Abstract
The aim of this study is to develop a secure, Google-based data-mining tool for radiology reports using free and open source technologies and to explore its use within an academic radiology department. A Health Insurance Portability and Accountability Act (HIPAA)-compliant data repository, search engine and user interface were created to facilitate treatment, operations, and reviews preparatory to research. The Institutional Review Board waived review of the project, and informed consent was not required. Comprising 7.9 GB of disk space, 2.9 million text reports were downloaded from our radiology information system to a fileserver. Extensible markup language (XML) representations of the reports were indexed using Google Desktop Enterprise search engine software. A hypertext markup language (HTML) form allowed users to submit queries to Google Desktop, and Google’s XML response was interpreted by a practical extraction and report language (PERL) script, presenting ranked results in a web browser window. The query, reason for search, results, and documents visited were logged to maintain HIPAA compliance. Indexing averaged approximately 25,000 reports per hour. Keyword search of a common term like “pneumothorax” yielded the first ten most relevant results of 705,550 total results in 1.36 s. Keyword search of a rare term like “hemangioendothelioma” yielded the first ten most relevant results of 167 total results in 0.23 s; retrieval of all 167 results took 0.26 s. Data mining tools for radiology reports will improve the productivity of academic radiologists in clinical, educational, research, and administrative tasks. By leveraging existing knowledge of Google’s interface, radiologists can quickly perform useful searches.
Key words: Google, data mining, reports, HIPAA, search engine
Background
Digital archival of information is one of the greatest outcomes of the computing revolution, allowing an unprecedented storage of and access to information.1 The ability to store and access radiologic data (images and text reports) has profoundly changed the practice of radiology during the last 30 years.2 Picture archiving and communication systems (PACS) have greatly improved our ability to retain and access prior imaging for clinical comparison,3 while radiology information systems (RIS)4 allow rapid access to text reports for clinical, billing, and research purposes. Continuing advances in these radiologic archives5 have played an important role in increasing the clinical productivity required of radiologists as demand for radiological examinations grows.6
As digital archives grow larger, the ability to access information within them becomes more difficult, and their value lies not in the ability to store larger amounts of information but in the ability to provide efficient access to relevant information. Search tools like PubMed for medical literature,7 as well as Entrez and Blast for genomic/proteomic data,8 make these vast repositories a source of discovery in clinical care and research. In a broader sense, online search engines like Google9 have allowed patients10 and physicians11 an efficient search tool for one of the world’s largest data repositories, the World Wide Web.
“Data mining” or “knowledge discovery in databases” has been defined as “the science of extracting useful information from large data sets or databases.”12 Data mining techniques have been applied to all of the activities of academic physicians, including clinical,13–15 educational,16–19 research,20–22 and administrative23 tasks. Though radiologic data (both image and text) account for a large proportion of patients’ electronic medical records within hospital databases, a relatively small set of tools exists to aid radiologists in extracting relevant information from hospital databases. The ability to data mine report information in an academic radiology department has enormous implications for the daily clinical, teaching, research, and administrative activities of radiologists. Our goals were to develop a tool to allow radiologists to directly and efficiently mine data from years of radiology reports while protecting patient privacy and to explore how such a tool can be used in an academic radiology department.
Methods
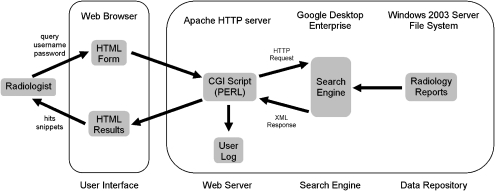
Radsearch, our HIPAA-compliant data repository search engine and user interface (Fig. 1) was created to facilitate treatment, health care operations, and reviews preparatory to research and is not “research” as defined by the United States Department of Health and Human Services.24 As such, the Institutional Review Board (IRB) waived the review of this project, and informed consent was not required.
Fig 1.
Radsearch schematic. A radiologist submits a query via an HTML form, which is interpreted by a PERL CGI script running on the web server. The CGI prepares an HTTP request to the search engine, and the search engine’s XML response is interpreted by the same script. The user information and search terms are logged, and the hits and snippets are returned to the radiologist’s web browser.
