Abstract
Diabetic cardiomyopathy is associated with increased risk of heart failure in type 1 diabetic patients. Mitochondrial dysfunction is suggested as an underlying contributor to diabetic cardiomyopathy. Cardiac mitochondria are characterized by subcellular spatial locale, including mitochondria located beneath the sarcolemma, subsarcolemmal mitochondria (SSM), and mitochondria situated between the myofibrils, interfibrillar mitochondria (IFM). The goal of this study was to determine whether type 1 diabetic insult in the heart influences proteomic make-up of spatially distinct mitochondrial subpopulations and to evaluate the role of nuclear encoded mitochondrial protein import. Utilizing multiple proteomic approaches (iTRAQ and two-dimensional-differential in-gel electrophoresis), IFM proteomic make-up was impacted by type 1 diabetes mellitus to a greater extent than SSM, as evidenced by decreased abundance of fatty acid oxidation and electron transport chain proteins. Mitochondrial phosphate carrier and adenine nucleotide translocator, as well as inner membrane translocases, were decreased in the diabetic IFM (P < 0.05 for both). Mitofilin, a protein involved in cristae morphology, was diminished in the diabetic IFM (P < 0.05). Posttranslational modifications, including oxidations and deamidations, were most prevalent in the diabetic IFM. Mitochondrial heat shock protein 70 (mtHsp70) was significantly decreased in diabetic IFM (P < 0.05). Mitochondrial protein import was decreased in the diabetic IFM with no change in the diabetic SSM (P < 0.05). Taken together, these results indicate that mitochondrial proteomic alterations in the type 1 diabetic heart are more pronounced in the IFM. Further, proteomic alterations are associated with nuclear encoded mitochondrial protein import dysfunction and loss of an essential mitochondrial protein import constituent, mtHsp70, implicating this process in the pathogenesis of the diabetic heart.
Keywords: diabetes, mitochondria
cardiovascular complications, including diabetic cardiomyopathy, are the leading cause of mortality among type 1 diabetic patients. A number of studies have indicated that mitochondrial dysfunction underlies and contributes centrally to the dysfunction seen in the type 1 diabetic heart (11, 13, 35, 38). Increasing evidence suggests that mitochondrial subcellular location influences the pathogenesis of dysfunctional mitochondria in the type 1 diabetic heart (11, 24). Cardiac mitochondria are characterized by different spatial location within the cell, including mitochondria located at the sarcolemma, subsarcolemmal mitochondria (SSM), and mitochondria situated between the myofibrils, interfibrillar mitochondria (IFM). We and others have shown that these two spatially distinct subpopulations of mitochondria are differentially affected with various pathological insults (11, 24, 26, 27, 32, 36, 42). Specifically, we have demonstrated that the IFM display greater dysfunctional profiles with type 1 diabetic insult, as evidenced by enhanced oxidative stress, diminished electron transport chain (ETC) function, and decreased cardiolipin content (11).
Proteomic evaluations have played an important role in furthering our understanding of mitochondrial dysfunction in the diabetic heart. Several proteomic studies have been undertaken in various diabetic models in an effort to clarify the nature of the proteomic changes associated with the diabetic heart (5, 15, 21, 38, 40). Turko and Murad (40) identified 30 altered mitochondrial proteins in isolated mitochondria from streptozotocin (STZ)-induced type 1 diabetic rat hearts, which included enhanced fatty acid oxidation (FAO) proteins and reduction of oxidative phosphorylation (OXPHOS) protein subunits. Utilizing an iTRAQ labeling method, Jüllig et al. (21) identified 65 proteins significantly changing in the STZ-induced type 1 diabetic rat heart compared with control, with the most significant changes to FAO enzymes and OXPHOS proteins. In a recent study using label-free proteome expression analyses, Bugger et al. (5) examined mitochondrial proteomes of several tissues from the Akita mouse (kidney, liver, brain, and heart). Their results indicate that FAO proteins were less abundant in liver mitochondria, whereas FAO protein content was induced in mitochondria from all other tissues. In addition, levels of OXPHOS subunits were coordinately increased in liver mitochondria, whereas mitochondria from other tissues were unaffected (5). Taken together, these data suggest that the cardiac mitochondrial proteome is impacted during type 1 diabetes mellitus, though a number of variables may contribute to conflicting results. To date, no one has examined proteomic differences in spatially distinct subpopulations of mitochondria in the type 1 diabetic heart, which may offer further insight into the nature of this dynamic process in the type 1 diabetic heart.
Studies examining mechanisms underlying or contributing to the proteomic alterations in the diabetic heart are limited (16). Potential mechanisms of proteomic dysregulation may include pathological alterations in gene expression, increases in posttranslational modifications (PTMs) of proteins, or upregulation of posttranslational regulators such as microRNAs (miRNAs). Alternatively, dysfunctional nuclear encoded mitochondrial protein import could also disturb the proteomic composition of the mitochondrion. Currently, there are ∼1,200 known proteins in the human mitochondrion (37) and an estimated 1,500 proteins (6). Of this number, only 13 proteins are transcribed and translated inside the organelle itself. As a result, the vast majority of proteins (∼99%) must be imported into the mitochondrion through a complex mechanism of translocation involving interaction between protein targeting signals and mitochondrial translocases (7). To date, no study has examined the impact of type 1 diabetes mellitus on spatially distinct mitochondrial proteomes in the diabetic heart or offered insight into the mechanisms influencing specific proteomic profiles. The goal of this study was to determine whether proteomic differences exist in subpopulations of mitochondria, as well as to evaluate the role of nuclear encoded mitochondrial protein import in the type 1 diabetic heart.
MATERIALS AND METHODS
Experimental Animals and Diabetes Induction
The animal experiments in this study conformed to National Institutes of Health (NIH) Guidelines for the Care and Use of Laboratory Animals and were approved by the West Virginia University Animal Care and Use Committee. Male FVB mice (Charles River Laboratories, Wilmington, MA) were housed in the West Virginia University Health Sciences Center animal facility. Mice were given unlimited access to a rodent diet and water. Type 1 diabetes mellitus was induced in 8-wk-old mice following the protocol of the Animal Models of Diabetic Complications Consortium using multiple low-dose STZ (Sigma, St. Louis, MO) injections. Injections of 50 mg/kg body wt STZ dissolved in sodium citrate buffer (pH 4.5) were performed daily for five consecutive days after 6 h of fasting. Mice that served as vehicle controls were given the same volume per body weight of sodium citrate buffer. One week postinjection, hyperglycemia was confirmed by measuring urinary glucose levels (Chemstrip 2GP Urine test strips; Roche Diagnostics, Indianapolis, IN). Five weeks after hyperglycemia onset, animals were killed for further experimentation. To characterize the diabetic phenotype, fasting blood glucose (Bayer, Mishawaka, IN) and plasma insulin (ALPCO, Salem, NH) levels were determined using commercially available kits.
Preparation of Individual Mitochondrial Subpopulations
At 5 wk posthyperglycemia onset, FVB mice and their littermate controls were killed, and the hearts were excised. Hearts were rinsed in PBS (pH 7.4), then blotted dry. SSM and IFM were isolated as previously described following the methods of Palmer et al. (31) with minor modifications (11, 12, 42). Protein concentrations were determined using the Bradford method with BSA as a standard (3).
iTRAQ Labeling
Pooled SSM and IFM subpopulations (n = 4) from diabetic and control hearts were lysed and precipitated overnight in acetone at −20°C, and pellets were resuspended in 20 μl of 0.5 M triethylammonium bicarbonate (TEAB; pH 8.5), as previously described (10). Protein contents were determined using a two-dimensional Quant Kit (Amersham, Piscataway, NJ), and 100 μg of each pooled sample was than denatured with 0.1% SDS and reduced with 5 mM tris-(2-carboxyethyl) phosphine. After incubation for 1 h at 60°C, cysteines were blocked with 10 mM methyl methane thiosulfonate (MMTS) in isopropanol, and the samples were incubated at room temperature for 10 min. The addition of 10 μl of sequencing-grade trypsin (Applied Biosystems, Foster City, CA) was added in a trypsin/protein ratio of 1:20, and the samples were incubated at 37°C overnight. Digested samples were labeled with the iTRAQ reagents following the protocol provided by the vendor (Applied Biosystems).
After digestion and iTRAQ labeling, the ultra-complex protein digests were combined to create a 400-μg pooled protein digest sample that contained equal fractions of each of the four labeled samples for subsequent multidimensional protein identification technology (MudPIT) analysis (28). After lyophilization, the digest mixture was reconstituted in strong cation exchange (SCX) loading buffer (5 mM ammonium formate in 20% acetonitrile; pH 3.0) to be fractionated with SCX SpinTips (Protea Biosciences, Morgantown, WV) per the manufacturer's protocol. Briefly, the sample solution was loaded centrifugally onto the SCX SpinTip. The nonadsorbing solution that passed through the SCX SpinTip was collected. Eight different elution solutions were used to fractionate the peptides (20, 60, 100, 150, 200, 250, 400, and 500 mM ammonium formate in 10% acetonitrile) in a step-wise manner, for a total of nine sample fractions. The collected fractions were cleaned by repeated lyophilization and reconstituted in a 0.1 M acetic acid solution, and then lyophilized to dryness. The fractions were then submitted for LC-MALDI TOF/TOF mass spectral analysis for protein identification, characterization, and differential expression analysis.
MS Analyses with iTRAQ Labeling
The LC-MALDI mass spectrometry system utilized was an ABI Tempo LC MALDI spotter with Tempo LC MALDI ver. 2.00.09 data acquisition and processing software. Lyophilized SCX sample fractions were reconstituted in LC aqueous run buffer (0.1% trifluoroacetic acid, 2% acetonitrile) and injected onto a Zorbax C18 chromatographic column, 150 × 0.3 mm (Agilent Technologies, Wilmington, DE). The peptides were eluted from the column using an acetonitrile/trifluoroacetic acid gradient (2–72% acetonitrile in 35 min) and spotted directly onto a MALDI plate in 6-s spot fractions. The MALDI spots were analyzed using an ABI 4800 MALDI TOF/TOF analyzer operated with 4000 Series Explorer software. The MS acquisition was in positive ion reflector mode with 400 laser shots per spectrum performed. The 15 strongest precursors per spot were chosen for MS/MS, and the MALDI spot was interrogated until at least 4 peaks in the MS/MS, spectra achieved a S/N ≥ 70.
