linkage process is largely made possible through the use of some combination of the three common identifiers used to store and identify PHI at OSUMC (and which are generally analogous to those used in most common clinical information systems): MRN (used to identify an individual patient), encounter number (ENC, used to identify a patient-specific encounter from which an item of data is derived), and accession number (ASC, used to identify the specific component or activity association with an encounter from which an item of data is derived). When a user-constructed query is executed, OTP returns only identifiers or deidentified pointers to the data, depending on the access privileges assigned to a user for a specific project. It is important to note that, at this stage, no other data than such identifiers is returned by OTP (e.g., no textual, structured, or image data is returned).
Data retrieval with OTP. Once the end user selects a patient or group of patients from which they wish to obtain further data, OTP manages the retrieval of all non-imaging data (with such image data retrieval being managed using PixelMed). All the data tables that need to be accessed with this framework are indexed using the three previously defined identifiers MRN, ENC, and ASC. Therefore, on-demand access to any of the text or structured data elements does not present significant performance implications.
Fig 6.
The Ontology Tools Package (OTP) allows users to interact with a local UMLS knowledge collection instance, diagnostic databases, and text report databases. Users can query OTP with a keyword or textual phrase, which is then mapped to one or more concept codes derived from text mining approaches informed by the UMLS and using MMTx. Once users finalize a set of targeted search concepts, text reports that contain those concepts are queried and returned. Users can also retrieve ICD9-CM codes that correspond to their query keywords and use those codes to query tables containing structured data.
Use of PixelMed. PixelMed provides open source Java libraries for reading, writing, manipulating, and communicating DICOM objects. In addition, PixelMed provides a simple PACS and WADO (web access to DICOM objects) server implementation. In our framework and model system, PixelMed is used for handling all operations related to DICOM objects as follows:
DICOM query construction and execution. Once a user identifies which patient cases he or she wants to review, it is then necessary to query for and retrieve corresponding images from the targeted PACS. Our PACS system (AGFA, Impax 5.2) can be queried by ASC. Here, the ASCs retrieved from the OTP are formed into a DICOM query, and those queries are subsequently executed through the use of standard DICOM messaging. Each ASC is mapped to a DICOM study object within the PACS, supporting the retrieval of (1) the entire imaging study, (2) any series contained in the study, or (3) any single image from any series.
DICOM object manipulations. When images are retrieved for display via the previously described web interface, they must be converted to a web compatible format such as JPEG or Bitmap (BMP). In addition, images may need to be deidentified based upon a user’s and/or project’s privileges. PixelMed provides functionality such as the conversion and deidentification of DICOM objects (based on DICOM 3.0 standards).
Grid Enablement. Grid enabling our model system allows us to support research collaborations involving institutionally and geographically disparate participants, a scenario that has motivated many recent infrastructural research and development programs, such as caBIG. In this project, we specifically have Grid enabled our system using the caBIG developed caGrid middleware. To implement this type of functionality, a wrapper application was created that supports Grid-compliant electronic data interchange with both OTP and PixelMed (thus, making them available as caGrid analytical services). As Grid services operate in a manner similar to web services or applications, implementing our Grid-based system within the institutional firewall did not require any specialized network configurations other than the placement of the presentation and application tiers within a demilitarized zone (DMZ) of our network, which for security purposes, incorporates an access control list to restrict connectivity to known hosts.
Data Sources Tier
Several key requirements influenced the design of the data sources tier, specifically, the needs to (1) ensure HIPAA-compliant deidentification of the data when necessary, (2) generate proxy data sources for efficient access, and (3) manage access privileges at multiple levels of granularity from projects to individuals. These requirements are described in detail in the following sections.
Deidentification. The need to deidentify data can be broadly separated into requirements that pertain to structured data, textual data, and image data:
Structured data. As structured data resides on relational databases, its deidentification is straightforward compared to other data types. This type of deidentification was achieved by removing and replacing patient-unique identifiers (using surrogate identifiers that we refer as deidentifiers).
