Abstract
This paper presents an adaptive attention window (AAW)-based microscopic cell nuclei segmentation method. For semantic AAW detection, a luminance map is used to create an initial attention window, which is then reduced close to the size of the real region of interest (ROI) using a quad-tree. The purpose of the AAW is to facilitate background removal and reduce the ROI segmentation processing time. Region segmentation is performed within the AAW, followed by region clustering and removal to produce segmentation of only ROIs. Experimental results demonstrate that the proposed method can efficiently segment one or more ROIs and produce similar segmentation results to human perception. In future work, the proposed method will be used for supporting a region-based medical image retrieval system that can generate a combined feature vector of segmented ROIs based on extraction and patient data.
Key words: Microscopic image, nuclei segmentation, region of interest (ROI), adaptive attention window (AAW), quad-tree, region-based image retrieval
Introduction
With the increase of genome projects that have decoded the genomes of several species, created genetic maps, and analyzed the order of the chromosome maps, various medical assistance systems such as the picture archiving communication system have been introduced that integrate information communication, computer networking, database management, digital imaging, and a user interface. These systems manage digital images acquired using various imaging modalities such as CT, X-ray, MRI, and PET. As such, a lot of research has recently been focused on efficient medical image retrieval, allowing target images to be found in a huge database. Traditionally, medical images have been indexed and retrieved using just text. Yet, traditional text-based retrieval can produce irrecoverable mismatches according to the subjectivity and viewpoint of the writer. Furthermore, this kind of retrieval is costly and time consuming.
Thus, to overcome these problems, various types of content-based image retrieval (CBIR)1–4 have been proposed over the last few decades. Unlike text-based retrieval, CBIR indexes images using color, texture, shape, and sound, which are then used for retrieval instead of keywords. Ideally, the goal is to create an interactive system for retrieving images that is semantically related to the user’s query from the database. More recent research has also focused on region-based retrieval5,6 that allows the user to specify a particular region or object in an image and request the system to retrieve similar images containing similar regions.
Most existing region-based image retrieval systems rely on image segmentation and require extraction of the region of interest (ROI), which occupies a large portion of the entire image. Therefore, semantic ROI segmentation is essential for efficient region-based image retrieval.
In the medical field, region-based image retrieval is also helpful for diagnostic purposes. For example, diagnosis systems based on cytology and histophysiology are used to analyze tissue specimens to detect lesions as an early signal of latent cancer. Plus, measuring the cell cycle using a diagnosis system can enhance the effectiveness of drug discovery and development. However, existing diagnosis systems are restricted when dealing with cells and due to the subjective variance of an observer. Therefore, an integrated diagnosis system7,8 with an automatic aid method was recently developed to assist with the detection of cancer.
For a semantic analysis, an automatic aid method requires an ROI-based approach rather than a pixel-based approach to detect an abnormal nucleus or lesion. In particular, the ROI segmentation is a crucial preprocess to enable successful cell classification or diagnosis.
Comaniciu and Meer9 developed the Image Guided Decision Support system to analyze tissue structures and organ states to support diagnosis and identify factors in clinical pathology. The system extracts an ROI within the attention scope using the mean-shift segmentation method. However, to extract the ROI before segmentation, the attention window must be defined by hand.
Chen et al.10 developed a method of image analysis to resolve the problems of touching cells and ambiguous correspondence, resulting in a computational bio image system that facilitates the automated segmentation, tracking, and classification of cancer cell nuclei in time-lapse microscopy images.
Tscherepanow et al.11 proposed a method for classifying segmented regions in bright field microscope images. However, since an active contour is used for the cell segmentation, the performance can deteriorate when an image contains cells with a complex structure.
Unlike general natural images, microscopic images have different characteristics with distinct meanings based on human estimation and a varying brightness according to the fluorescence staining. For example, salient parts such as cell nuclei tend to be brighter, while the remaining parts have a more monotonous appearance. All existing medical image-segmentation methods segment regions from an image regardless of the meaning of the ROIs, meaning that exact ROI segmentation is impossible without human interaction. Thus, for semantic ROI segmentation such as salient cells, knowledge of the exact positions of relevant ROIs is crucial.
Accordingly, this paper presents an adaptive attention window (AAW)-based ROI microscopic cell image-segmentation method. For semantic AAW detection, an initial attention window (IAW) is created using a luminance map, then the IAW is reduced close to the size of the ROI cell using a quad-tree. The purpose of the AAW is to determine the rough position of relevant ROIs, thereby reducing the amount of processing time for segmenting ROIs. Finally, region-level segmentation is performed within the AAW, along with background removal and region clustering to segment only the ROIs.
The remainder of this paper is organized as follows. “Adaptive Attention Window Generation” explains the AAW generation based on human perception, then “ROI Segmentation Within the AAW” describes the ROI segmentation within the AAW. “Experimental Results” evaluates the accuracy and applicability of the proposed ROI segmentation based on experiments, and some final conclusions and areas for future work are presented in “Conclusion”.