The resulting MS/MS spectra were analyzed using ABI Protein ProteinPilot software 2.0 (Applied Biosystems). The spectral data were searched against the mouse protein database (NCBI nr.fasta database customized to select for all mouse proteins) for identification of the peptides and corresponding proteins. In ProteinPilot, the sample type was selected as iTRAQ 4Plex for retrieval of the isotopic tag information from the mass spectra. After database correlation analysis, the proteins were grouped, scored, and normalized against one of four isotope correction factors. The Pro Group algorithm of ProteinPilot generated a protein score (ProtScore) that is a cumulative score from each of the peptides used by the algorithm in the protein identification. ProtScores above 2.0, 1.0, and 0.47 expressed the percent confidence levels of >99, >90, and >66%, respectively. Each peptide match showed the iTRAQ isotopic labels, MMTS-labeled cysteines, and other PTMs present as mass spectral shifts identified during the database correlation analysis. Each protein identified also showed the differential protein expression compared against the other iTRAQ-labeled samples for relative quantitation.
Two-Dimensional-Differential In-Gel Electrophoresis
Sample preparation.
One-hundred micrograms of isolated mitochondrial subpopulations from control and diabetic hearts were placed in lysis buffer (7 M urea, 2 M thiourea, 30 mM Tris, 5 mM magnesium acetate, 4% CHAPS, and 58 mM DTT). Protein concentration of the samples was determined using a two-dimensional (2D) quant kit (Amersham Biosciences, Piscataway, NJ).
2D-Differential In-Gel Electrophoresis and Differential-Display Proteome Analyses
One-third of each lysed sample was removed and combined into a separate tube to serve as an internal standard. Fifty micrograms of the remaining samples were labeled separately with 400 pmol of Cy3 or Cy5 NHS-ester minimal labeling reagents for 30 min on ice and in the dark. The samples were then quenched with 2 μl of 10 mM lysine for 10 min on ice in the dark. Fifty micrograms of the combined sample was also labeled with 400 pmol of Cy2 and than quenched with 10 μl of 10 mM lysine. Individual Cy3- and Cy5-labeled samples were combined with the same amount of the Cy2-labeled internal standard. Each gel contained one control and one diabetic sample from individual mitochondrial subpopulations, and the Cy3 and Cy5 dyes were alternated to account for dye-labeling variability. The samples were separated by standard 2D gel electrophoresis utilizing a manifold-equipped IPGphor first-dimension isoelectric focusing unit and 24 cm 3–10 immobilized pH gradient strips. The initial separation was followed by a second-dimension 12% SDS-PAGE homogenous on hand-cast gels. The Cy2 (mixed standard), Cy3, and Cy5 components of each gel were visualized separately at 100-μm resolution with mutually exclusive excitation/emission wavelengths using a Typhoon 9400 Variable Mode Fluorescence Imager (GE Healthcare, Piscataway, NJ). A Sypro Ruby protein poststain (Invitrogen, Carlsbad, CA) was used to extract protein from the gels. Gel images were submitted to Ludesi 2D Analysis (Ludesi, Malmo, Sweden; http://www.ludesi.com) for spot detection, matching, and analysis. Control:standard and diabetic:standard normalized volume ratios were calculated for each protein on every gel, and the internal standards were used to normalize and compare these ratios across the 10 2D-DIGE gels. This method allowed for the calculation of average abundance changes and Student's t-test P values for the variance of these ratios for each protein-pair across all 10 individual gels.
Mass Spectrometry and Database Analysis
Proteins that were changing were excised and digested in gel with trypsin protease and the resulting peptides analyzed using a Micromass MALDI micro MX TOF spectrometer (Waters, Milford, MA). The resulting peptide mass maps were compared with sequences present in the SWISS-PROT and NCBInr databases to generate statistically significant identifications of proteins using GPS Explorer software (Applied Biosystems) running the MASCOT search algorithm. Searches were performed without constraining protein molecular weight or isoelectric point, with complete carbamidomethylation of cysteine, partial oxidation of methionine residues, and one missed cleavage, also allowed in the search parameters. Molecular weight search scores from database interrogation above 58 (P < 0.05), number of matched ions, number of matching sequence coverage, and correlation of gel region with predicted MW and pI were considered for each protein identification.
Protein Import
Plasmid construction.
The fusion protein construct pAcGFP1-Mito (Clontech Laboratories, Mountain View, CA) containing the precursor subunit VIII of human cytochrome-c oxidase and the green fluorescent protein from Aequorea coerulescens (AcGFP1) was cloned into pIVEX2.3d (Roche Applied Science, Indianapolis, IN) at restriction sites Not I and blunted Nco I/Nhe I creating pMITOGFP1 (Supplemental Fig. S1). The correct sequence was confirmed by dideoxynucleotide sequencing. pMITOGFP1 was grown and isolated using mini-prep plasmid DNA isolation to a concentration of 500 ng/μl (Qiagen, Valencia, CA).
In Vitro Synthesis of Mitochondrial Protein
In vitro transcription and translation of MitoGFP1 was performed using the S30 T7 protein expression system (Promega, Madison, WI), as per the manufacturer's protocol. Additionally, fluorescent labeling of MitoGFP1 was performed using the FluoroTect GreenLys tRNA in vitro labeling system (Promega, Madison, WI), as per the manufacturer's protocol. MitoGFP1 and fluorescent MitoGFP1 lysates were subsequently used as substrates for the in vitro protein import process.
Mitochondrial Protein Import
The mitochondrial protein import procedure was performed following the protocol from Stojanovski et al. (39) with modifications. Briefly, 40 μg of mitochondria was resuspended in 100 μl of import buffer (250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 2 mM KH2PO4, 10 mM MOP-KOH, at pH 7.2) with addition of 2 mM ATP and 10 mM Na-succinate. Lysate (1–5 μl) containing MitoGFP1 protein was added, and protein import was performed at increasing time intervals of 2, 5, and 10 min at 25°C. Valinomycin (2 μM) was added to stop the import reaction, and samples were centrifuged at 12,000 g for 5 min at 4°C. The supernatant was discarded, and the pellet was resuspended in SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOP-KOH, at pH 7.2) containing trypsin, on ice. Trypsin was subsequently inhibited by protease inhibitor cocktail (BioVision, Mountain View, CA). Samples were centrifuged again at 12,000 g for 5 min at 4°C. Pellets were then resuspended in lysis buffer (BioVision), subjected to SDS-PAGE, and subsequent Western blot analyses performed. Quantification of blots was performed using Pierce ECL Western blotting substrate (Pierce Biotech, Rockford, IL). The primary antibody was an anti-GFP monoclonal antibody raised in a mouse host (Clontech Laboratories, Mountain View, CA). The secondary antibody was an anti-mouse IgG horseradish peroxidase conjugate (Sigma, St. Louis, MO). Quantification of chemiluminescent signals were detected using a G:BOX (Syngene, Frederick, MD), and data were expressed as arbitrary optical density units. Additionally, visualization of imported fluorescent MitoGFP1 was performed using a Typhoon 9400 Variable Mode Fluorescence Imager (GE Healthcare, Piscataway, NJ) with a 532-nm excitation. Control for protein loading was confirmed using Ponceau staining.
Western Blot Analyses
SDS-PAGE was run on 4–12% gradient gels, as previously described (25, 42) with equal amounts of protein loaded for each study treatment. Relative amounts of subpopulation-specific mitochondrial heat shock protein 70 (mtHsp70), translocases of the outer membrane 40 and 20 (Tom40 and Tom20), translocases of the inner membrane 23 and 44 (Tim23 and Tim44), were analyzed using the following primary antibodies; anti-mtHsp70 mouse antibody (product no. SPA-825; Stressgen, Ann Arbor, MI), anti-Tom40 goat antibody (product no. sc-11025; Santa Cruz Biotechnology, Santa Cruz, CA), anti-Tom20 mouse antibody (product no. 612278; BD Biosciences, San Jose, CA), anti-Tim23 mouse antibody (product no. 611222; BD Biosciences), and anti-Tim44 mouse antibody (product no. 612582; BD Biosciences). The secondary antibodies used in the analyses were donkey anti-goat IgG HRP conjugate (product no. sc-2020; Santa Cruz Biotechnology) for Tom40, and goat-anti mouse conjugate (product no. 31430; Pierce Biotechnology, Rockford, IL) for mtHsp70, Tom20, Tim23, and Tim44. Detection of signal was performed according to the manufacturer's instructions as above. Quantification of chemiluminescent signals and data analysis were performed as above. Control for protein loading was confirmed using Ponceau staining.
mRNA Analyses
Thirty milligrams of tissue was excised from control and diabetic hearts for mRNA extraction using an RNeasy mini kit, per manufacturer's protocol (Qiagen, Valencia, CA) and reverse transcribed. Equal amounts of cDNA from the hearts of control and diabetic mice were subjected to real-time PCR. Custom primers were designed for target genes adenine nucleotide translocator 1 (ANT1), ATP synthase subunit alpha (ATP5A1), and ATP synthase subunit beta (ATP5B). SYBR Green I was used for quantification of respective cDNA replicates (Qiagen). Data were normalized by gene expression relative to the levels of GAPDH.
Mitochondrial Membrane Potential
Mitochondrial membrane potential (ΔΨm) was measured by flow cytometry using the ratiometric dye 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazol carbocyanine iodide (JC-1) (Molecular Probes, Carlsbad, CA), which is a lipophilic cation that enters selectively into mitochondria, as previously described (42). Values are expressed as the mean orange fluorescence divided by the mean green fluorescence of 100,000 mitochondrial events per individual mitochondrial sample.
Statistics
Means ± SE were calculated for all data sets. Data were analyzed with a one-way ANOVA method to evaluate the main treatment effect, diabetes induction (GraphPad Software, La Jolla, CA). Fisher's least significant difference (LSD) post hoc tests were performed to determine the significant differences among means. When appropriate a Student's t-test was employed. P < 0.05 was considered significant.