Text data. Within the OSUMC IW, all text reports are pre-processed to generate distinct, deidentified versions of the original source text, which were used in this project.
Image data. PixelMed allows DICOM objects to be separated into metadata and image components and provides total programmatic control over the metadata section through its Java-based API. Hence, by rewriting and manipulating the metadata of the DICOM objects, images are deidentified or anonymized according to the HIPAA guidelines.
Proxy Data Sources. To prevent computational performance issues pertaining to the use of operational data repositories, we implemented proxy relational databases and proxy PACS as part of our data sources tier. Research images and data are moved to these proxy resources in a time-sensitive, batch operation. To support regulatory compliance, research datasets are also deidentified during this batch oration. The two specific components of our proxy data source are:
Proxy relational database. Within our framework this is a physically and logically distinct Oracle instance. Oracle enables tables and views based on remote data sources; however, in our case, this proxy database only holds deidentified summary tables and materialized views that are based on our operational databases. User accounts created for accessing this proxy database only have view-only privileges for that data.
Proxy PACS. The proxy PACS is an instance of PixelMed’s PACS server. Images that have been approved for research use are moved to this PACS in deidentified form. The use of this proxy PACS allows for more frequent and large-scale queries to be executed as would be possible if searching the operational PACS used for clinical care purposes, owing to performance degradation concerns in such an alternative scenario.
RESULTS
To evaluate the feasibility and performance of our model system implementation, we performed an evaluation of the system using the previously introduced motivating use case applied to 30 months of patient data stored in the OSUMC IW, which comprised 1,373,194 radiology reports and 304,212 pathology reports that corresponded to 4,753,985 patient encounters. Our example searches focused on two main ICD9 code categories: lung cancer (162, malignant neoplasm of trachea, bronchus, and lung) and coagulation defects (286, coagulation defects). Whereas our example searches involve both radiology and pathology reports for the first category (lung cancer), searches for the second category (coagulation defects) were applied to radiology reports only.
Lung Cancer Searches
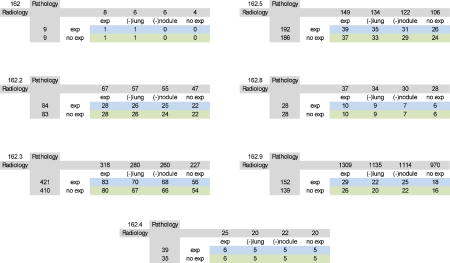
Under this category, seven different ICD9-CM codes were used for query construction: trachea (162.0), main bronchus (162.2), upper lobe (162.3), middle lobe (162.4), lower lobe (162.5), other parts of bronchus or lung (162.8), and bronchus and lung, unspecified (162.9). For our search on radiology reports, the concept “lung nodule” was expanded to include “thoracic,” “chest” for “lung,” and “lesion” for “nodule.” For our search on pathology reports, the keyword “carcinoma” was expanded by the keyword “neoplasm.” Table 1 depicts the effects of the thesaurus-based conceptual expansions for each lung cancer ICD9 code.
Table 1.
Combining ICD9 Codes for “Malignant Neoplasm of Trachea, Bronchus, and Lung” with Radiology Reports that Contain Concept Corresponding to “Lung Nodules” along with Pathology Reports that Contain Concept Corresponding “Carcinoma”
In some cases, the use of knowledge-anchored query expansion captures more relevant data than would be possible in queries without such expansion. Column descriptions: Exp Both terms “lung” and “nodule” are expanded during the search on radiology reports; (-) Lung the term “lung” is not expanded during the search on radiology reports; (-) Nodule the term “nodule” is not expanded during the search on radiology reports; No exp no UMLS MT expansions were applied during the search on radiology reports. Row descriptions: Exp With aid of UMLS MT, both keywords “carcinoma” and “neoplasm” are used during the search on pathology reports; No exp no UMLS MT expansions were applied during the search on pathology reports.