Data Repository
A new protected health information (PHI) repository was registered with our institution to contain the text of all radiology reports created at our medical center. The repository consisted of a fileserver (Dell PowerEdge 2950; Dell Computer Corporation, Round Rock, TX, USA) with Intel Xeon 5160 (Core 2 Duo) processors (Intel Corporation, Santa Clara, CA, USA) and 2 GB of random access memory. We installed the Microsoft Windows Server 2003 operating system (Microsoft Corporation, Redmond, WA, USA). Disk storage consisted of two redundant arrays of independent disks (RAID-1) containers. The fileserver was networked to a secure, firewalled intranet connection for accessing electronic PHI, allowing free access from computers within the medical center and secure encrypted access via virtual private networks (VPN) from outside the institution.
We populated this repository with radiology reports in text file format obtained from our IdxRad RIS (General Electric Healthcare, Burlington, VT, USA). Typically, radiology reports for a given 1-month interval were obtained as a single text file (↭50,000 reports comprising ↭100 megabytes of disk space). The document was parsed into individual reports using a practical extraction and report language (PERL) script and interpreter (ActiveState Software Incorporated, Vancouver, BC, Canada). The document was also parsed into extensible markup language (XML) representations with another PERL script (Fig. 2). The PERL interpreter is available free of charge (ActivePerl, version 5.8.8.817, 2006; http://www.activestate.com).
Fig 2.
Text and XML radiology reports. Text radiology reports (a) are converted to XML representations (b) prior to indexing by the search engine. By placing field tags (e.g., “<exam>”) adjacent to terms (e.g., “chest”) within the XML documents, text within different fields can be identified independently. A query of “<exam> chest” would yield documents where the examination was a chest X-ray, whereas a query of “chest” would identify documents where the word chest appeared anywhere in the report (e.g., “chest pain”). To maintain patient confidentiality within this figure, PHI has been anonymized (shaded in gray).
Search Engine
To make the documents searchable, we indexed the XML representation of radiology reports on the fileserver using Google Desktop Enterprise software (Google Corporation, Mountain View, CA, USA), which we used in accordance with their end-user license agreement. We specified that only folders on the fileserver that contain radiology reports should be indexed. We disabled other features of Google Desktop not relevant to the project (e.g., Google Gadgets, sharing across computers, e-mail indexing, web history indexing). The search engine is available free of charge (Google Desktop Enterprise, version 4; http://desktop.google.com/enterprise).
User Interface
To allow users to submit queries to the search engine, we created a web form (Fig. 3), which is served to client machines using the open-source Apache hypertext transfer protocol (HTTP) server (The Apache Software Foundation, Forest Hill, MD, USA). The form contains the following fields, which are submitted to the webserver: user name, password, query, reason for search, and display mode. To submit each query, users must select one or more of the approved reasons for searching: clinical care, education/training, management, quality assurance, administration, and reviews preparatory to research. A disclaimer on the Radsearch homepage states the following: “The use of this tool for research without prior IRB approval, waiver, or exemption is forbidden.”
Fig 3.
Radsearch user interface. a Search form. To perform a search, a radiologist must fill in the search terms, username, password, and reason for search. Users can specify which results to show (e.g., patient records, contact info, presentations) as well as output format (snippets or list). b Results display. The number of results, duration of search, links to matching radiology reports, and snippets are displayed for each search. Additional links allow for highlighting, anonymization, and display of XML documents. To maintain patient confidentiality within this figure, PHI has been anonymized (shaded in gray).
After submitting the form, a common gateway interface (CGI) script written in PERL authenticates the username and password, interprets the query, and submits an HTTP request to Google Desktop. The same CGI receives the XML response from Google Desktop and formats the output into hypertext markup language (HTML). The output includes a list of “hits,” which are hyperlinks to radiology reports matching the search query, ordered by relevancy. Depending on the output format, each hit can feature a “snippet,” which is a short sample of text from the related document, as well as links that display the original XML, highlight search terms, and anonymize PHI of the document. The webserver can be obtained free of charge (Apache HTTP server, version 2.2.3; http://httpd.apache.org).
User Log
As part of HIPAA compliance for access to a PHI repository, we created an audit trail that tracks all user interactions with Radsearch, including the date, time, search terms, and reason for search. Because the snippets that are returned after a search may contain PHI, we log the name and birthdate of the patients from the reports where the snippets originated. If a radiologist follows a link to a document, the date and time of the access, as well as the patient’s name and birthdate are logged.