RESULTS
Diabetes Mellitus Phenotype
The diabetes mellitus phenotype was characterized 5 wk after STZ treatment by assessing fasting blood glucose and plasma insulin levels. Blood glucose levels were significantly elevated in diabetic mice compared with control (Control: 140 ± 5 mg/dl vs. Diabetic: 548 ± 22 mg/dl; P < 0.05). Further, plasma insulin levels were significantly reduced in diabetic mice compared with control (Control: 1.92 ± 0.17 ng/ml vs. Diabetic: 0.47 ± 0.06 ng/ml).
Proteomic Analyses
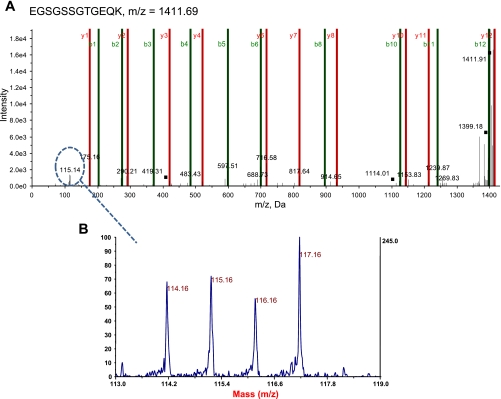
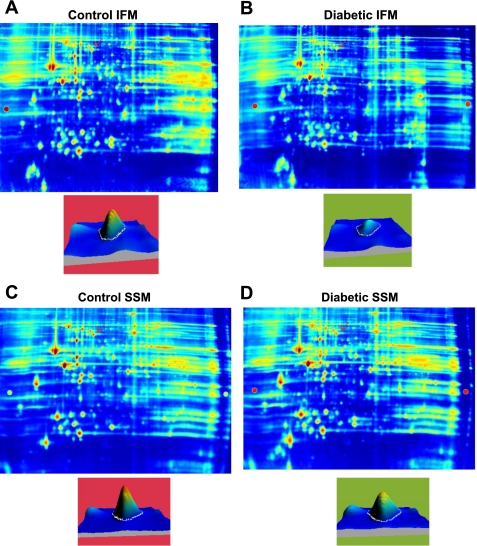
To determine whether type 1 diabetes mellitus differentially impacts spatially distinct mitochondrial subpopulation proteomes, we utilized both iTRAQ and 2D-DIGE approaches. iTRAQ enables simultaneous identification and relative quantification of mitochondrial proteins through isobaric peptide tagging, while 2D-DIGE analysis is a quantitative technique that enables multiple protein samples to be separated on the same 2D gel, thereby greatly reducing the introduction of potential artifacts due to gel-to-gel variations. Figure 1 is an example of an MS/MS fragment ion (spectra) for a peptide that was used to identify a protein, mtHsp70. Located below the spectra is an example of an iTRAQ tag mass spectral signature (m/z 114, 115, 116, and 117) used to make a relative quantification of mtHsp70 within the four labeled sample groups (see Fig. 1 legend for groups). Figures 2, A–D are representative 2D gels from four mitochondrial samples: control IFM (A), diabetic IFM (B), control SSM (C), and diabetic SSM (D), labeled with different Cyanine dyes. In this example, an individual spot, identified as mtHsp70, is indicated by the red arrows, and the quantitative differences are assessed by examination of peak densities, which are shown in each individual representative gel image (Fig. 2, A–D). MtHsp70 peak density was significantly decreased (P < 0.02) in the diabetic IFM compared with control IFM, with no changes in diabetic SSM compared with control SSM. For all proteomic evaluations, only those proteins that showed statistically significant changes as a result of diabetes mellitus were reported in the tables.
Fig. 1.
Representative iTRAQ spectra. Isolated mitochondrial subpopulations from control and diabetic hearts were labeled with iTRAQ reagents 114 [diabetic subsarcolemmal mitochondria (SSM)], 115 [diabetic interfibrillar mitochondria (IFM)], 116 (control SSM), and 117 (control IFM); then, they were combined for analysis with mass spectrometry. A: representative spectra of simultaneous quantitation of a mtHsp70 peptide in control and diabetic mitochondrial subpopulations. B: MS/MS spectra for the reporter groups of the iTRAQ reagents (114, 115, 116, and 117) from a mtHsp70 peptide. These spectra were used along with other peptides to simultaneously quantify mtHsp70 control and diabetic mitochondrial subpopulations.
Fig. 2.
Representative two-dimensional-differential in-gel electrophoresis (2D-DIGE) gels. Samples were labeled with either Cy3 or Cy5, alternating between control and diabetic SSM and IFM. A control and treated mitochondrial sample was run on the same gel, and an internal standard was labeled with Cy2 and run on every gel for gel-to-gel comparisons. A: representative gel showing Cy3-labeled control IFM. B: same gel as A showing Cy5-labeled diabetic IFM. C: representative gel showing Cy3-labeled control SSM. D: same gel as C showing Cy5-labeled control SSM. Below the gel images are individual spots that were identified as mtHsp70 (indicated by the red arrow), and quantitative differences were assessed by examination of peak density.
Proteins of FAO.
FAO proteins were significantly altered in both diabetic SSM and diabetic IFM compared with control using both iTRAQ and 2D-DIGE methods. In contrast to our previous study in a type 2 diabetes mellitus model (10), iTRAQ analysis revealed 8 of 10 proteins involved in FAO, which were decreased in the diabetic IFM, while 4 of 10 were decreased in the diabetic SSM (Table 1). Interestingly, carnitine palmitoyltransferase-1 (CPT-1), an outer mitochondrial membrane protein that mediates transport of long-chain fatty acids across the membrane, was significantly increased in the diabetic SSM, while diminished in the diabetic IFM (Table 1). Further, 2D-DIGE analysis identified six FAO proteins that were also significantly decreased in the diabetic IFM, while two of six were decreased in the diabetic SSM (Table 2). Taken together, both proteomic analyses suggest that FAO proteins are impaired in both diabetic SSM and IFM, with the effect being greatest in the IFM.
Table 1.
iTRAQ Proteomic analysis of mitochondrial subpopulations from control and diabetic hearts
| Protein Name | SSM Diabetic/SSM Control | IFM Diabetic/IFM Control |
|---|---|---|
| Mitochondrial fatty acids β-oxidation | ||
| Hydroxyacyl-coenzyme A dehydrogenase 3 ketoacyl coenzyme A | 0.69 | 0.59 |
| Mitochondrial trifunctional protein, β subunit | 0.61 | 0.72 |
| Acetyl-coenzyme A acyl transferase 2 | NS | 0.85 |
| Acyl coenzyme A dehydrogenase, medium chain | NS | 0.87 |
| Acyl Coenzyme A dehydrogenase, very long chain | 0.76 | 0.77 |
| 2,4-dienoyl CoA reductase 1 | 0.76 | 0.71 |
| Enoyl coenzyme hydratase 1 | NS | 0.79 |
| Carnitine palmitoyltransferase 1 | 1.37 | 0.57 |
| Mitochondrial respiratory chain | ||
| ATP synthase, H+ transporting, mitochondrial F1 complex, Beta subunit | NS | 0.66 |
| ATP synthase, H+ transporting, F0 complex subunit F | 1.36 | 0.60 |
| ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit | NS | 0.88 |
| ATP synthase, H+ transporting, mitochondrial F0, subunit b, isoform 1 | NS | 0.80 |
| NADH dehydrogenase ubiquinone flavoprotein 1 | NS | 0.83 |
| NADH dehydrogenase ubiquinone 1 α subcomplex 8 | 1.21 | NS |
| NADH dehydrogenase (ubiquinone) 1 α subcomplex 12 | NS | 0.71 |
| Ubiquinol cytochrome-c reductase, Rieske iron sulfur unit polypeptide 1 | NS | 1.20 |
| Cytochrome c-1 | 1.36 | NS |
| Cytochrome-c oxidase subunit IV isoform 1 | 1.18 | NS |
| Cytochrome-c oxidase subunit VIb polypeptide 1 | NS | 0.70 |
| Electron transfer flavoprotein-ubiquinone oxidoreductase | NS | 0.87 |
| Electron transfer flavoprotein subunit α | NS | 0.74 |
| Electron transfer flavoprotein β polypeptide | 1.19 | 0.85 |
| TCA cycle | ||
| Aconitase 2 | NS | 0.91 |
| Malate dehydrogenase 2, NAD | NS | 1.07 |
| Isocitrate dehydrogenase 2 (NADP+) | 1.16 | 0.75 |
| Pyruvate dehydrogenase (lipoamide) β | 1.28 | NS |
| Creatine kinase | 1.46 | NS |
| Transport proteins | ||
| Adenine nucleotide translocator, member 4 | NS | 0.70 |
| Vdac (Vdac-1) | NS | 1.15 |
| Solute carrier family 25, member 3 phosphate carrier | NS | 0.67 |
| mtHsp70 (Grp75) | NS | 0.80 |
| Structural | ||
| Inner membrane protein mitofilin (immt) | NS | 0.64 |
| Miscellaneous | ||
| Myoglobin | NS | 1.51 |
| Es 1 protein | NS | 0.80 |
| Multimerin 2 | NS | 0.88 |
| Hydroxysteroid dehydrogenase like 2, isoform CRA_c | 0.60 | 0.79 |
| Mlrq-like protein | NS | 0.66 |
iTRAQ analysis of proteins identified as significantly changing; they are categorized into groups consisting of fatty acid oxidation (FAO), mitochondrial respiratory chain, citric acid cycle (TCA), transport proteins, structural and miscellaneous proteins in isolated subsarcolemmal mitochondria (SSM) and interfibrillar mitochondria (IFM) from control and diabetic hearts. All values presented indicate a significant difference of at least P < 0.05 for control vs. diabetic groups, while NS represents no significant differences between any groups; n = 4 for each group.
Table 2.