Coagulation Defects Searches
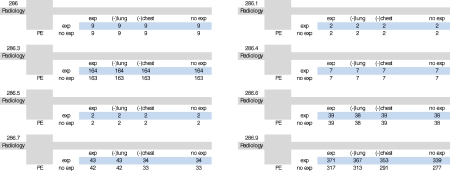
Under this category, nine different ICD9-CM codes were used for query construction: “congenital factor VIII disorder” (286.0), “congenital factor IX disorder” (286.1), “congenital factor XI deficiency” (286.2), “congenital deficiency of other clotting factors” (286.3), “von Willebrand’s disease” (286.4), “hemorrhagic disorder due to intrinsic circulating anticoagulants” (286.5), “defibrination syndrome” (286.6), “acquired coagulation factor deficiency” (286.7), and “other and unspecified coagulation defects” (286.9). For our search on radiology reports the keyword “pulmonary embolism” was expanded as follows: “lung,” “chest,” also synonym for pulmonary embolism “PE.” Table 2 depicts the effects of the thesaurus-based conceptual expansions for each coagulation defects ICD9 code.
Table 2.
Combining ICD9 Codes for “Coagulation Defects” with Radiology Reports that Contain Concept Corresponding to “Pulmonary Embolism”
Column descriptions: Exp Both terms “lung” and “chest” are expanded during the search on radiology reports; (-) Lung the term “lung” is not expanded during the search on radiology reports; (-) Chest the term “chest” is not expanded during the search on radiology reports; No exp no UMLS MT expansions were applied during the search on radiology reports. Row descriptions: Exp With aid of UMLS MT, the additional synonym “PE” for the concept of “pulmonary embolism” was used for expansion; No exp no synonyms were added during the search on radiology reports.
In our “lung cancer” cases, query expansion returned 43% more reports on average than without such expansion when applied to radiology reports (Table 1). However, in the context of pathology reports, this increase in returned reports was only 4%. In “coagulation defects” cases, similar query expansion yielded 8% additional reports than a query without such expansion (Table 2). Considering our earlier example on the keyword expansion of “melanocarcinoma” with “melanoma” where we demonstrate much greater (0 to 2706) expansion, these results demonstrate the impact on overall performance of the initial keywords used to pose a query.
The execution times associated with our model system were evaluated in two stages: (1) The first stage focused on the average time required to identify and retrieve images, which was found to range between 2–10 s; (2) the second stage focused on the average time required to retrieve structured and free-text data corresponding to a given image or image series, which was found to range between 4–10 s. These measurements were derived by 30 repeated measurements of the elapsed time required to retrieve and view the first eight images of a CT or MRI series for a given patient, and then to retrieve three on average related text reports and structured data elements for that study via the previously introduced web interface.
DISCUSSION
The retrieval of image data in support of research requirements is usually more meaningful when patient-derived phenotypic context data accompanies such images. Such phenotypic data can be derived from multiple sources, including free-text data such as radiology or pathology reports, and structured data such as diagnosis codes. As we have demonstrated, by applying integrative knowledge-anchored strategies, conceptual searches spanning all of the preceding data types are possible and, in some cases, can generate larger amounts of data meeting the criteria used to define a motivating use case and its associated data query requirements than is otherwise possible. We have also demonstrated that, in those instances where the deidentification of imaging and corresponding phenotypic data is needed to satisfy regulatory and patient confidentiality requirements, such deidentification can be performed in such a manner that overall context and the ability to recreate the linkage between data elements is maintained. Whereas such an approach by necessity introduced additional technical challenges to the proper deidentification of data, the programmatic utilization of the open-source PixelMed PACS API within our framework allows us to replace identifiers within metadata for images with deidentifiers, which are consistent with other deidentified phenotypic data, thus, demonstrating one possible solution to such challenges.
Interoperability is a major requirement for sharing data and collaborative work in a research environment, especially when that environment spans institutional and geographically distributed investigators and research participants. By adapting to the caGrid middleware in our framework and model system, we are able to facilitate such interoperability and both the syntactic and semantic levels—thus, addressing the prior requirement. In particular, the multi-tier architecture we have adopted simplifies deployment of new technology such as caGrid within our framework. This multi-tiered framework has additional benefits, including more efficient control of and access to multiple underlying data sources and the ability to mitigate potential performance concerns in operational systems through the utilization of appropriate proxy data sources.