Results
Repository
The Radsearch repository currently contains 2,944,740 associated reports dating from January 1, 2001 to October 31, 2006. The total size of the files is 7.9 GB, which occupies 12.9 GB on the fileserver. Initial indexing of the data set took 90 h. Indexing speed peaks early at over 100,000 records per hour, slowing to an average speed of about 25,000 records per hour. Approximately 50,000 new records are added each month, and monthly indexing of new records takes about 1 h.
Usage
We created 171 accounts for every attending radiologist, fellow, and resident in our department. In the first 2 months of operation, 84 members of the department used the system: 43 residents (51%), 16 fellows (19%), and 25 attending radiologists (30%). Radsearch usage was highest among residents, with 43 of 71 residents (60%) using Radsearch, followed by fellows (16 of 33, 48%) and attending radiologists (25 of 67, 37%).
In the first 2 months of operation, 4,224 queries were performed (Table 1). A total of 3,146 queries (88%) were performed for education/training, followed by quality assurance (230 queries, 6%), reviews preparatory to research (108 queries, 3%), clinical care (81 queries, 2%), administration (26 queries, <1%), and management (1 query, <1%). To describe the variety of user queries performed, we list 20 actual searches performed using Radsearch (Table 2). A total of 2,674 searches (64%) were performed during the clinical workday (7 a.m.–5 p.m.), with the remaining 1,550 (36%) performed after hours. Four hundred thirty-eight queries (10%) were performed during weekends.
Table 1.
Radsearch Statistics: First 2 Months of Operation
| Parameter | No. of Searches |
|---|---|
| Reason for search | |
| Education/training | 3,146 (88) |
| Quality assurance | 230 (6) |
| Review preparatory to research | 108 (3) |
| Clinical care | 81 (2) |
| Administration | 26 (<1) |
| Management | 1 (<1) |
| Time search was performed | |
| Clinical workday (7 am-5 pm) | 2,674 (64) |
| After hours (5 pm-7 am) | 1,550 (36) |
| Search results | |
| Searches resulting in 0 hits | 632 (15) |
| Searches resulting in 1 or more hits | 3,592 (85) |
| 1–9 | 608 (16) |
| 10–99 | 1,078 (30) |
| 100–999 | 938 (26) |
| 1,000–9,999 | 462 (13) |
| >10,000 | 506 (14) |
Data in parentheses are percentages.
Table 2.
Radsearch: Selected Queries Performed by Radiologists
| Search terms |
|---|
| Education/training |
| Esophageal cancer barium esophagram |
| Hysterosalpingogram bicornuate |
| “<exam> MR” “ventricular septal defect” |
| “Normal MRA of the neck” |
| “Tetralogy of fallot” CT |
| Clinical |
| Flexor carpi ulnaris ultrasound |
| Chest wall desmoid |
| Hyperostosis frontalis interna CT 2006 |
| Aortogram injury transaction |
| Elastofibroma |
| Reviews preparatory to research |
| Endometriosis MRI “<year> 2006” |
| “Anomalous coronary arteries” |
| Pulmonary hypertension CT |
| Hypertensive encephalopathy MRI hemorrhage |
| Pseudotumor of liver |
| Quality assurance/administration/management |
| “Dr Smith”* arterial stent |
| Rheumatoid shoulder ultrasound |
| “Dr Jones”* complication |
| Functional brain MR |
| “Dr Brown”* shoulder MR 2006 |
To maintain patient confidentiality within this table, PHI has been anonymized (noted with asterisks).
Queries
An average of 76 ± 64 (SD) queries was performed each day (range, 1–290). A total of 3,592 (85%) yielded one or more results. Six hundred thirty-two (15%) failed to yield any results, usually due to improper search syntax or misspelling of search terms. Of the 3,592 positive search results, 608 (16%) yielded one to nine results, 1,078 (30%) yielded ten to 99 results, 938 (26%) yielded 100–999 results, 462 (13%) yielded 1,000–9,999 results, and 506 (14%) yielded greater than 10,000 results.