2D-DIGE proteomic analysis of mitochondrial subpopulations from control and diabetic hearts
| Protein Name | SSM Diabetic/SSM Control | IFM Diabetic/IFM Control |
|---|---|---|
| Mitochondrial fatty acid β-oxidation | ||
| Very long chain-specific acyl-CoA dehydrogenase | 0.87 | 0.80 |
| Long-chain-specific acyl-CoA dehydrogenase | NS | 0.82 |
| Isoform 2 of 2-oxoglutarate dehydrogenase E1 complex | NS | 0.78 |
| Medium-chain-specific acyl-coA dehydrogenase | 0.69 | 0.76 |
| Mitochondrial acyl-CoA thioesterase 1 | NS | 0.91 |
| Delta(3,5)-delta(2,4)-dienoyl-CoA isomerase | 2.02 | 0.90 |
| Mitochondrial respiratory chain | ||
| Isoform 1 of NADH dehydrogenase (ubiquinone) flavoprotein 2 | 1.25 | 0.84 |
| NADH dehydrogenase iron-sulfur protein 2 | NS | 0.86 |
| NADH ubiquinone oxidoreductase, 75-kDa subunit | 0.46 | 0.76 |
| NADH dehydrogenase ubiquinone 1 alpha subcomplex subunit 10 | NS | 0.81 |
| NADH dehydrogenase ubiquinone iron-sulfur protein 8 | NS | 0.86 |
| NADH dehydrogenase (ubiquinone) iron-sulfur protein 3 | NS | 0.85 |
| NADH dehydrogenase (ubiquinone) iron-sulfur protein 4 | NS | 0.91 |
| Electron transfer flavoprotein-ubiquinone oxidoreductase | 2.83 | 0.85 |
| Ubiquinol cytochrome-c reductase core protein 1 | NS | 0.86 |
| Cytochrome-b-cl complex c | NS | 0.85 |
| Cytochrome-b-c1 complex subunit Rieske | NS | 0.75 |
| Ubiquinone biosynthesis protein COQ9 | 0.95 | 0.81 |
| Succinate dehydrogenase (ubiquinone) iron-sulfur unit | NS | 0.59 |
| ATP synthase subunit β | NS | 0.80 |
| ATP synthase subunit O | 1.13 | 0.60 |
| ATP5a1 ATP synthase subunit α | NS | 0.82 |
| TCA cycle | ||
| Aldehyde dehydrogenase | 1.15 | 0.87 |
| Pyruvate dehydrogenase E1 component subunit β | NS | 0.77 |
| Dihydrolipoyllysine-residue acetyltransferase | 1.50 | 0.80 |
| Malate dehydrogenase | 2.30 | 0.75 |
| Isocitrate dehydrogenase NAD subunit α | NS | 0.68 |
| Aconitase hydratase | NS | 0.71 |
| Amino acid metabolism | ||
| Aspartate aminotransferase | NS | 1.96 |
| Ubiquitin protein ligase E3C | 1.34 | 0.43 |
| Oxidative stress related | ||
| Prdx3 | NS | 0.81 |
| Transport proteins | ||
| Heat shock protein 60 | 0.71 | 0.66 |
| mtHsp70 (Grp75) | NS | 0.73 |
| Prohibitin-2 | NS | 0.72 |
| Structural | ||
| IMMT mitofilin | NS | 0.75 |
2D-DIGE analysis of proteins identified and significantly changing, categorized into groups consisting of fatty acid oxidation (FAO), mitochondrial respiratory chain, citric acid cycle (TCA), amino acid metabolism, oxidative stress related, transport proteins, and structural proteins in isolated SSM and IFM from control and diabetic hearts. All values presented indicate a significant difference of at least P < 0.05 for control vs. diabetic groups, while NS represents no significant differences between any groups. n = 4 for each group.
Proteins of the mitochondrial respiratory chain.
Utilizing both iTRAQ and 2D-DIGE, a number of proteins of the mitochondrial respiratory chain were significantly altered in both diabetic SSM and diabetic IFM compared with control. With iTRAQ, 10 of 14 respiratory chain proteins identified were significantly decreased in the diabetic IFM compared with control, while no respiratory chain proteins identified were decreased in expression in the diabetic SSM compared with control (Table 1). Interestingly, 5 of 14 respiratory chain proteins identified were increased in the SSM compared with control. In the diabetic IFM, all four proteins identified from complex V (ATP synthase) were decreased, while two of six proteins from complex I, and one of four from complex IV, were decreased (Table 1). In the diabetic SSM, one protein each from complexes I, III, IV, and V, respectively, were significantly increased compared with control SSM (Table 1). Similar to the iTRAQ analysis, 2D-DIGE analysis revealed 16 proteins of the respiratory chain decreased in the diabetic IFM, with three proteins increased in the diabetic SSM (Tables 1 and 2). Proteins that were decreased in the diabetic IFM were from all respiratory complexes, including complexes I, II, III, IV, and V, with the greatest number of decreased proteins occurring at complex I. Similar to the iTRAQ analysis, several respiratory proteins were increased in the diabetic SSM, suggesting a possible compensatory mechanism to prevent dysfunction.
Proteins of the citric acid cycle (TCA).
iTRAQ and 2D-DIGE analyses of proteins involved in the TCA revealed significant changes in both the diabetic SSM and IFM compared with control. In the diabetic IFM, two of five TCA proteins were decreased, while three of five proteins were increased in the diabetic SSM (Table 1). Of particular interest was the observation that isocitrate dehydrogenase 2 (NADP+) displayed increased content in the diabetic SSM yet decreased content in the diabetic IFM (Table 1). Similar to the iTRAQ analysis, 2D-DIGE analysis reported that six TCA proteins decreased in the diabetic IFM, and three proteins increased in the diabetic SSM (Table 2). These data suggest again that TCA proteins and other proteins necessary for energy production are increased in the diabetic SSM, yet decreased in the diabetic IFM.
Transport proteins.
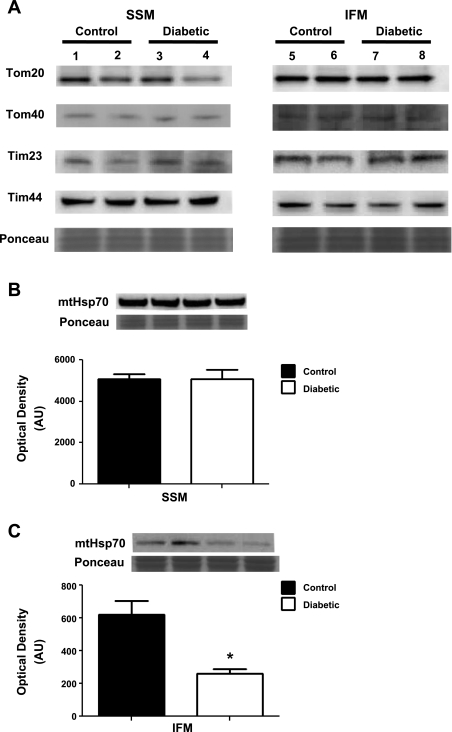
Proteins involved in transport of proteins and or substrates across the inner mitochondrial membrane (IMM) were preferentially decreased in the diabetic IFM compared with control IFM, as evidenced by a decreased content in three of four proteins, with no changes in the SSM (Table 1). Mitochondrial phosphate carrier, a protein that is involved in transport of phosphate groups from the cytosol to the mitochondrial matrix, was significantly decreased in diabetic IFM compared with control (Table 1). Another translocase required for exchange of ADP and ATP across the IMM, adenine nucleotide translocator 1 (ANT1), was also significantly decreased in the diabetic IFM compared with control (Table 1). Interestingly, mtHsp70 (also known as glucose-regulated protein 75; Grp75), a heat shock protein and a constituent of the mitochondrial import process essential for import of proteins into mitochondria, was also significantly decreased only in diabetic IFM compared with control (Tables 1 and 2). Western blot analyses of mtHsp70 (Fig. 5C), provided confirmation of both the iTRAQ and 2D-DIGE results observed. Another heat shock protein involved in folding and protein import into the mitochondrion, Hsp60, was also significantly decreased only in the diabetic IFM, as revealed by 2D-DIGE analysis (Table 2).
Fig. 5.
Western blot analysis of protein import constituents. Key proteins involved in mitochondrial protein import were assessed using Western blot analysis. A: control SSM (lanes 1 and 2), diabetic SSM (lanes 3 and 4), control IFM (lanes 5 and 6), and diabetic IFM (lanes 7 and 8) were analyzed for protein expression in translocases of the outer mitochondrial membrane (Tom20 and Tom40) and inner mitochondrial membrane (Tim23 and Tim44). Representative Western blots and densitometry analysis for mtHsp70 protein expression in SSM (B), and IFM (C) from control and diabetic hearts. Values are presented as means ± SE; *P < 0.05 for control vs. diabetic. Control for protein loading was confirmed with Ponceau staining; n = 5 for each group.
Structural proteins.
With both iTRAQ and 2D-DIGE analyses, alterations to proteins involved in mitochondrial structure were apparent in only the diabetic IFM compared with control with no changes in diabetic SSM. In particular, mitofilin, a transmembrane IMM protein that regulates mitochondrial cristae morphology (cristae density) (19), was significantly decreased only in diabetic IFM as indicated by both iTRAQ and 2D-DIGE analysis (Tables 1 and 2). Further, prohibitin-2, a protein involved in mitochondrial chaperone functions, which may impact mitochondrial morphology, as well as maintenance of mitochondrial DNA (1), was significantly decreased in the diabetic IFM with 2D-DIGE analysis (Table 2). These data are consistent with our previously published data, suggesting that mitochondrial internal complexity is decreased in the diabetic IFM (11).
For a general comparison highlighting the similarities and differences in proteomic responses between type 1 and type 2 diabetes mellitus, see Supplemental Table 2S.
Posttranslational Modifications
Utilizing MudPIT technology to assess posttranslational modifications (PTM), we identified 43 proteins as having a PTM in the diabetic IFM, while 22 proteins were identified in the diabetic SSM (Table 3). Interestingly, 11 of the 43 proteins in the diabetic IFM contained oxidations on various amino acid residues. In particular, three proteins of complex III, two proteins of complex V, and one protein of complex I were oxidized in the diabetic IFM. Further, mtHsp70 and mitofilin were only oxidized in the diabetic IFM. Additionally, 13 proteins in the diabetic IFM and 8 proteins in the diabetic SSM involved in the ETC presented with some sort of PTM, including acetylations, deamidations, and methylations. Of greatest interest was that the majority of the modified proteins were IMM proteins, suggesting that this locus may be specifically impacted and, as a result, play a key role in the pathogenesis of cardiac dysfunction associated with type 1 diabetes mellitus.