There are several limitations with our framework and model system that should be noted including (1) our relatively simple approach to text mining does not exploit more advanced semantic interpretation of clinical narrative text nor does it allow for the detection and reasoning upon negation within that text, which could effect the recall and precision of information retrieval tasks (however, an analysis of the recall and precision of the text mining process was infeasible during this study because of limited scope); (2) the query expansion techniques employed by virtue of the previously noted simple approaches to text mining do not lend themselves to fully assisting end users in identifying optimal descriptors or codes to be used during the query formulation process, and thus, the efficacy of our queries are, in part, reliant on the domain expertise and heuristic knowledge of our end users; (3) the text data deidentification scheme employed relies on preexisting deidentification processes external to our framework and model system; and (4) our evaluation of the described software framework and model system is limited to a single instance and basic scope, owing to the preliminary status of our work as a model formulation effort.
CONCLUSION
We have described a model and associated software framework with promising and unique combination of components that are capable of providing translational research users with an integrative query and information retrieval tool that spans multiple, critical biomedical information sources including structured data, narrative text, and images. Furthermore, the inclusion of both deidentification mechanisms and standard-compliant electronic data interchange modalities within our system have significant potential to address inherent challenges to the conduct of multi-center or cross-disciplinary translational research in the modern regulatory environment. Our future plans for this project include the continued evaluation of the framework, with specific emphasis on the types of novel hypotheses that can be addressed using such a knowledge-anchored, integrative query platform, as well as its applicability to other usage scenarios. We fully anticipate that our system, with its focus on satisfying a critical translational research information need, will continue to develop into an operational platform for use by researchers at OSUMC that will also be extensible to the broader informatics and research communities.
Acknowledgments
Authors would like to thank Jason Buskirk, Felix Liu, Scott Silvey, Tremayne Smith, Ty Tolley and Herb Smaltz.
References
- 1.Cimino JJ. From data to knowledge through concept-oriented terminologies: experience with the Medical Entities Dictionary. J Am Med Inform Assoc. 2000;7(3):288–297. doi: 10.1136/jamia.2000.0070288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sujansky W. Heterogeneous database integration in biomedicine. J Biomed Inform. 2001;34(4):285–298. doi: 10.1006/jbin.2001.1024. [DOI] [PubMed] [Google Scholar]
- 3.Kamal J, et al. Information warehouse as a tool to analyze computerized physician order entry order set utilization: opportunities for improvement. AMIA Annu Symp Proc. 2003;2003:336–340. [PMC free article] [PubMed] [Google Scholar]
- 4.Prather JC, et al. Medical data mining: knowledge discovery in a clinical data warehouse. Proc AMIA Annu Fall Symp. 1997;1997:101–105. [PMC free article] [PubMed] [Google Scholar]
- 5.Brown M, et al. CAD in clinical trials: current role and architectural requirements. Comput Med Imaging Graph. 2007;31(4–5):332–337. doi: 10.1016/j.compmedimag.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 6.Kamauu AW, et al. Informatics in radiology (infoRAD): vendor-neutral case input into a server-based digital teaching file system. Radiographics. 2006;26(6):1877–1885. doi: 10.1148/rg.266065707. [DOI] [PubMed] [Google Scholar]