Adaptive Attention Window Generation
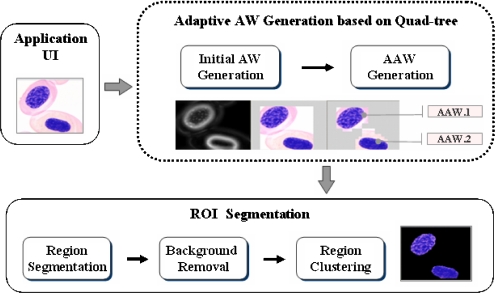
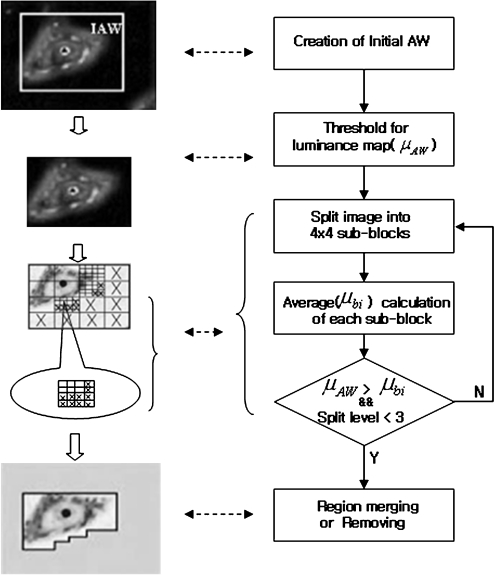
For the semantic segmentation of ROIs, the positions of the considered ROIs must first be identified within an image. Therefore, this paper proposes an AAW method for ROI extraction. The coarse positions of the considered ROIs are determined using the proposed AAW, which changes adaptively according to the location of the ROIs within an image. Figure 1 shows the architecture of the proposed AAW-based segmentation. The AAW generation is based on human perception in the preprocessing step. Then, as a result of the proposed method, only the ROIs remain in the segmentation step.
Fig 1.
Architecture of proposed method.
Luminance Map Generation
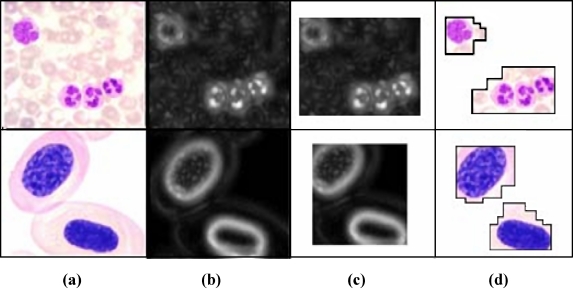
Human perception plays an important role in computer vision and pattern recognition, and many studies have attempted to use it to analyze the semantic meaning within an image. For example, Itti et al.12 proposed a saliency-based visual attention model based on color, luminance, and orientation, then selected the most salient area based on a winner-take-all competition. However, in this paper, the IAW within an image is detected using a luminance map and quad-tree split based on human perception during the image-segmentation preprocessing. In medical images, since the intensity is the unique component, a luminance feature map is used to detect the IAW, rather than a color feature map, as the color can change according to the dye used. Figure 2 shows the AAW generation process within an image.
Fig 2.
AAW generation process. a Input image, b luminance map, c IAW, and d final AAWs.
First, to generate the luminance map (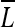 ), two different sized filters (7 × 7 and 13 × 13) are applied to a 1/2 down-sampled gray image (L′(s)) against the original image and the luminance contrast computed. As such, the filters estimate the center-surround differences between the center point and the surrounding points within the filter scale s, and these differences produce the feature map.
), two different sized filters (7 × 7 and 13 × 13) are applied to a 1/2 down-sampled gray image (L′(s)) against the original image and the luminance contrast computed. As such, the filters estimate the center-surround differences between the center point and the surrounding points within the filter scale s, and these differences produce the feature map.
When the center surround difference is large, the most active location stands out, and the map is strongly promoted. Conversely, when the difference is small, the map contains nothing unique and is suppressed. Thus, when using Eq. 1, the sum yields two different feature maps from the two filters.
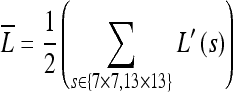 |
1 |
These maps are then summed and normalized into one luminance feature map 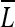 , which is up-sampled to the size of the original image and smoothed with a Gaussian filter to eliminate any pixel-level noise and highlight the neighborhood of influence for the output map. Examples of luminance maps are shown in Figure 2b.
, which is up-sampled to the size of the original image and smoothed with a Gaussian filter to eliminate any pixel-level noise and highlight the neighborhood of influence for the output map. Examples of luminance maps are shown in Figure 2b.
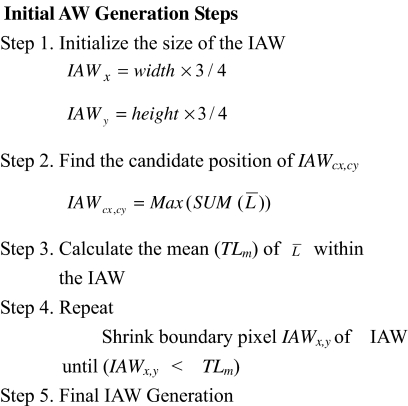
Initial AW Generation Using Luminance Map
After generating the luminance map, the IAW is detected to remove useless regions such as background, thereby reducing the amount of processing time required for ROI segmenting and improving the extraction performance. ROIs are generally located near the center area; however, since this is not always true, the size of the IAW is very important.