Table 3.
Posttranslational modifications (PTM) in mitochondrial subpopulations from control and diabetic hearts
| Protein Diabetic IFM | Peptide Sequence Diabetic IFM | Posttranslational Modification Diabetic IFM |
|---|---|---|
| ATP synthase, H+ transporting mitochondrial F1 complex, β subunit | ALVYGQMNEPPGAR | Oxidation(M)@7; Cation:K(E)@9 |
| Ubiquinol cytochrome-c reductase core protein 2 | NALANPLYCPDYR | Carbamidomethyl(C)@9; Dioxidation(Y)@12; Arg->GluSA(R)@13 |
| Solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 [Mus musculus] | GADIMYTGTLDCWR | Carbamidomethyl(C)@12; Dioxidation(W)@13; Arg->GluSA(R)@14 |
| Ubiquinol-cytochrome-c reductase core protein 1 | VYEEDAVPGLTPCR | Oxidation(P)@8; Oxidation(P)@12; Carbamidomethyl(C)@13; Arg->GluSA(R)@14 |
| Ubiquinol-cytochrome-c reductase core protein 1 | NALVSHLDGTTPVCEDIGR | Dioxidation(P)@12; Carbamidomethyl(C)@14; Arg->GluSA(R)@19 |
| Acetyl-coenzyme A acetyltransferase 1 | ENGTITAANASTLNDGAAALVLMTAEAAQR | Oxidation(N)@2; Deamidated(N)@9 |
| mtHsp70 (glucose-regulated protein 75) | MKETAENYLGHTAK | Oxidation(M)@1 |
| ATP synthase, H+ transporting, mitochondrial F0 complex, subunit b, isoform 1 | QIQDAIDMEK | Oxidation(M)@8 |
| Titin (connectin) | KMEAPPPKAPKKR | Dioxidation(P)@5 |
| NADH dehydrogenase (ubiquinone) 1β subcomplex 8 | VEDYEPYPDDGMGYGDYPMLPNR | Oxidation(D)@3 |
| Innermembrane protein, mitochondrial (mitofilin) | RVAQDWLKEAR | Deamidated(Q)@4; Oxidation(W)@6 |
| Dodecenoyl-coenzyme A δ isomerase | SLHMYLEK | Oxidation(M)@4 |
| ATP synthase, H+ transporting mitochondrial F1 complex, β subunit | TREGNDLYHEMIESGVINLK | Deamidated(N)@5 |
| ATP synthase, H+ transporting mitochondrial F1 complex, β subunit | GSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSR | Acetyl@N-term; Deamidated(Q)@7; Carbamidomethyl(D)@14 |
| ATP synthase, H+ transporting, mitochondrial F1 complex, α subunit, isoform 1 | VVDALGNAIDGK | Deamidated(N)@7 |
| 3-ketoacyl-CoA thiolase, mitochondrial (β-ketothiolase) | AALSAGKVPPETIDSVIVGNVMQSSSDAAYLAR | Deamidated(N)@20 |
| Mitochondrial trifunctional protein, β | DNGIRPSSLEQMAK | Deamidated(N)@2 |
| Mitochondrial trifunctional protein, β | LKPAFIKPYGTVTAANSSFLTDGASAMLIMSEDR | Deamidated(N)@16 |
| Glutamate oxaloacetate transaminase 2, mitochondrial [Mus musculus] | DDNGKPYVLPSVR | Deamidated(N)@3 |
| Glutamate oxaloacetate transaminase 2, mitochondrial [Mus musculus] | KMNLGVGAYRDDNGKPYVLPSVR | Deamidated(N)@13 |
| Succinate dehydrogenase, Fp subunit [Mus musculus] | NTVIATGGYGR | Deamidated(N)@1 |
| Solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator) | GDQALSFLKDFLAGGIAAAVSK | Protein Terminal Acetyl@N-term |
| Solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator) | GDQALSFLK | Protein Terminal Acetyl@N-term |
| Ubiquinol-cytochrome-c reductase core protein 1 | TATFAQALQSVPETQVSILDNGLR | Deamidated(N)@21 |
| Ubiquinol-cytochrome-c reductase core protein 1 | EVESIGAHLNAYSTR | Deamidated(N)@10 |
| Cytochrome-c oxidase, subunit Va | LNDFASAVR | Deamidated(N)@2 |
| Cytochrome-c oxidase, subunit Va | RLNDFASAVR | Deamidated(N)@3; Dehydrated(D)@4 |
| Acyl-coenzyme A dehydrogenase, long-chain [Mus musculus] | SPAHGISLFLVENGMK | Deamidated(N)@13 |
| mtHsp70 (glucose-regulated protein 75) | ASNGDAWVEAHGK | Deamidated(N)@3 |
| mtHsp70 (glucose-regulated protein 75) | STNGDTFLGGEDFDQALLR | Deamidated(N)@3 |
| Fumarate hydratase 1 | YPIEHGIITNWDDMEK | Methyl(H)@5 |
| Fumarate hydratase 1 | LNDHFPLVVWQTGSGTQTNMNVNEVISNR | Deamidated(Q)@17 |
| ATP synthase, H+-transporting, mitochondrial F0 complex, subunit F | RQASGGPVDIGPEYQQDLDRELYK | Deamidated(Q)@2; Dehydrated(S)@4 |
| Pyruvate dehydrogenase E1 α 1 [Mus musculus] | TREEIQEVRSKSDPIMLLK | Deamidated(R)@9 |
| Superoxide dismutase 2, mitochondrial [Mus musculus]; Sod2 protein [Mus musculus] | FNGGGHINHTIFWTNLSPK | Deamidated(N)@2 |
| Carnitine O-acetyltransferase (carnitine acetylase) (CAT) | IWNSSLQSNKEPVGILTSNHR | Deamidated(N)@3 |
| NADH dehydrogenase (ubiquinone) 1β subcomplex, 9 [Mus musculus] | EAEEEFWQNQHPQPYIFPDSPGGTSFER | Deamidated(N)@9 |
| 3-hydroxyacyl CoA dehydrogenase | LDKFAAEHTIFASNTSSLQITNIANATTR | Deamidated(N)@14 |
| Aldehyde dehydrogenase family 6, subfamily A1 | NHGVVMPDANKENTLNQLVGAAFGAAGQR | Deamidated(N)@10 |
| Aldehyde dehydrogenase family 6, subfamily A1 | IVNDNPYGNGTAIFTTNGATAR | Deamidated(N)@9; Deamidated(N)@17 |
| Dihydrolipoamide S-succinyltransferase | NDVITVQTPAFAESVTEGDVR | Deamidated(N)@1 |
| NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2 [Mus musculus] | MMNGRPGHEPLK | Protein Terminal Acetyl@N-term; Deamidated(N)@3 |
| Isocitrate dehydrogenase 3 (NAD+), γ | KAVLASMDNENMHTPDIGGQGTTSQAIQDII | Deamidated(N)@9 |
| ATP synthase, H+ transporting mitochondrial F1 complex, β subunit | IPSAVGYQPTLATDMGTMQER | Oxidation(P)@2 |
| Acetyl-coenzyme A acyltransferase 2 | VPPETIDSVIVGNVMQSSSDAAYLAR | Oxidation(P)@2 |
| Ubiquinol-cytochrome-c reductase core protein 1 | VYEEDAVPGLTPCR | Dioxidation(P)@12; Carbamidomethyl(C)@13; Arg->GluSA(R)@14 |
| Ubiquinol-cytochrome-c reductase core protein 1 | TATFAQALQSVPETQVSILDNGLR | Oxidation(F)@4; Deamidated(Q)@6; Deamidated(N)@21 |
| NADH dehydrogenase (ubiquinone) 1β subcomplex 8 [Mus musculus] | VEDYEPYPDDGMGYGDYPMLPNR | Oxidation(D)@3 |
| ATP synthase, H+ transporting, mitochondrial F1 complex, α subunit, isoform 1 | VVDALGNAIDGK | Deamidated(N)@7 |
| Enoyl-coenzyme A hydratase (trifunctional protein), α subunit | SLNSEMDNILANLRLPAKPEVSSDEDVQYR | Deamidated(N)@3 |
| Glutamate oxaloacetate transaminase 2, mitochondrial [Mus musculus] | ILIRPLYSNPPLNGAR | Deamidated(N)@13 |
| Glutamate oxaloacetate transaminase 2, mitochondrial [Mus musculus] | MNLGVGAYRDDNGKPYVLPSVR | Deamidated(N)@12 |
| ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d | NIIPFDQMTIDDLNEIFPETKLDKK | Deamidated(N)@1 |
| Electron transfer flavoprotein-ubiquinone oxidoreductase | IPVPILPGLPMNNHGNYIVR | Deamidated(N)@12 |
| Oxoglutarate dehydrogenase (lipoamide) | SSLATMAHAQSLVEAQPNVDKLVEDHLAVQSLIR | Deamidated(Q)@10 |
| Ubiquinol cytochrome-c reductase core protein 2 [Mus musculus] | LPNGLVIASLENYAPLSR | Deamidated(N)@3 |
| Ubiquinol cytochrome-c reductase core protein 2 [Mus musculus] | RGNNTTSLLSQSVAK | Deamidated(N)@3 |
| Ubiquinol cytochrome-c reductase core protein 2 [Mus musculus] | TSAAPGGVPLQPQDLEFTKLPNGLVIASLENYAPLSR | Deamidated(Q)@13; Deamidated(N)@22 |
| Fumarate hydratase 1 | LNDHFPLVVWQTGSGTQTNMNVNEVISNR | Deamidated(Q)@17 |
| Cytochrome-c-1 | AANNGALPPDLSYIVR | Deamidated(N)@4 |
| Carnitine palmitoyltransferase 2 | LSAVSGPAEYLQHSIVPTMHYQDSLPR | Deamidated(Q)@12 |
| NADH dehydrogenase (ubiquinone) 1α, subcomplex 8 | TDRPLPENPYHSR | Dehydrated(S)@12; Deamidated(R)@13 |
| Pyruvate dehydrogenase E1 α1 | KEIEDAAQFATADPEPPLEELGYHIYSSDPPFEVR | Deamidated(Q)@8 |
| Pyruvate dehydrogenase E1 α1 | MVNSNLASVEELKEIDVEVR | Deamidated(N)@3 |
| Pyruvate dehydrogenase E1 α1 | TREEIQEVRSKSDPIMLLK | Deamidated(R)@9 |
Multidimensional protein identification technology (MudPIT) was used to identify posttranslational modifications (PTM) of proteins from SSM and IFM from control and diabetic mitochondria. Peptides presented represent only those peptides that have PTMs in the diabetic SSM or diabetic IFM and not present in the control group.