- 7.NCIA: Reference Image Database to Evaluate Response (RIDER). Available at http://ncia.nci.nih.gov/ncia/collections. Cited 2007
- 8.Sigal R: PACS as an e-academic tool International Congress series 2005, 1281, CARS 2005: Computer Assisted Radiology and Surgery, pp. 900–904
- 9.Boochever SS. HIS/RIS/PACS integration: getting to the gold standard. Radiol Manage. 2004;26:16–24. [PubMed] [Google Scholar]
- 10.Gruber TR. Toward principles for the design of ontologies used for knowledge sharing. In: Guarino N, Poli R, editors. Formal Ontology in Conceptual Analysis and Knowledge Representation. Norwell: Kluwer; 1993. [Google Scholar]
- 11.Joseph P, Bruce GB: Ontology-guided knowledge discovery in databases. In: Proceedings of the international conference on knowledge capture. Victoria, British Columbia, Canada: ACM Press, 2001
- 12.Smith B, Kumar A. On controlled vocabularies in bioinformatics: a case study in the gene ontology. Biosilico: Drug Discovery Today. 2004;2(1):246–252. doi: 10.1016/S1741-8364(04)02424-2. [DOI] [Google Scholar]
- 13.Gurcan MN, et al. Lung nodule detection on thoracic computed tomography images: preliminary evaluation of a computer-aided diagnosis system. Med Phys. 2002;29(11):2552–2558. doi: 10.1118/1.1515762. [DOI] [PubMed] [Google Scholar]
- 14.Ebbert JO, Dupras DM, Erwin PJ. Searching the medical literature using PubMed: a tutorial. Mayo Clin Proc. 2003;78(1):87–91. doi: 10.4065/78.1.87. [DOI] [PubMed] [Google Scholar]
- 15.Olson GM, et al: Collaboratories to support distributed science: the example of international HIV/AIDS research. In: Proceedings of SAICSIT, South Africa. Victoria, British Columbia, Canada: ACM Press, 2002
- 16.Butler D. Data, data, everywhere. Nature. 2001;414(6866):840–841. doi: 10.1038/414840a. [DOI] [PubMed] [Google Scholar]
- 17.Marks RG, Conlon M, Ruberg SJ. Paradigm shifts in clinical trials enabled by information technology. Stat Med. 2001;20(17–18):2683–2696. doi: 10.1002/sim.736. [DOI] [PubMed] [Google Scholar]
- 18.Payne PR, Greaves AW, Kipps TJ. CRC clinical trials management system (CTMS): an integrated information management solution for collaborative clinical research. AMIA Annu Symp Proc. 2003;2003:967. [PMC free article] [PubMed] [Google Scholar]
- 19.Kuchenbecker J, et al. Use of internet technologies for data acquisition in large clinical trials. Telemed J E Health. 2001;7(1):73–76. doi: 10.1089/153056201300093976. [DOI] [PubMed] [Google Scholar]
- 20.Marks L, Power E. Using technology to address recruitment issues in the clinical trial process. Trends Biotechnol. 2002;20(3):105–109. doi: 10.1016/S0167-7799(02)01881-4. [DOI] [PubMed] [Google Scholar]
- 21.Bates DW, et al. A proposal for electronic medical records in U.S. primary care. J Am Med Inform Assoc. 2003;10(1):1–10. doi: 10.1197/jamia.M1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sung NS, et al. Central challenges facing the national clinical research enterprise. JAMA. 2003;289(10):1278–1287. doi: 10.1001/jama.289.10.1278. [DOI] [PubMed] [Google Scholar]
- 23.Bates DW, et al. Effect of computerized physician order entry and a team intervention on prevention of serious medication errors. JAMA. 1998;280(15):1311–1316. doi: 10.1001/jama.280.15.1311. [DOI] [PubMed] [Google Scholar]
- 24.Huang H, et al. Picture archiving and communication systems (PACS) in medicine. New York: Springer; 1991. [Google Scholar]
- 25.Duerinckx AJ, Pisa EJ. Filmless picture archiving and communication system (PACS) in diagnostic radiology. Proc SPIE. 1982;318:9–18. [Google Scholar]