Therefore, this paper proposes a top-down IAW shrinking method that uses the created luminance map. The initial rectangular IAW is three quarters the size of the image and reduced until it meets predefined conditions. This size was determined by experiments and the analysis of reference images in a database where the largest cell size as the ROI in the experimental database set was found to be less than three quarters the size of the image.
To determine the proper location (AWcx,cy) and size (AWx,y) of the IAW, the window that includes the maximum magnitude from the luminance map (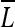 ) for the full image is initially chosen, then the size of the IAW is shrunk to the approximate size of the ROIs. Figure 3 shows the shrinking steps and conditions that are presented as a pseudo code for the IAW generation, while Figure 2c shows some examples of IAWs.
) for the full image is initially chosen, then the size of the IAW is shrunk to the approximate size of the ROIs. Figure 3 shows the shrinking steps and conditions that are presented as a pseudo code for the IAW generation, while Figure 2c shows some examples of IAWs.
Fig 3.
Initial AW generation.
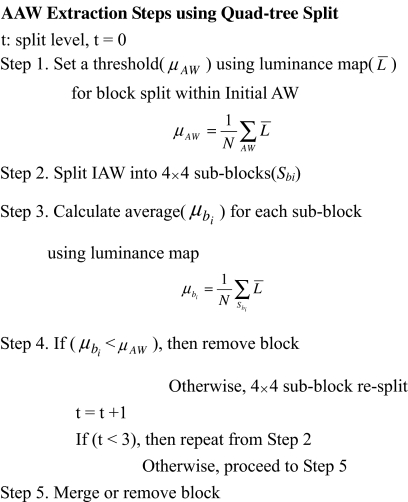
AAW Extraction Using Quad-tree Split
After the IAW is selected, the IAW needs to be shrunk to the most approximate size of the salient ROIs. The existing split method for extracting close-shape ROIs initially considers the image as one region, then iteratively splits according to a homogeneity criterion into smaller and smaller regions. This split method is realized according to geometrical structures such as squares, triangles, and polygons13. To split into real ROIs and reduce the processing time, the proposed method extracts an AAW with a size close to the real ROI within the limited IAW using a quad-tree that splits based on a square structure. Unlike previous research14,15, the IAW is shrunk using the luminance feature map as the split condition within the IAW. Figure 4 shows the splitting steps for extracting the AAW.
Fig 4.
Pseudo code for AAW extraction.
The shrinking steps and conditions are as follows. First, the average (μAW) of the luminance map is calculated within the IAW, and the IAW is split into 4 × 4 scale 1 sub-blocks. The average is then used as a threshold to further divide the sub-blocks. As such, for every sub-block, if the average (μAW) of the luminance map within a sub-block is above the threshold, that sub-block is split again into 4 × 4 scale 2 sub-blocks; otherwise, it is removed.
This process of removing sub-blocks is based on the assumption that humans usually concentrate on special regions that have a high luminance contrast12. That is, if the average of the luminance map is low, this means that the sub-block has a lower probability of containing salient regions; alternatively, if the average is high, this means the sub-block has a higher probability of containing salient regions. Therefore, a sub-block with a higher luminance feature map compared to the predefined threshold is split again to detect close ROIs. This process is repeated to scale 3. Finally, the scale 3 sub-blocks are either merged into adjacent large sub-blocks or removed if they are far away from major sub-blocks. Figure 5 shows the proposed AAW extraction processes.
Fig 5.
AAW extraction processes.
ROI Segmentation Within the AAW
While object-based image segmentation is useful in many applications, it is still generally beyond current computer vision techniques due to the uncontrolled nature of the available images and requirement of much processing time. Since an object is generally a group of related regions, the present study proposes a way of segmenting an image into regions, then merging these regions into an ROI. Thus, after the AAW is created, the regions within the AAW are classified as ROIs or background regions. To do this, the AAW is segmented into several regions that are then clustered into ROIs according to the proposed algorithm.
In the ROI segmentation step, the AAW is filtered and segmentation performed based on region merging and labeling using each channel. The boundary regions are then removed as background regions, which have a relatively low importance. Finally, the major regions with the highest luminance are selected from among the segmented regions, except for the background regions, and the adjacent regions merged into the major regions to produce the final segmented ROIs. As the result, the proposed method allows multiple salient cell regions to be segmented as ROIs according to the image characteristics.
Region Segmentation and Background Removal
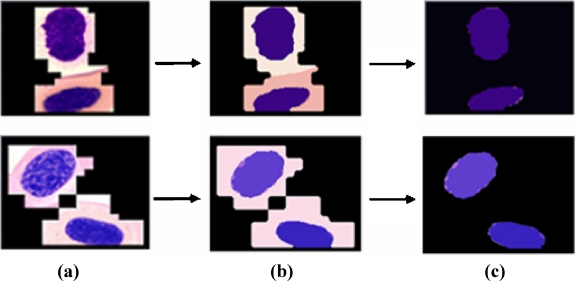
As mentioned in the introduction, the image-segmentation technique of Ko et al.6 is applied in the proposed method for semantic region segmentation where three types of adaptive circular filter are used to segment an image based on the amount of image texture information and Bayes’s theorem. After an image is segmented within the AAW, the segmented regions still consist of ROIs and some meaningless regions as shown in Figure 6b. Thus, for semantic ROI extraction, these meaningless regions need to be removed as background.