The average query took 1.56 ± 1.4 s to complete. The number of hits did not correlate with the time of the query. Rather, the number of hits was more dependent on the number of terms searched and on the “uniqueness” of the terms within the dataset. For example, searching for a rare term like “hemangioendothelioma” took 0.23 s to retrieve the ten most relevant results of 167 total results; to retrieve all 167 results took 0.26 s. Searching for a common term like “pneumothorax” took 1.36 s to retrieve the ten most relevant results of 705,550 total results; to retrieve the first 1,000 results took 1.7 s. A search including two common terms such as “pneumothorax AND tumor” took 1.14 s to retrieve the ten most relevant results of 4,907 total results; to retrieve the first 1,000 results took 1.52 s. A search including two rare terms such as “epithelial AND hemangioendothelioma” took 0.23 s to retrieve the ten most relevant results of 14 total results; to retrieve all of the results took 0.23 s.
Discussion
We have described methods for developing a secure, HIPAA-compliant data mining tool for radiology reports based on the Google search engine. We employed the Google Desktop application programmers interface (API), which allows utilization of Google’s core search technologies, while retaining the ability to customize the application for use in radiology. By employing Google search algorithms and a Google-like interface,9 we offer radiologists the ability to quickly leverage their existing knowledge of Google’s interface, query protocol, and relevancy ranking system to quickly perform useful searches.
Whereas a typical search engine protects the user’s privacy, Radsearch is designed to protect the patient’s privacy. HIPAA allows disclosure of a patient’s PHI for several specific purposes without prior specific written authorization including treatment, payment, and health care operations purposes.25 HIPAA’s definition of health care operations includes education/training, quality assurance, management, and administration.26 Thus, although PHI is visible to Radsearch users, when usage is limited to the above HIPAA authorized purposes, IRB approval is not required before each search. To prevent inappropriate searching, each event where a radiologist encounters PHI is logged to create an audit trail to maintain HIPAA compliance. This surveillance and logging method rather than access restriction is the same method used in our institution’s hospital clinical information system to protect PHI from being used for research by physicians who are authorized to use the very same PHI for treatment, payment, or operations. Radsearch logs every user interaction, including those after the display of links (such as opening an individual radiology report), an uncommon practice when compared to most search engines. Logs are reviewed periodically in accordance with HIPAA to identify patterns of inappropriate usage.
In contrast, for research purposes, data mining tools such as Radsearch cannot be used on a PHI repository without IRB approval, waiver, or exemption.27 HIPAA defines research as “a systematic investigation, including research development, testing, and evaluation, designed to develop or contribute to generalizable knowledge.”24 Thus, creation of a PHI repository, a search engine for that repository, and a user interface for that search engine is not, in and of itself, an act of research. To insure that our data mining tool and repository is not used for research, each user must stipulate that their interaction with the repository fits within HIPAA-allowed uses pertaining to treatment, payment, or health care operations (e.g., education/training, quality assurance, management, administration). Although HIPAA prohibits “research” (as defined above) conducted using a PHI repository without prior authorization, HIPAA allows the use of PHI without prior authorization for “reviews preparatory to research.”28 Reviews preparatory to research have a very narrow definition, described under HIPAA in two instances: (a) the use or disclosure of PHI solely to prepare a research protocol or (b) the use or disclosure of PHI solely to identify prospective research participants for purposes of seeking an authorization.
At this time, we have not allowed Radsearch to be used for research purposes, even for investigators who have prior IRB approval, because we currently do not have a mechanism to restrict access only to the subset of PHI within the repository for which the investigator has prior authorization. We are currently investigating various procedures to allow for the use of Radsearch for research purposes. One proposed methodology would create a human “record gatekeeper” who would serve as a liaison to the researcher who has received IRB authorization to specific PHI. The liaison could conduct research searches on the researcher’s behalf. The researcher would only be provided with the PHI for which prior authorization had been obtained. This additional safeguard would provide a check on unauthorized searches, while giving the researcher the ability to conduct searches that are within the scope of their IRB approval. Another methodology that we are investigating to facilitate data mining research is “real-time anonymization.” Rather than creating a separate, anonymized repository with no link to the original PHI, which can typically be used for research without prior authorization,29 we are exploring secure anonymization that occurs at the time of search. In this way, a researcher might be able to search for and view anonymized records to be used for research from the same data repository that contains unanonymized records that a clinician uses for treatment, payment, or operations.