Mitochondrial Protein Import Construct
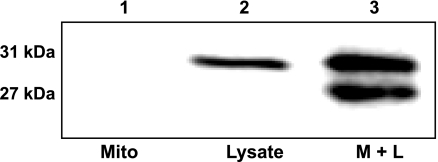
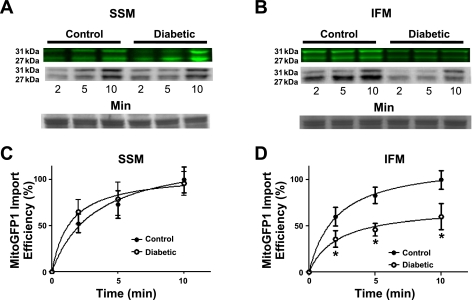
The MITOGFP1 cDNA was inserted as a Not I, Nhe I blunt fragment into the PIVEX2.3d plasmid (Supplemental Fig. S1). PIVEX2.3d has all necessary components for efficient protein production in a cell-free protein expression lysate system, including a T7 promoter, ribosomal binding site, and poly A tail. The MITOGFP1 gene consists of an N-terminal mitochondrial targeting sequence derived from cytochrome-c oxidase subunit VIII (targeted to the matrix) fused with AcGFP. Translated MitoGFP1 protein was incubated with isolated mitochondria to allow for proper import. Mitochondria were subsequently cleaned of nonimported protein and lysed; a Western blot analysis was performed probing for AcGFP. Isolated cardiac mitochondria alone do not produce a signal on the blot (Fig. 3, lane 1), whereas pure protein lysate from the expression system produces the full-length 31-kDa band containing the N-terminal presequence (Fig. 3, lane 2). The addition of total mitochondria and lysate together produces a similar 31-kDa band, which is the precursor protein in transit through the mitochondrial outer and inner membrane yet to be completely imported into the mitochondrial matrix. Additionally, there is a 27-kDa band representing the mature MitoGFP1 protein, in which the precursor sequence has been cleaved and properly imported into the mitochondrial matrix (Fig. 3, lane 3). Presequence cleavage of our target protein by mitochondrial processing peptidase will not occur until the protein is pulled into the matrix. Therefore, the smaller mature sequence was quantified for import analysis.
Fig. 3.
MITOGFP1 protein import. Western blot analysis of MitoGFP1 protein import into isolated cardiac mitochondria probed for AcGFP in samples containing 40 μg of mitochondria only (lane 1), 0.5 μl of MitoGFP1 protein lysate (lane 2), or combined mitochondria and lysate (3 μl) (lane 3). The upper 31-kDa band represents the full-length precursor MitoGFP1 in transit and yet to be processed, while the lower 27-kDa band represents a mature cleaved MitoGFP1 protein residing within the mitochondrial matrix.
Mitochondrial Protein Import
Western blot analyses in Fig. 4, A and C show increasing amounts of protein abundance at 2, 5, and 10 min, respectively, of the mature 27-kDa MitoGFP1 in the control SSM and diabetic SSM subpopulations, with no statistical difference between the samples. Conversely, a significant decrease in the amount of MitoGFP1 imported into the diabetic IFM compared with control IFM is shown at all three time points (Fig. 4, B and D; P < 0.05 for all three). Additionally, fluorescent labeling and subsequent import of MitoGFP1 was performed to confirm our Western blot findings (Figs. 4, A and B). The 31-kDa band is also decreased in the diabetic IFM compared with control denoting lesser amounts of precursor proteins in transit to the matrix.
Fig. 4.
Mitochondrial protein import. Effect of type 1 diabetes mellitus on MitoGFP1 import in SSM and IFM subpopulations. Cardiac mitochondrial subpopulations from control and diabetic mice were isolated and incubated with 3 μl of MitoGFP1 protein lysate at 2-, 5-, and 10-min time points. Representative Western blots and fluorescent images from SSM (A) and IFM (B) protein import assay. Graphical representation of mitochondrial protein import performed in SSM (C) and IFM (D) control and diabetic subpopulations. The relative amount of imported MitoGFP1 was determined by densitometry. Values were calculated based upon a 100% scale, and presented as mean ± SE; *P < 0.05 for control vs. diabetic subpopulations. Control for protein loading was confirmed by Ponceau staining; n = 8 per group.
Protein Import Machinery
Several outer and inner mitochondrial membrane translocases essential for proper mitochondrial protein import were also analyzed by Western blot analysis. There were no significant differences observed in the protein expression of Tom20, Tom40, Tim23, or Tim44 in either the diabetic SSM or diabetic IFM compared with control (Fig. 5A). It should be noted that Tim44 protein content showed a trend for decrease in the diabetic IFM, but the difference was not significant (P = 0.07). In contrast, the essential mitochondrial protein import motor, mtHsp70, was significantly decreased in only the diabetic IFM subpopulation compared with control (Fig. 5C), with no change in the diabetic SSM compared with control (Fig. 5B). These results indicate that of the several mitochondrial protein import constituents examined, only mtHsp70 is significantly decreased in abundance during type 1 diabetic insult, and this phenomenon is specific to the IFM subpopulation.
mRNA Analyses
Expression of ANT1, ATP5B, and ATP5A1, transcripts was evaluated to determine whether proteomic alterations (Tables 1 and 2) were preceded by transcriptional dysregulation in the diabetic heart. Our results revealed no significant decreases in the gene expression of these three specific proteins (ANT1, ATP5A1, and ATP5B) in diabetic vs. control hearts (Table 4). These results suggest that diabetes-induced proteomic decreases to these IFM proteins were not a consequence of decreased mRNA presence.
Table 4.
Relative mRNA expression from control and diabetic hearts
| Gene/Protein | mRNA (Diabetic/Control) | Protein (IFM Diabetic/IFM Control) |
|---|---|---|
| Adenine nucleotide translocator 1 (ANT1) | 1.27 | 0.70* |
| ATP5B ATP synthase F1 subunit β(ATP5B) | 0.94 | 0.66* |
| ATP5a1 ATP synthase subunit α (ATP5A1) | 1.03 | 0.82* |
Myocardial mRNA expression of ANT1, ATP5B, and ATP5A1 in control and diabetic mice as compared to respective protein expression levels in IFM. Values represent cellular mRNA transcript levels and IFM protein expression in diabetic compared to control. mRNA expression was normalized to GAPDH. Values are presented as means;
P < 0.05 for control vs. diabetic; n = 4 for each group.
Mitochondrial Membrane Potential
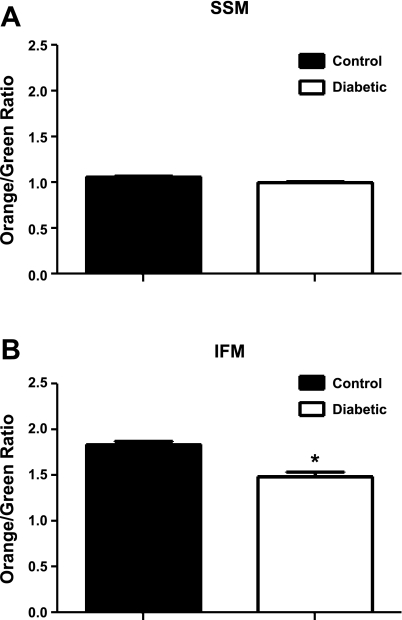
Mitochondrial membrane potential (ΔΨm) is essential for protein translocation across the IMM. Therefore, ΔΨm was analyzed in freshly isolated mitochondrial subpopulations by flow cytometry using JC-1. No significant differences in ΔΨm were observed in SSM control vs. diabetic (Fig. 6A). However, there was a 19% reduction in diabetic IFM ΔΨm compared with control (Fig. 6B). Additionally, baseline ΔΨm was higher in IFM compared with SSM, which is in agreement with previous reports (42). These data suggest that IFM ΔΨm is decreased relative to control during diabetic insult, whereas SSM ΔΨm remains unaffected.
Fig. 6.
Mitochondrial subpopulation membrane potential. Isolated mitochondrial subpopulations were incubated with JC-1, and 100,000 gated events were analyzed per sample. Control and diabetic SSM (A) and IFM (B) samples were analyzed. Values are represented as means ± SE. *P < 0.05 for control vs. diabetic; n = 4 for each group.
DISCUSSION
A significant amount of data supports the notion that mitochondrial dysfunction plays a critical role in the pathogenesis of diabetic cardiomyopathy. Despite the growing evidence for mitochondrial abnormalities underlying diabetic cardiomyopathy, the mechanisms involved in this dysfunction are not entirely clear. Evidence suggests that mitochondrial dysfunction varies depending on subcellular location, with those mitochondria located between the myofibrils being affected to a greater extent in the type 1 diabetic heart (11, 42). Data also suggest that dysfunction manifests via disturbance to the IMM as evidenced by diminished ETC complex activities, an enhanced oxidative environment, and depletion of cardiolipin, an essential phospholipid required for proper mitochondrial function (11, 33). The goal of this study was to build upon our previous findings and determine whether mitochondrial subpopulation-specific proteomes are altered with type 1 diabetes mellitus in the heart. In an effort to elucidate the mechanism, we examined whether mitochondrial import of nuclear-encoded proteins is decreased resulting in mitochondrial proteome dysregulation.