Fig 6.
Background region removal. a AAW, b segmentation results, and c background removal.
In conventional background removal methods such as Malpica et al.16 and Otsu17, a threshold is used to separate ROIs, like cells, from background regions such as cytoplasm. Although background removal using a global threshold is simple and efficient as it uses a histogram to create a bi-modal representation of an image, this separation method is not always suitable as a histogram does not necessarily provide a bi-modal representation and can fail to accurately segment ROIs. Therefore, this paper proposes a background removal method using the boundaries of the detected AAW and segmented regions.
If region segmentation is performed as shown in Figure 6b, boundary regions are defined as background within the AAW and removed. This definition is based on the following assumption: The major region is most likely to exist inside the AAW, while the background regions are more likely to exist at the AAW boundaries.
Therefore, the ratio for each region that connects with the AAW boundary is estimated using the Eq. 2.
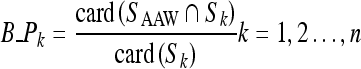 |
2 |
Where SAAW and Sk represent the boundary coordinate set for the AAW and kth region, respectively, card (A) represents the cardinality of each set, and the symbol B_Pk indicates the ratio of the kth region that connects with the AAW boundary. These ratios are then compared with a predefined threshold and, if larger, that region is regarded as a background region and removed. To determine the threshold for background region removal, the ratios for regions that connected with the AAW boundaries for database images were estimated and the average ratio taken as the threshold. Thus, in this paper, the threshold for background removal was set at 2.3, which demonstrated the best boundary removal performance based on several experiments using database images. As such, this threshold means that 2.3% of all boundary pixels in the kth region connect with the AAW boundary.
Figure 6c shows the ROIs separated when using the proposed background removal algorithm where nuclei are the major regions and cytoplasm is the background, which consists of monotonous colors.
Region Clustering
After removing the background regions, some regions may still not be an ROI. Thus, for exact segmentation of the ROIs, the segmented regions have to be clustered as major regions. The clustering algorithm used in this study is updated version of Ko and Nam’s15 region clustering algorithm as the shape and size or connectivity of regions is used to deal with the problem of over-segmentation.
After the background region removal, the major regions are selected for clustering, whereby similar adjacent regions are merged sequentially if the adjacent regions satisfy a few conditions. The merging steps according to clustering conditions are based on Eq. 3. For the regions segmented within the AAW, the size of each region is calculated, except for the removed background regions. Then, n seed regions with a saliency above a predetermined threshold and larger size compared to the other regions are selected as the major regions. In this paper, the number of seed regions was fixed at five based on the finding that the cell images in the experimental database tended to include fewer than five nuclei. Second, the ratio that the adjacent region is connected with the boundary or belongs to the seed region is calculated. Starting from each seed region sequentially, two regions are merged if the two regions satisfy the following conditions:
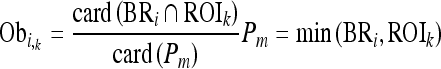 |
3 |
where BRi is the set of boundary pixels in the ith comparison region, ROIk denotes the set for the kth seed region, Pm denotes the set for the relatively smaller region between BRi and ROIk, the symbol card (A) represents the cardinality of set A, and Obi,k denotes the ratio of the inclusion relation between the kth ROIk and ith BRi. The symbol Obi,k becomes zero when the BRi region is not connected and exists outside of ROIk and becomes one when the BRi region surrounds ROIk. Finally, if Obi,k is above 0, BRi is merged with ROIk, and the area of Obi,k is updated. However, if Obi,k is 0, which means that BRi is located outside Obi,k, BRi is removed and merged with the background region according to Eq. 4.
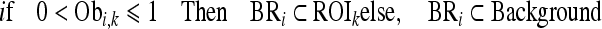 |
4 |
Figure 7 shows the ROI segmentation results when using the proposed algorithm.
Fig 7.
ROI segmentation results.
This step is repeated until no more regions exist. If the initial seed ROIk has no other regions to merge with and the final size of the clustered ROIk is below the predefined threshold, it is regarded as noise and merged with the background region.