Usage of Radsearch has been particularly strong among trainees; in fact, 70% of users are residents or fellows, and 88% of the searches have been reported as performed for “education/training.” Inspection of the usage logs of trainees suggests several patterns of use. First year trainees often perform queries to identify a normal model report to use as a basis for dictation (e.g., “normal computed tomography of the chest”). Trainees also search for studies previously dictated by an attending radiologist to assist in generating a report that includes all of the elements and style that the attending radiologist prefers (e.g., “interstitial lung disease Dr Smith”). In unusual cases of entities with which the trainee may be unfamiliar, the trainee can search for a radiology report of a different patient with similar indications and/or findings (e.g., “carcinoid CT Abdomen 2006”), rather than searching for similar cases on the internet.11 In contrast to trainees, the majority of faculty has used Radsearch for reviews preparatory to research (e.g., “hypertensive encephalopathy MRI”) and for quality assurance (e.g., “Dr Jones CT guided biopsy”), which may suggest facilitation of their academic and administrative activities. Despite minimal instructions, we have witnessed rapid adoption of Radsearch within the clinical workflow in our department, with 64% of searches performed during the clinical workday. Although we did not formally survey users to ascertain how effectively Radsearch performed in delivering relevant reports, informal discussions with users were overwhelmingly positive, both in the effectiveness of the search engine, as well as its utility in helping radiologists pursue their various academic missions.
In previous years at our institution, searches of the RIS for academic purposes were performed by members of our Information Systems group. These searches required a custom-designed SQL database query of the RIS by a database programmer. The output listed matching patient names, birthdates, and complete reports in chronologic order, which then required the radiologist to inspect each matched record to determine its relevance. It would typically take several weeks to a month to obtain results from the programmer for a single query. The current Radsearch system offers several key advantages over the previous one. First, time is saved, as searches conducted via Radsearch take one to two orders of magnitude less time to perform than SQL queries on the RIS. Second, immediate feedback from search results allows for rapid refinement of search terms. In addition, snippets of the relevant text allow rapid identification of relevant records.
Google Desktop uses a complex, non-relational approach to index text within documents. This provides great flexibility when indexing text radiology reports that have a variety of formats and structures.30 However, since the resulting index is not relational, the ability to identify particular fields within a text report is lost. Thus, in a strictly non-relational search, searching for “Dr Brown AND December” would result in hits where “Dr Brown” could be the referring physician, radiologist, or a physician to whom results were reported. Likewise, searching for “December” might yield examinations performed in December, or examinations where comparison studies were performed in December. To overcome this limitation, we indexed XML representations of the reports (Fig. 2) instead of the actual report, resulting in indexing of a text word and its adjacent field tag (e.g., “<month> December” or “<radiologist> Dr Brown”) rather than of indexing the text word alone (e.g., “December” or “Dr Brown”). By indexing the field tags, as well as the text, a user can (1) search for the term “Dr Brown” and find XML documents where the term is found in any field or (2) search for the term “<radiologist> Dr Brown” and find only XML documents where Dr Brown is the radiologist. In this way, despite using non-relational text indexing, we retain the ability to search for specific fields within the radiology report.31 Therefore, we achieve the benefits of a fast, non-relational free text search of the report while maintaining the ability to identify keywords along with their related structured field identity.32
Google does not provide documentation on the exact algorithm that Google Desktop uses in ranking a page retrieved by a search. However, by repeated searching of combinations of terms, it can be inferred that the Google Desktop algorithm considers the number of times a search term is found in the document, as well as the proximity of the search terms to each other in the document. Unlike a typical webpage, each radiology report in the repository contains only text, with no links to other documents. Thus, Google’s PageRank algorithm for webpages, which also considers the number of links to a document as part of its ranking, is not applied. Future radiology search engines might apply a Google PageRank approach. For example, a document in the repository might contain the text of the report, as well as link to other reports of the same patient and/or links to reports generated during the patients particular hospital visit. By applying the PageRank approach, when a set of terms was searched, patients who have undergone multiple examinations would have a higher ranking. This method would be beneficial if a search were performed to generate a teaching file, where patients with the same finding on multiple examinations within a hospital visit is desired.