Proteomic analyses have played an integral role in furthering our understanding of mitochondrial dysfunction in the diabetic heart. Several proteomic studies, in a variety of diabetic models, have been employed to assess proteomic changes associated with the diabetic heart (5, 10, 15, 21, 38, 40). However, to date, no examination of subpopulation-specific proteomic make-up has been performed in a type 1 diabetic model. Proteomic profile comparisons of mitochondrial subpopulations from diabetic and control mouse hearts revealed that the majority of proteins changing were in the IFM and that almost all of these changes were decreases in protein abundances. Utilizing iTRAQ and 2D-DIGE methodologies afforded a number of advantages over the use of a single proteomic methodology, including the ability to confirm experimental results and accuracy by essentially comparing results from each method. By using MudPIT analysis combined with iTRAQ labeling, the limitations associated with 2D-DIGE, such as solubility and separation, were bypassed. Further, the iTRAQ approach enabled comparison of total protein levels, which can be more difficult using 2D-DIGE separation, where PTMs such as phosphorylation can alter pIs and hence gel migration (21). Utilizing 2D-DIGE allowed us to examine proteins that may be in less abundance and, as a result, less likely to show up with iTRAQ analyses. Because each method has its own unique set of advantages, the combined use of both techniques enhanced our level of confidence in the reported findings.
Most of the proteins identified as changing with both 2D-DIGE and iTRAQ approaches in the diabetic IFM were FAO and ETC proteins. In contrast to previous studies (21, 40), proteins involved in FAO (transport and/or utilization of fatty acids) were decreased in abundance in the diabetic IFM to a greater extent than in diabetic SSM. It is not entirely clear why the disparity in findings was observed. One could speculate that differences in animal models employed (mouse vs. rat), timing of examinations, STZ protocols utilized (single dose vs. multiple dose), mitochondria examined (total vs. subpopulations), or a combination of these variations in study designs may have contributed to the dissimilarity in findings. It is important to point out that though distinct sets of FAO proteins were identified with each proteomic platform (iTRAQ and 2D-DIGE), the results observed seemed to point in the same direction. Nevertheless, the findings are interesting and warrant further investigation. In addition, though only a few fatty acid utilization enzymes were decreased in diabetic SSM, CPT-1, a transporter of fatty acids into the mitochondrion, was upregulated, which is consistent with other proteomic studies utilizing an STZ-induced diabetic model (21). One could hypothesize that the diminished ability to utilize fatty acids within the diabetic IFM may have hampered its ability to produce ATP which, in turn, contributed to an overall decrease in cardiac contractile function as reported previously (11, 41). Additionally, ETC complexes and translocases within the IMM act as key contributors to overall oxidative phosphorylation and energy production within the mitochondrion. In total, we identified 9 ETC proteins from iTRAQ analyses and 16 ETC proteins from 2D-DIGE analyses as being decreased in the diabetic IFM. We did not observe changes to any mitochondrial genome-encoded proteins in the current study. It is unclear whether such a finding was the result of the proteomic methods employed or a reflection of the biological impact of diabetes mellitus on these specific proteins. Future examination focusing on this topic is warranted. Jüllig et al. (21) reported decreases in ETC constituents that are involved in oxidative phosphorylation in total mitochondria, which is somewhat different from our current study that examined mitochondrial subpopulations. Further, translocases located within the IMM that are essential for energy production through synthesis (mitochondrial phosphate carrier) and transport of ATP (ANT1) were also significantly decreased only in the diabetic IFM. Interestingly, Jüllig et al. (21) did not report changes in ANT1 or the mitochondrial phosphate carrier, which may have resulted from their analyses being performed on total mitochondria as opposed to our study, which examined subpopulations of mitochondria. Our proteomic data are consistent with previous reports of decreased respiratory capacity in the diabetic heart, as well as functional data indicating downregulation of ETC activities, specifically in the IFM (11, 24, 38). Downregulation of multiple components of the oxidative phosphorylation machinery and FAO proteins suggests that mitochondria in the type 1 diabetic mouse heart, specifically IFM, may be less efficient in producing ATP required for contraction and calcium homeostasis.
Mitochondrial morphological abnormalities have been reported with both type 1 and type 2 diabetes mellitus (4, 23, 38). Previous reports suggest that IFM morphology is specifically impacted by diabetes mellitus (11, 23). We reported previously that IFM were smaller and less complex in the type 1 diabetic heart with no changes in the SSM (11). Both 2D-DIGE and iTRAQ analyses identified mitofilin, a protein critical for proper cristae morphology (19), as being decreased only in the diabetic IFM. It is conceivable that diminished mitofilin, along with our previous reports of depleted cardiolipin, may have been a contributing factor to the altered IFM morphology reported previously (11).
To date, no attempts have been made to define the mechanisms accounting for mitochondrial proteomic dysregulation in the diabetic heart. Many potential mechanisms of proteomic dysregulation exist, including alterations in gene expression, enhanced PTMs, or upregulation of regulators, such as miRNAs (22, 30). To start to dissect some of these potential mechanisms, we examined a small subset of mRNAs encoding for nuclear encoded mitochondrial proteins whose contents were decreased in diabetic IFM. Our results indicate that despite significant decreases in the IFM protein contents for these proteins (ANT1, ATP5A1, and ATP5B), no significant decreases in mRNAs encoding for these proteins were observed. Though the subset is small, our results suggest that decreased mRNA abundance does not necessarily account for the decreased IFM protein contents observed and suggest that other mechanisms may be involved, such as protein import deficiencies and/or enhanced PTMs. In the present study, PTMs identified included oxidations, deamidations, deacetylations, and methylations, with oxidations and deamidations being the most prevalent. Interestingly, the diabetic IFM displayed the greatest number of oxidatively modified proteins. These findings complement previous studies, suggesting that enhanced mitochondrial reactive oxygen species formation during type 1 diabetic insult may precipitate oxidative damage to mitochondrial constituents (2, 11, 38). Also in agreement are the results from our previous study reporting increased superoxide formation in the type 1 diabetic IFM and the associated enhancement of protein oxidation (nitrotyrosine) within the mitochondrion (11). Another PTM that was prevalent in the diabetic IFM was deamidation. It has been suggested that deamidation of asparaginyl and glutaminyl residues within proteins causes structural and biological alterations to peptide and protein structure (34). Although the contribution of deamidation and oxidation to protein dysfunction is somewhat unclear, these PTMs may alter protein structure/function, making the protein more susceptible for degradation by the proteasome system (17, 34). In addition, several proteins that were decreased in the diabetic IFM also contained oxidations and deamidations within their structure. Interestingly, mtHsp70, mitofilin, and several components of complex V were decreased in abundance with both proteomic technologies and contained oxidations and deamidations within their structures. It is plausible that these modifications may have contributed to altered functionality and subsequent proteasomal degradation, all of which may have contributed to the morphological and oxidative phosphorylation deficiencies present in the type 1 diabetic heart.
Given that the majority of mitochondrial proteins are nuclear encoded and must be imported into the mitochondrion, it is possible that defects in this complicated process may underlie proteomic dysregulation in the diabetic heart. Nuclear encoded mitochondrial proteins constitute 99% of the organelle's proteome (7). These proteins are translated outside the mitochondrion and must be imported into the mitochondrion via an intricate mechanism involving coordination between the outer and inner mitochondrial membranes (8). The two membranes come together to form a “supercomplex,” which creates a singular avenue for translocation of the imported protein into the IMM and matrix (8). Given the considerable importance of this process, protein import decrements resulting from pathological conditions could represent a mechanism for mitochondrial dysfunction in the diabetic heart. Hence, a major goal of this study was to gain insight into whether diabetes mellitus causes protein import deficiencies in the mitochondrion and determine whether this phenomenon manifests differently in mitochondrial subpopulations. Utilizing a mitochondrial protein import methodology that eliminated the use of protein radiolabeling, we observed dysfunction of the IFM protein import process without effect to the SSM. Previous literature examining mitochondrial protein import in the skeletal muscle of aging mice revealed enhanced cytoplasmic protein degradation, as opposed to protein import decrements as the contributing source of the mitochondrial dysfunction (16). It is unclear why the differences were observed, but they may be due, in part, to the different pathologies and tissue types examined. In addition, movement of preproteins into the IMM is aided by ΔΨm. Our results indicate that a 5-wk diabetic insult caused a modest decrease (19%) in IFM ΔΨm, suggesting that mitochondrial protein import deficiencies in diabetic IFM could be at least partially due to mitochondrial ΔΨm deficiencies. However, previous studies examining aged skeletal muscle have indicated that as much as a 50% decrease in ΔΨm had no impact on mitochondrial protein import mechanics (16). Therefore, it's possible that the reduced ΔΨm, as reported in our study, may still be sufficient to allow for proper mitochondrial protein import.
Mitochondrial protein import dysfunction has been attributed in part to cardiolipin loss (20). Cardiolipin is known to surround and stabilize protein import complexes, including Tim23, the major pore-forming unit essential for protein translocation. Studies examining a mutation of cardiolipin that leads to loss of the phospholipid, caused reduced ΔΨm and decreased mitochondrial protein import (18). Another potential mechanism of mitochondrial import dysfunction is PTMs to proteins essential for protein translocation. As indicated above, we observed multiple modifications to mtHsp70 in the diabetic IFM subpopulation. MtHsp70 is a heat shock protein and the central subunit of the presequence translocase-associated motor complex that binds to a translocating polypeptide chain and drives its movement through the IMM into the matrix by a reaction cycle that requires hydrolysis of ATP (7). Previous literature examining inactivation of yeast mtHsp70 revealed a near-complete inhibition of oxidase assembly 1 protein translocation through the IMM (14). Additionally, cardiac mitochondria in senescent mice were shown to have elevated mtHsp70 protein content, as well as increased protein import of a mitochondrial matrix protein, malate dehydrogenase (9). In our study, we show a marked decrease in mtHsp70 protein content coinciding with decreased protein import in the diabetic IFM subpopulation, suggesting that mtHsp70 protein content is associated with protein import capacity. Our examination of several other key translocases that play a major role in mitochondrial protein import, including translocase of the outer membrane Tom20, outer membrane pore Tom40, inner membrane pore Tim23, and docking subunit Tim44 indicated that none of these mitochondrial protein import constituents was significantly decreased in the diabetic heart. Although not significant, Tim44 protein content revealed a trend for decrease in the diabetic IFM. This result is of interest in that previous studies have suggested a protective role for Tim44 overexpression in multiple tissue types following diabetic insult (29, 43). Additional experimentation regarding Tim44 and the diabetic heart are warranted.