Experimental Results
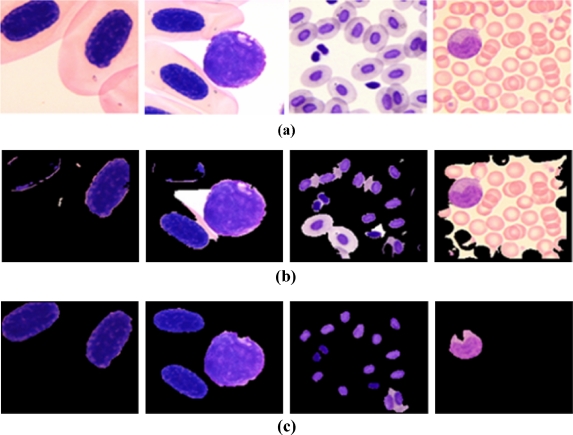
The experiments conducted to evaluate the proposed method were performed on a Pentium PC single-CPU 2.8 GHz with a Windows operating system. The proposed method was implemented using Visual C++ 6.0 language and 180(W) × 140(H) reference images from a database used for experiments. In this paper, a set of 200 cell images that included plant cells, white blood cells, and red blood cells were used where the histology was taken from a microscope. The image set consisted of 106 images of normal cells (fewer than five cells) and 94 images of numerous cells and abnormal cells (fewer than four cells).
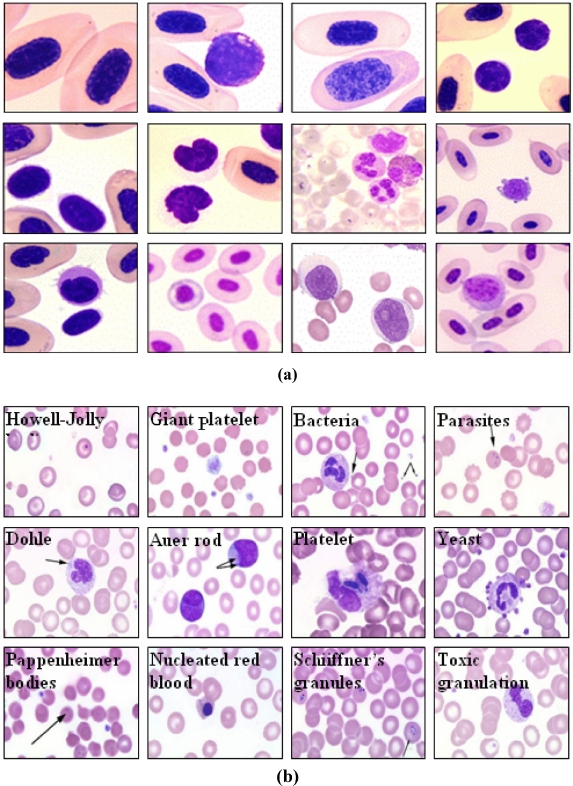
Figure 8 shows some examples of the cell images used for the experiments.
Fig 8.
Examples of cell images of a normal cells and b abnormal cells.
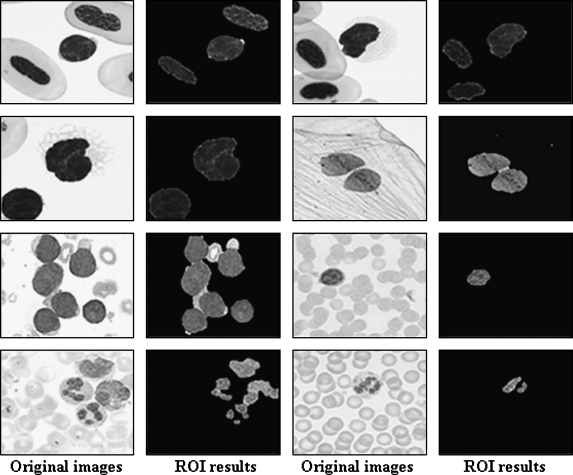
Experimental comparison of ROI segmentation is generally very difficult as there are few if any related works using cell images with AW. Thus, to validate the effectiveness of the proposed approach, the segmentation performance of the proposed system was compared with that of Comaniciu and Meer’s9 algorithm where the user creates the initial AW by hand and the segmentation is then performed using a mean-shift algorithm that is normally used for natural image segmentation. Figure 9 shows the ROI segmentation results for both ROI-based extraction methods.
Fig 9.
ROI segmentation results. a Original images. b Segmentation results by Comaniciu and Meer. c Segmentation results by proposed method.
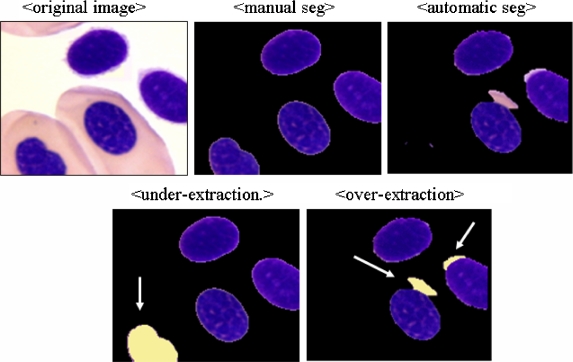
To evaluate the performance of the ROI extraction, the evaluation method proposed by Kim et al.18 was modified. First, three different pathologists were asked to crop the ROI from each image using a graphic tool, and only ROIs were used for comparison when at least two pathologists were in agreement. Figure 10 shows the under- and over-extraction errors marked by arrows for the automatically extracted ROIs when compared with the manually selected ROIs.
Fig 10.
Under- and over-segmentation errors.