Our Google-based data mining technique has several limitations. Our application is a text word search, and only exact matches are identified as hits. Although phrase, Boolean “AND”, and negation searches are available, searches aided by wildcards, stemming, or spelling suggestions have not been implemented in the current version of Google Desktop. Synonyms and subsumption, concepts central to natural language processing (NLP), are also absent, requiring the user to be precise regarding the terms over which searches are conducted. For example, although “CT,” “CAT scan,” and “computed tomography” are synonyms and refer to the same idea, a search for one of these terms will not yield documents that contain the other terms. Ongoing research and implementation of natural language processing algorithms will aid search applications in this difficulty by mapping keywords to ontologies, allowing the search of ideas represented by search terms within a document rather than the search terms themselves.33 In addition, the current Google Desktop API does not provide all of the features found in the Google or other web search engines. Notably, neither the “OR” Boolean operator nor grouping operators (e.g., parentheses) currently exist. Google Desktop’s “date range” function cannot be applied to our repository, as Google stores a document’s index date rather than its creation date. By using XML tags for “month” and “year,” we currently implement limited date functionality. Based on previous updates of Google Desktop, it is likely that future versions of the software will add features that already exist in the Google web search. That notwithstanding, we are beginning to explore a more general approach to overcoming many of these challenges by replacing Radsearch’s Google-based search engine with a custom search application built with Lucene, a free, open source, full-featured search engine class library written in Java (Apache Lucene, version 2.0.0; http://lucene.apache.org).
Conclusion
We have described a method for creating a HIPAA-compliant, Google-based data mining tool for radiology reports using free and open-source technologies. Following the introduction of Radsearch, we have witnessed a change in the approach to clinical, educational, research, and administrative problem solving that occurs during the daily activities of members of the department. We anticipate that the use of radiology report data mining tools like Radsearch will become an integral part of the daily workflow of academic radiologists.
Contributor Information
Joseph P. Erinjeri, Phone: +1-212-7462600, FAX: +1-212-7468463, Email: erinjeri@gmail.com
Daniel Picus, Phone: +1-314-3622978, FAX: +1-314-7474671.
Fred W. Prior, Phone: +1-314-3622978, FAX: +1-314-7474671
David A. Rubin, Phone: +1-314-3622978, FAX: +1-314-7474671
Paul Koppel, Phone: +1-314-3622978, FAX: +1-314-7474671.
References
- 1.Iwata S, Chen RS. Science and the digital divide. Science. 2005;310:405. doi: 10.1126/science.1119500. [DOI] [PubMed] [Google Scholar]
- 2.Thrall JH. Reinventing radiology in the digital age: part I. The all-digital department. Radiology. 2005;236:382–385. doi: 10.1148/radiol.2362050257. [DOI] [PubMed] [Google Scholar]
- 3.Hynes DM, Stevenson G, Nahmias C. Towards filmless and distance radiology. Lancet. 1997;350:657–660. doi: 10.1016/S0140-6736(97)08157-9. [DOI] [PubMed] [Google Scholar]
- 4.Tamm EP, Kawashima A, Silverman P. An academic radiology information system (RIS): a review of the commercial RIS systems, and how an individualized academic RIS can be created and utilized. J Digit Imaging. 2001;14:131–134. doi: 10.1007/s10278-001-0012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thrall JH. Reinventing radiology in the digital age. Part II. New directions and new stakeholder value. Radiology. 2005;237:15–18. doi: 10.1148/radiol.2371050258. [DOI] [PubMed] [Google Scholar]
- 6.Meghea CI, Sunshine JH. Who’s overworked and who’s underworked among radiologists? An update on the radiologist shortage. Radiology. 2005;236:932–938. doi: 10.1148/radiol.2363041885. [DOI] [PubMed] [Google Scholar]
- 7.Steinbrook R. Searching for the right search—reaching the medical literature. N Engl J Med. 2006;354:4–7. doi: 10.1056/NEJMp058128. [DOI] [PubMed] [Google Scholar]