- 26.Gurcan MN, et al. GridImage: a novel use of grid computing to support interactive human and computer-assisted detection decision support. J Digit Imaging. 2007;20:160–171. doi: 10.1007/s10278-007-9020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Craver JM, Gold RS. Research collaboratories: their potential for health behavior researchers. Am J Health Behav. 2002;26(6):504–509. doi: 10.5993/ajhb.26.6.11. [DOI] [PubMed] [Google Scholar]
- 28.Kukafka R, et al. Grounding a new information technology implementation framework in behavioral science: a systematic analysis of the literature on IT use. J Biomed Inform. 2003;36(3):218–227. doi: 10.1016/j.jbi.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Johnson MS, Gonzales MN, Bizila S. Responsible conduct of radiology research part V. The health insurance portability and accountability act and research. Radiology. 2005;237(3):757–764. doi: 10.1148/radiol.2373041148. [DOI] [PubMed] [Google Scholar]
- 30.Liu BJ, Zhou Z, Huang HK. A HIPAA-compliant architecture for securing clinical images. J Digit Imaging. 2006;19(2):172–180. doi: 10.1007/s10278-005-9248-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amendolia SR, et al: MammoGrid: a service oriented architecture based medical grid application. In: 3rd International Conference on Grid and Cooperative Computing, Wuhan, China, 2004
- 32.Blanquer I, et al: A Middleware grid for storing, retrieving and processing DICOM medical images. In: Workshop on Distributed Databases and Processing in Medical Image Computing (DIDAMIC), Rennes, France, 2004
- 33.Espert IB, Garcaa VH, Quilis JD. An OGSA middleware for managing medical images using ontologies. J Clin Monit Comput. 2005;19(4–5):295–305. doi: 10.1007/s10877-005-0675-0. [DOI] [PubMed] [Google Scholar]
- 34.Montagnat J, et al: Medical image content-based queries using the grid. In: HealthGrid’03, France, Lyon, 2003
- 35.Power D, et al: A relational approach to the capture of DICOM files for Grid-enabled medical imaging databases. In: ACM symposium on applied computing, Cyprus, Nicosia, 2004, pp 272–279
- 36.Foster I, Kesselman C. The Grid 2: blueprint for a new computing infrastructure. 2. New York: Morgan Kaufman; 2003. p. 748. [Google Scholar]
- 37.Payne PR, et al. Conceptual knowledge acquisition in biomedicine: a methodological review. J Biomed Inform. 2007;40:582–602. doi: 10.1016/j.jbi.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.NLM: Unified Medical Language System. Available at http://www.nlm.nih.gov/research/umls/meta2.html. Cited 2007
- 39.Bodenreider O. Using UMLS semantics for classification purposes. Proc AMIA Symp. 2000;2000:86–90. [PMC free article] [PubMed] [Google Scholar]
- 40.Campbell KE, et al. Representing thoughts, words, and things in the UMLS. J Am Med Inform Assoc. 1998;5(5):421–431. doi: 10.1136/jamia.1998.0050421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas BJ, et al. Automated computer-assisted categorization of radiology reports. Am J Roentgenol. 2005;184(2):687–690. doi: 10.2214/ajr.184.2.01840687. [DOI] [PubMed] [Google Scholar]
- 42.Tsui F-C, et al. Value of ICD-9-coded chief complaints for detection of epidemics. J Am Med Inform Assoc. 2002;9:S41–S47. doi: 10.1197/jamia.M1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedman C, et al. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. 2004;11(5):392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srinivasan S, et al: Finding UMLS Metathesaurus concepts in MEDLINE. In: American Medical Informatics Association Annual Symposium, 2002, pp 727–731 [PMC free article] [PubMed]
- 45.Taira RK, Soderland SG, Jakobovits RM. Automatic structuring of radiology free-text reports. Radiographics. 2001;21:237–245. doi: 10.1148/radiographics.21.1.g01ja18237. [DOI] [PubMed] [Google Scholar]
- 46.Zou Q, et al: IndexFinder: a method of extracting key concepts from clinical texts for indexing. In: American Medical Informatics Association Annual Symposium, 2003, pp 763–767 [PMC free article] [PubMed]