The segmentation results for each method were compared with the manually extracted segmentation result and the error ratio estimated using Eq. 5.
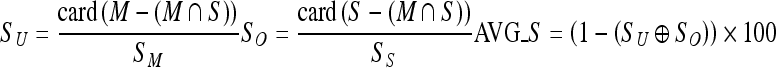 |
5 |
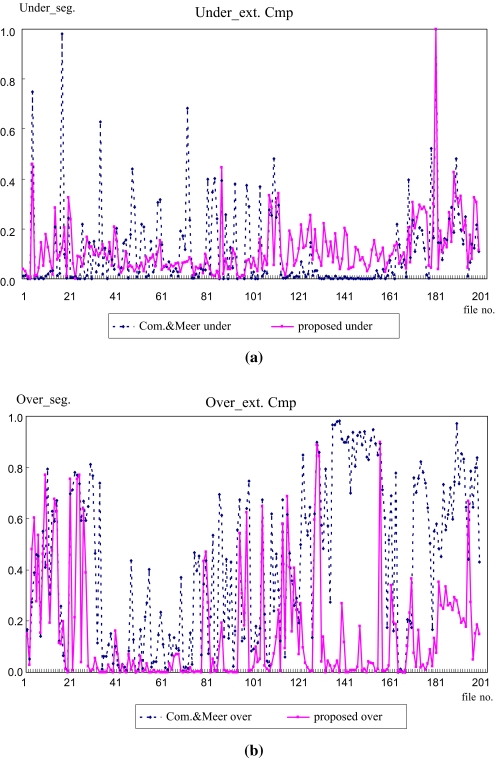
Where M represents a manually extracted ROI and S represents an automatically extracted ROI using each segmentation method, SM and SS denote the total size of the pixels of an extracted ROI, SU and SO represent the inaccuracy of the under-extraction ratio and over-extraction ratio, respectively, and AVG_S represents the accuracy between a manually cropped ROI and a systematically extracted ROI where a number closer to 100 represents a lower error and higher accuracy, while a number closer to 0 represents a higher error and lower reliability. Figure 11 shows the performance evaluation results using Eq. 5.
Fig 11.
Segmentation error evaluation results. a Under-segmentation error comparison. b Over-segmentation error comparison.
When compared to Comaniciu and Meer’s method, the proposed method showed a similar under-segmentation ratio, as shown in Figure 11. However, the proposed method had a lower over-segmentation ratio of 14.8% compared to Comaniciu and Meer’s segmentation method at 43.1%, as shown in Figure 11 and Table 1, respectively.
Table 1.
Error Comparison
| Method | Under-segmentation | Over-segmentation | Average accuracy (%) | Standard Deviation (under) | Standard Deviation (over) |
|---|---|---|---|---|---|
| Comaniciu and Meer | 0.109 | 0.431 | 46.0 | 0.165 | 0.317 |
| Proposed method | 0.116 | 0.148 | 73.6 | 0.112 | 0.214 |
From the experimental results in Figure 11, since Comaniciu and Meer’s method segmented the ROIs within a static rectangle region defined by the user, this led to a high over-extraction error ratio and also created many fragments as useless regions or ROIs. In addition, the over-extraction error with Comaniciu and Meer’s method was higher for the images between 106 and 185 that included numerous normal cells and abnormal cells, as the window was almost the same as the original image. In contrast, as the proposed method created an AAW with a form close to the ROIs and the segmentation was performed within the created AAW, this enabled the position of the ROIs to be detected accurately. As such, the proposed method had a lower over-extraction error ratio and high accuracy AVG_S at 73.6%. Therefore, the proposed segmentation method produced a better performance than the comparative algorithm. Furthermore, the standard deviation for the under- and over-extraction error ratio represented a regular rate with the proposed segmentation.