- 8.Birney E, Bateman A, Clamp ME, Hubbard TJ. Mining the draft human genome. Nature. 2001;409:827–828. doi: 10.1038/35057004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giustini D. How Google is changing medicine. BMJ. 2005;331:1487–1488. doi: 10.1136/bmj.331.7531.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Connor JB, Johanson JF. Use of the Web for medical information by a gastroenterology clinic population. JAMA. 2000;284:1962–1964. doi: 10.1001/jama.284.15.1962. [DOI] [PubMed] [Google Scholar]
- 11.Greenwald R. And a diagnostic test was performed. N Engl J Med. 2005;353:2089–2090. doi: 10.1056/NEJM200511103531923. [DOI] [PubMed] [Google Scholar]
- 12.Hand DJ, Mannila P, Smyth P. Principle of Data Mining. Cambridge, MA: MIT; 2001. [Google Scholar]
- 13.Mullins IM, Siadaty MS, Lyman J, et al. Data mining and clinical data repositories: insights from a 667,000 patient data set. Comput Biol Med. 2006;36:1351–1377. doi: 10.1016/j.compbiomed.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Nigrin DJ, Kohane IS. Data mining by clinicians. Proc AMIA Symp. 1998;1998:957–961. [PMC free article] [PubMed] [Google Scholar]
- 15.Prather JC, Lobach DF, Goodwin LK, Hales JW, Hage ML, Hammond WE. Medical data mining: knowledge discovery in a clinical data warehouse. Proc AMIA Annu Fall Symp. 1997;1997:101–105. [PMC free article] [PubMed] [Google Scholar]
- 16.Ananiadou S, Kell DB, Tsujii JI. Text mining and its potential applications in systems biology. Trends Biotechnol. 2006;24:571–579. doi: 10.1016/j.tibtech.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Cohen AM, Hersh WR. A survey of current work in biomedical text mining. Brief Bioinform. 2005;6:57–71. doi: 10.1093/bib/6.1.57. [DOI] [PubMed] [Google Scholar]
- 18.Heinze DT, Morsch ML, Holbrook J. Mining free-text medical records. Proc AMIA Symp. 2001;2001:254–258. [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts PM. Mining literature for systems biology. Brief Bioinform. 2006;7:399–406. doi: 10.1093/bib/bbl037. [DOI] [PubMed] [Google Scholar]
- 20.Bekhuis T. Conceptual biology, hypothesis discovery, and text mining: Swanson’s legacy. Biomed Digit Libr. 2006;3:2. doi: 10.1186/1742-5581-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scherf M, Epple A, Werner T. The next generation of literature analysis: integration of genomic analysis into text mining. Brief Bioinform. 2005;6:287–297. doi: 10.1093/bib/6.3.287. [DOI] [PubMed] [Google Scholar]
- 22.Schonbach C, Nagashima T, Konagaya A. Textmining in support of knowledge discovery for vaccine development. Methods. 2004;34:488–495. doi: 10.1016/j.ymeth.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Sokol L, Garcia B, Rodriguez J, West M, Johnson K. Using data mining to find fraud in HCFA health care claims. Top Health Inf Manage. 2001;22:1–13. [PubMed] [Google Scholar]
- 24.Definitions: research. Title 45 Code of Federal Regulation, Pt. 46.102(d), 2000
- 25.Use and Disclosure for Treatment, Payment and Health Care Operations. Title 45 Code of Federal Regulation, Pt. 164.506, 2000
- 26.Definition: health care operations. Title 45 Code of Federal Regulation, Pt. 164.501(2), 2000
- 27.IRB review of research. Title 45 Code of Federal Regulation, Pt. 46.109, 2000
- 28.Reviews Preparatory to Research. Title 45 Code of Federal Regulation, Pt. 164.512(h)(i)(1)(ii), 2000
- 29.De-identification of protected health information. Title 45 Code of Federal Regulation, Pt. 164.514(a), 2000
- 30.Magos A, Gambadauro P. Desktop search engines: a modern way to hand search in full text. Lancet. 2005;366:203–204. doi: 10.1016/S0140-6736(05)66906-1. [DOI] [PubMed] [Google Scholar]
- 31.Smith AC. Effect of XML markup on retrieval of clinical documents. AMIA Annu Symp Proc. 2003;2003:614–618. [PMC free article] [PubMed] [Google Scholar]
- 32.Hulse NC, Rocha RA, Bradshaw R, Del Fiol G, Roemer L. Application of an XML-based document framework to knowledge content authoring and clinical information system development. AMIA Annu Symp Proc. 2003;2003:870. [PMC free article] [PubMed] [Google Scholar]
- 33.Hripcsak G, Austin JH, Alderson PO, Friedman C. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology. 2002;224:157–163. doi: 10.1148/radiol.2241011118. [DOI] [PubMed] [Google Scholar]