- 47.Alonso O, et al: Oracle text white paper. Available at http://www.oracle.com/technology/products/text/index.html. Cited 2006
- 48.International Business Machines Corporation: DB2 text extender. Available at ftp://ftp.software.ibm.com/software/data/db2/extenders/text/db2tewkspecsheet.pdf. Cited 2002
- 49.Microsoft Corporation: SQL Server 2000 full-text search deployment white paper. Available at http://www.support.microsoft.com/kb/323739. Cited 2004
- 50.Ferrucci D, Lally A. Building an example application with the unstructured information management architecture. IBM Syst J. 2004;43(3):455–475. doi: 10.1147/sj.433.0455. [DOI] [Google Scholar]
- 51.Baecker R, Small I, Mander R: Bringing icons to life. Proceedings of the SIGCHI conference on human factors in computing systems: reaching through technology. New Orleans, Louisiana, USA: ACM Press, 1991, pp 1–6
- 52.NEMA: Digital imaging and communications in medicine. Available at http://www.medical.nema.org/. Cited 2007
- 53.Armato SG, III, et al. Lung image database consortium: developing a resource for the medical imaging research community. Radiology. 2004;232:739–748. doi: 10.1148/radiol.2323032035. [DOI] [PubMed] [Google Scholar]
- 54.Sigal R: PACS as an e-academic tool. In CARS 2005: computer assisted radiology and surgery. 2005
- 55.Toms AP, et al. Building an anonymized catalogued radiology museum in PACS: a feasibility study. Br J Radiol. 2006;79:661–671. doi: 10.1259/bjr/11861604. [DOI] [PubMed] [Google Scholar]
- 56.Cohen S, Gilboa F, Uri S. PACS and electronic health records. San Diego, CA: SPIE; 2002. [Google Scholar]
- 57.Lehmann T, Wein B, Greenspan H: Integration of content-based image retrieval to picture archiving and communication systems. In: Medical Informatics Europe Conference, 2003
- 58.Traina A, Rosa NA, Traina C. Integrating images to patient electronic medical records through content-based retrieval techniques. In: 16th IEEE Symposium on Computer-Based Medical Systems, 2003
- 59.Leoni L, et al: A virtual data grid architecture for medical data using SRB. In: EuroPACS-MIR 2004, Trieste, Italy, 2004
- 60.Erdal S, et al: Flexible patient information search and retrieval framework: pilot implementation. In: Proceedings of the SPIE Medical Imaging, San Diego, CA, 2007
- 61.Erdal S, et al: Information warehouse application of caGrid: a prototype implementation. In: caBIG 2007 Annual Meeting, Washington, DC, 2007
- 62.Erdal S, et al: Integrating a PACS system to grid: a de-identification and integration framework. In: Annual Meeting of the Society for Imaging Informatics in Medicine (SIIM) 2007, Providence, RI, 2007
- 63.Lindberg C. The unified medical language system (UMLS) of the national library of medicine. J Am Med Rec Assoc. 1990;61(5):40–42. [PubMed] [Google Scholar]
- 64.Lindberg DA, Humphreys BL, McCray AT. The unified medical language system. Methods Inf Med. 1993;32(4):281–291. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cancer Biomedical Informatics Grid (caBIGä). Available ttps://cabig.nci.nih.gov/workspaces/Architecture/caGrid/, https://cabig.nci.nih.gov/workspaces/Architecture/caGrid/. Cited 2006
- 66.Eckerson WW. Three tier client/server architecture: achieving scalability, performance, and efficiency in client server applications. Open Inf Syst. 1995;10:1–12. [Google Scholar]
- 67.Gallaugher J, Ramanathan S: Choosing a client/server architecture. A comparison of two-tier and three-tier systems. Inf Syst Manage Mag 13(2):7–13
- 68.Clunie DA. DICOM structured reporting. Bangor, Pennsylvania: PixelMed; 2000. [Google Scholar]
- 69.Payne PR, et al. Breaking the translational barriers: the value of integrating biomedical informatics and translational research. J Investig Med. 2005;53(4):192–200. doi: 10.2310/6650.2005.00402. [DOI] [PubMed] [Google Scholar]