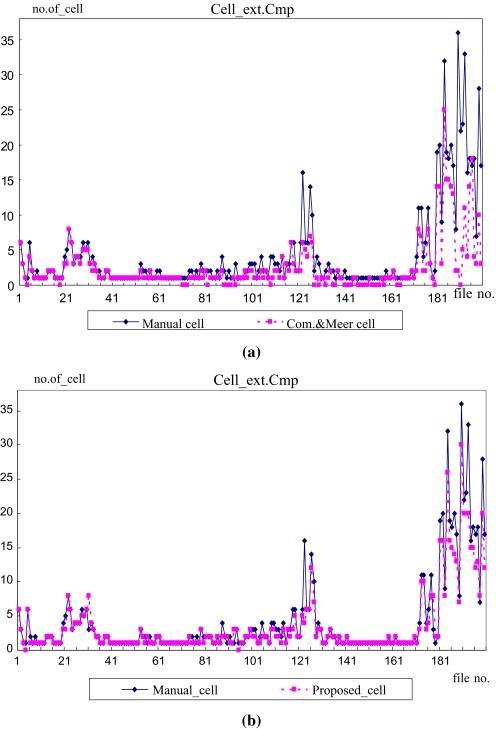
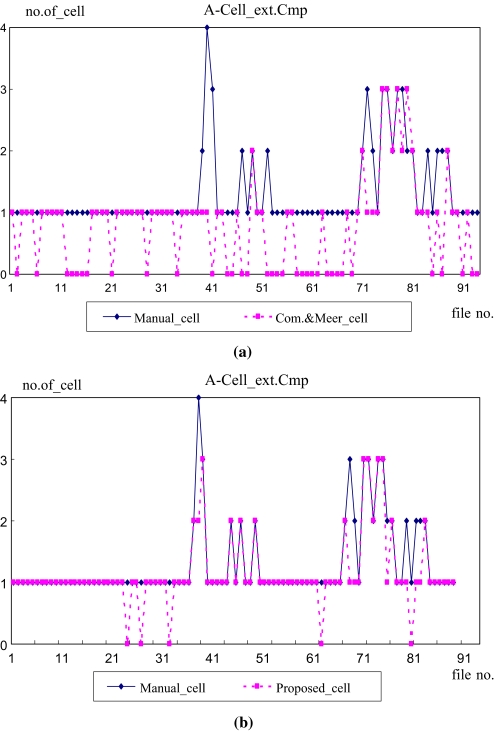
Next, to verify the applicability of the proposed method as a medical diagnosis aid system, the importance of the regions segmented using each method was compared with the manually cropped regions. Figure 8b shows examples of the abnormal cell images according to various diseases. For the evaluation, a count was made of the number of cells segmented from the total 200 images and number of abnormal cells segmented from the 94 images that coincided with the manually cropped ROIs. Table 2 compares the number of manually segmented cells with the number of cells automatically segmented by the two methods, while the graphs in Figures 12 and 13 show the number of cells extracted from the 200 images and number of abnormal cells extracted from the 94 images, respectively.
Table 2.
Comparison of ROI Segmentation
| Method | No. of ROIs | No. of abnormal ROIs |
|---|---|---|
| Manually segmented ROIs | 829 | 122 |
| Comaniciu and Meer method | 475 | 74 |
| Proposed method within AAW | 703 | 108 |
Fig 12.
Comparison of segmented cells. a ROI segmentation comparison between manually segmented cells with segmented cells using Comaniciu and Meer’s method. b ROI segmentation comparison between manually segmented cells with segmented cells using the proposed method.
Fig 13.
Comparison of segmented abnormal cells. a ROI segmentation comparison between manually segmented cells with segmented cells using Comaniciu and Meer’s method. b ROI segmentation comparison between manually segmented cells with segmented cells using the proposed method.
As shown in Table 2 and Figures 12 and 13, since Comaniciu and Meer’s method had a high over-segmentation error ratio and was unable to exactly segment the ROIs, it had a very low segmentation number. In the case of the abnormal cells, it also had a very low segmentation number at 74 compared with the number of manually segmented abnormal cells at 122. However, the proposed method segmented 703 cells, resulting in an 89% segmentation ratio, which was very close to the manually segmented results. Furthermore, in the case of the abnormal cells, the proposed method had a relatively high segmentation number at 108 out of the total 122 abnormal cells.
Since the salient parts in cell images, such as normal and abnormal cell nuclei, tend to be relatively bright against the background according to the dye material, the proposed method utilized this characteristic by splitting the images into small sub-regions with ROIs using a luminance contrast feature map to create AAWs and then segmented the exact ROIs within the AAWs. As such, the proposed method was able to segment the abnormal cells very efficiently. Thus, the experimental results confirmed the effectiveness of the proposed method as a diagnostic aid in relation to detecting abnormal cells.
Conclusion
Most existing CBIR systems supporting region-level retrieval use an image-segmentation method and require a ROI segmentation, which occupies a large portion of the entire image. Therefore, semantic ROI segmentation is essential for efficient region-based image retrieval. Also, an automatic aid method requires a region-based approach for ROIs, such as abnormal nuclei or lesions, rather than a pixel-based approach. Yet, this requires several techniques such as ROI segmentation, region classification, and cell-cycle tracking. In particular, ROI segmentation is crucial preprocess that then enables the other techniques to perform successfully.
Unlike natural images, microscopic cell images have different meanings according to human estimation and a varying brightness according to the fluorescence staining. For example, salient parts such as cell nuclei tend to be bright, while other parts have a more monotonous appearance. Most conventional medical image-segmentation methods segment regions from the complete image regardless of the ROIs. Thus, for the semantic segmentation of ROIs such as salient cells, the exact positions of the considered ROIs need to be known and segmented within an image.
Accordingly, this paper proposed an AAW-based microscopic cell image-segmentation method for ROI-based medical image retrieval and clinical diagnosis. The proposed method creates an IAW based on human perception using a luminance contrast map, then extracts an AAW that is similar to the size of the real ROIs using quad-tree split. Thereafter, region-level segmentation is performed within the AAW and the final ROIs segmented using background removal and region clustering. The segmentation within the extracted AAW not only reduces the amount of processing time for region segmenting but also extracts really exact ROIs. Experimental results confirmed that the proposed method could efficiently segment multiple ROIs and produced similar segmentation results to human perception. Its effectiveness as a medical diagnosis aid system was also demonstrated.
In future work, the proposed method will be used to develop a content-based medical image retrieval system that can generate a combined feature vector of segmented ROIs based on extraction and patient data. Furthermore, the proposed segmentation method will be applied to an automatic medical diagnosis aid system.
Acknowledgement
This work was supported by the grant No. RTI04-01-01 from the Regional Technology Innovation Program of the Ministry of Commerce, Industry and Energy (MOCIE).
Contributor Information
ByoungChul Ko, Phone: +82-53-5805235, FAX: +82-53-5806275, Email: niceko@kmu.ac.kr.
MiSuk Seo, Phone: +82-53-5805235, FAX: +82-53-5806275, Email: forever1004@kmu.ac.kr.
Jae-Yeal Nam, Phone: +82-53-5805235, FAX: +82-53-5806275, Email: jynam@kmu.ac.kr.
References
- 1.Chio H, Hwang H, Kim M, Kim J: Design of the breast carcinoma cell bank system. Enterprise Networking and Computing in Healthcare Industry 88–91, 2004
- 2.Wong K, Cheung K, Po L. MIRROR: an interactive content based image retrieval system. Proceedings of the IEEE International Conference on Circuits and Systems. 2005;2:1541–1544. [Google Scholar]
- 3.Kuo P, Aoki T, Yasuda H: PARIS: A personal archiving and retrieving image system. Proceedings of the Asia-Pacific Symposium on Information and Telecommunication Technologies 122–125, 2005
- 4.Distasi R, Nappi M, Tucci M, Vitulano S: Context: A technique for image retrieval integrating CONtour and TEXTure Information. Proceedings of the IEEE International Conference on Image Analysis and Processing 224–229, 2001
- 5.Hoiem D, Sukthankar R, Schneiderman H, Huston L. Object-based image retrieval using the statistical structure of images. Proceedings of the IEEE International Conference on Computer Vision and Pattern Recognition. 2004;2:490–497. [Google Scholar]
- 6.Ko B, Byun H. Frip: A region-based image retrieval tool using automatic image segmentation and stepwise boolean and matching. IEEE Trans Multimedia. 2005;7(1):105–113. doi: 10.1109/TMM.2004.840603. [DOI] [Google Scholar]
- 7.Comaniciu D, Meer P, Foran D. Shape-based image indexing and retrieval for diagnostic pathology. Proceedings of the International Conference on Pattern Recognition. 1998;1:902–904. [Google Scholar]
- 8.Ropers S-O, Bell AA, Wurflinger T, Bocking A, Meyer-Ebrecht D. Automatic scene comparison and matching in multimodal cytopathological microscopic images. Proceedings of the IEEE International Conference on Image Processing. 2005;1:1145–1148. [Google Scholar]
- 9.Comaniciu D, Meer P. Mean-shift: a robust approach toward feature space analysis. IEEE Transactions on Pattern Recognition and Machine Intelligence. 2002;24:603–619. doi: 10.1109/34.1000236. [DOI] [Google Scholar]
- 10.Chen X, Zhou X, Wong STC. Automated segmentation, classification, and tracking of cancer cell nuclei in time-lapse microscopy. IEEE Trans Biomed Eng. 2006;53:762–766. doi: 10.1109/TBME.2006.870201. [DOI] [PubMed] [Google Scholar]
- 11.Tscherepanow M, Zollner F, Kummert F. Classification of segmented regions in brightfield microscope images. Proceedings of the IEEE International Conference on Pattern Recognition. 2006;3:972–975. [Google Scholar]
- 12.Itti L, Koch C, Niebur E. A model of saliency-based visual attention for rapid scene analysis. IEEE Trans PAMI. 1998;20:1254–1259. [Google Scholar]
- 13.Barhoumi W, Zagrouba E: Towards a standard approach for medical image segmentation. Proceedings of the ACS/IEEE International Conference on Computer Systems and Applications 130–133, 2005
- 14.Kwak S., Ko B, Byun H. Automatic salient-object extraction using the contrast map and salient points. Proceedings of the Pacific-Rim Conference on Multimedia. 2004;3332:138–145. [Google Scholar]
- 15.Ko B, Nam J. Object-of-interest image segmentation using human attention and semantic region clustering. Journal of Optical Society of America A: Optics, Image Science, and Vision. 2006;23:2462–247. doi: 10.1364/JOSAA.23.002462. [DOI] [PubMed] [Google Scholar]
- 16.Malpica N, Solorzano C, Vaquero JJ, Santos A, Vallcorba I, Sagredo JG, Pozo F. Applying watershed algorithms to the segmentation of clustered nuclei. Cytometry. 1997;28:289–297. doi: 10.1002/(SICI)1097-0320(19970801)28:4<289::AID-CYTO3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 17.Otsu N. A threshold selection method from gray level histograms. IEEE Trans Syst, Man Cybern. 1979;9:62–66. doi: 10.1109/TSMC.1979.4310076. [DOI] [Google Scholar]
- 18.Kim S, Park S, Kim M. Central object extraction for object-based image retrieval. Proceedings of the International Conference on Image and Video Retrieval. 2003;2728:39–49. doi: 10.1007/3-540-45113-7_5. [DOI] [Google Scholar]