Perspectives and Significance
With the dramatic increase in diabetes mellitus incidence, elucidation of the mechanisms contributing to the pathogenesis of diabetic cardiomyopathy is critical. An increasing amount of evidence suggests that mitochondrial dysfunction in the diabetic heart does not manifest in a homogeneous fashion. Rather, mitochondrial dysfunction occurs in a heterogeneous fashion with specific impact on mitochondria located in distinct subcellular locales and dependent upon the pathology (type 1 diabetes mellitus vs. type 2 diabetes mellitus). In the current study, we observed that type 1 diabetic insult differentially influenced spatially distinct cardiac mitochondrial proteomes, with the greatest impact occurring to those mitochondria located between the myofibrils (IFM). Further, changes in IFM proteomic make-up were associated with dysfunction to the nuclear-encoded mitochondrial protein import process, which may account, in part, for the more pronounced loss of proteins in this specific mitochondrial subpopulation. Finally, loss of key mitochondrial protein import constituents in the IFM may underlie protein import deficiencies during type 1 diabetic insult in the heart. The results support a paradigm in which changes in mitochondrial subpopulation-specific proteomic make-up underlie mitochondrial dysfunction, necessitating the study of the underpinnings of the complex and coordinated process of nuclear encoded mitochondrial protein import during diabetes mellitus. In addition, these findings continue to emphasize the importance of incorporating subcellular spatial locale into the study of diabetic mitochondria, which may be of particular relevance for the future development of diagnostics and therapeutic interventions.
GRANTS
This work was supported by the National Institutes of Health from the National Institutes of Diabetes and Digestive and Kidney Diseases (Grant DP2DK083095). This work was also supported by a Grant-In-Aid from the American Heart Association (0855484D). Erinne Dabkowski is a recipient of an American Heart Association Predoctoral Fellowship (0815406D). Walter Baseler is a recipient of an NIH Predoctoral Fellowship (T32HL090610) and American Heart Association Predoctoral Fellowship (10PRE3420006). Flow cytometry studies were supported in part by grants (RR020866) and (RR16440).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
ACKNOWLEDGMENTS
We would like to thank Dr. Christopher Cuff and the contributions from the West Virginia University Flow Cytometry Core facility.
REFERENCES
- 1. Artal-Sanz M, Tavernarakis N. Prohibitin and mitochondrial biology. Trends Endocrinol Metab 20: 394–401, 2009 [DOI] [PubMed] [Google Scholar]
- 2. Baynes JW. Role of oxidative stress in development of complications in diabetes. Diabetes 40: 405–412, 1991 [DOI] [PubMed] [Google Scholar]
- 3. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254, 1976 [DOI] [PubMed] [Google Scholar]
- 4. Bugger H, Boudina S, Hu XX, Tuinei J, Zaha VG, Theobald HA, Yun UJ, McQueen AP, Wayment B, Litwin SE, Abel ED. Type 1 diabetic akita mouse hearts are insulin sensitive but manifest structurally abnormal mitochondria that remain coupled despite increased uncoupling protein 3. Diabetes 57: 2924–2932, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bugger H, Chen D, Riehle C, Soto J, Theobald HA, Hu XX, Ganesan B, Weimer BC, Abel ED. Tissue-specific remodeling of the mitochondrial proteome in type 1 diabetic akita mice. Diabetes 58: 1986–1997, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Calvo S, Jain M, Xie X, Sheth SA, Chang B, Goldberger OA, Spinazzola A, Zeviani M, Carr SA, Mootha VK. Systematic identification of human mitochondrial disease genes through integrative genomics. Nat Genet 38: 576–582, 2006 [DOI] [PubMed] [Google Scholar]
- 7. Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell 138: 628–644, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chacinska A, Rehling P, Guiard B, Frazier AE, Schulze-Specking A, Pfanner N, Voos W, Meisinger C. Mitochondrial translocation contact sites: separation of dynamic and stabilizing elements in formation of a TOM-TIM-preprotein supercomplex. EMBO J 22: 5370–5381, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Craig EE, Hood DA. Influence of aging on protein import into cardiac mitochondria. Am J Physiol Heart Circ Physiol 272: H2983–H2988, 1997 [DOI] [PubMed] [Google Scholar]
- 10. Dabkowski ER, Baseler WA, Williamson CL, Powell M, Razunguzwa TT, Frisbee JC, Hollander JM. Mitochondrial dysfunction in the type 2 diabetic heart is associated with alterations in spatially distinct mitochondrial proteomes. Am J Physiol Heart Circ Physiol 299: H529–H540, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dabkowski ER, Williamson CL, Bukowski VC, Chapman RS, Leonard SS, Peer CJ, Callery PS, Hollander JM. Diabetic cardiomyopathy-associated dysfunction in spatially distinct mitochondrial subpopulations. Am J Physiol Heart Circ Physiol 296: H359–H369, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dabkowski ER, Williamson CL, Hollander JM. Mitochondria-specific transgenic overexpression of phospholipid hydroperoxide glutathione peroxidase (GPx4) attenuates ischemia/reperfusion-associated cardiac dysfunction. Free Radic Biol Med 45: 855–865, 2008 [DOI] [PubMed] [Google Scholar]
- 13. Flarsheim CE, Grupp IL, Matlib MA. Mitochondrial dysfunction accompanies diastolic dysfunction in diabetic rat heart. Am J Physiol Heart Circ Physiol 271: H192–H202, 1996 [DOI] [PubMed] [Google Scholar]
- 14. Frazier AE, Chacinska A, Truscott KN, Guiard B, Pfanner N, Rehling P. Mitochondria use different mechanisms for transport of multispanning membrane proteins through the intermembrane space. Mol Cell Biol 23: 7818–7828, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hamblin M, Friedman DB, Hill S, Caprioli RM, Smith HM, Hill MF. Alterations in the diabetic myocardial proteome coupled with increased myocardial oxidative stress underlies diabetic cardiomyopathy. J Mol Cell Cardiol 42: 884–895, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang JH, Joseph AM, Ljubicic V, Iqbal S, Hood DA. Effect of age on the processing and import of matrix-destined mitochondrial proteins in skeletal muscle. J Gerontol A Biol Sci Med Sci 65: 138–146, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iwai K, Drake SK, Wehr NB, Weissman AM, LaVaute T, Minato N, Klausner RD, Levine RL, Rouault TA. Iron-dependent oxidation, ubiquitination, and degradation of iron regulatory protein 2: implications for degradation of oxidized proteins. Proc Natl Acad Sci USA 95: 4924–4928, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiang F, Ryan MT, Schlame M, Zhao M, Gu Z, Klingenberg M, Pfanner N, Greenberg ML. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J Biol Chem 275: 22387–22394, 2000 [DOI] [PubMed] [Google Scholar]
- 19. John GB, Shang Y, Li L, Renken C, Mannella CA, Selker JM, Rangell L, Bennett MJ, Zha J. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol Biol Cell 16: 1543–1554, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Joshi AS, Zhou J, Gohil VM, Chen S, Greenberg ML. Cellular functions of cardiolipin in yeast. Biochim Biophys Acta 1793: 212–218, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jüllig M, Hickey AJ, Middleditch MJ, Crossman DJ, Lee SC, Cooper GJS. Characterization of proteomic changes in cardiac mitochondria in streptozotocin-diabetic rats using iTRAQ™ isobaric tags. Proteomics Clin Appl 1: 565–576, 2007 [DOI] [PubMed] [Google Scholar]
- 22. Kane LA, Van Eyk JE. Post-translational modifications of ATP synthase in the heart: biology and function. J Bioenerg Biomembr 41: 145–150, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51: 2944–2950, 2002 [DOI] [PubMed] [Google Scholar]
- 24. King KL, Young ME, Kerner J, Huang H, O'Shea KM, Alexson SE, Hoppel CL, Stanley WC. Diabetes or peroxisome proliferator-activated receptor alpha agonist increases mitochondrial thioesterase I activity in heart. J Lipid Res 48: 1511–1517, 2007 [DOI] [PubMed] [Google Scholar]
- 25. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685, 1970 [DOI] [PubMed] [Google Scholar]
- 26. Lesnefsky EJ, Gudz TI, Moghaddas S, Migita CT, Ikeda-Saito M, Turkaly PJ, Hoppel CL. Aging decreases electron transport complex III activity in heart interfibrillar mitochondria by alteration of the cytochrome c binding site. J Mol Cell Cardiol 33: 37–47, 2001 [DOI] [PubMed] [Google Scholar]
- 27. Lesnefsky EJ, Slabe TJ, Stoll MS, Minkler PE, Hoppel CL. Myocardial ischemia selectively depletes cardiolipin in rabbit heart subsarcolemmal mitochondria. Am J Physiol Heart Circ Physiol 280: H2770–H2778, 2001 [DOI] [PubMed] [Google Scholar]
- 28. Lin D. Multidimensional protein identification technology as an effective tool for proteomics. Am Genom Proteom Technol 1: 38–46, 2001 [Google Scholar]
- 29. Matsuoka T, Wada J, Hashimoto I, Zhang Y, Eguchi J, Ogawa N, Shikata K, Kanwar YS, Makino H. Gene delivery of Tim44 reduces mitochondrial superoxide production and ameliorates neointimal proliferation of injured carotid artery in diabetic rats. Diabetes 54: 2882–2890, 2005 [DOI] [PubMed] [Google Scholar]
- 30. Nishi H, Ono K, Iwanaga Y, Horie T, Nagao K, Takemura G, Kinoshita M, Kuwabara Y, Mori RT, Hasegawa K, Kita T, Kimura T. MicroRNA-15b modulates cellular ATP levels and degenerates mitochondria via Arl2 in neonatal rat cardiac myocytes. J Biol Chem 285: 4920–4930, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Palmer JW, Tandler B, Hoppel CL. Biochemical properties of subsarcolemmal and interfibrillar mitochondria isolated from rat cardiac muscle. J Biol Chem 252: 8731–8739, 1977 [PubMed] [Google Scholar]
- 32. Palmer JW, Tandler B, Hoppel CL. Heterogeneous response of subsarcolemmal heart mitochondria to calcium. Am J Physiol Heart Circ Physiol 250: H741–H748, 1986 [DOI] [PubMed] [Google Scholar]
- 33. Paradies G, Petrosillo G, Paradies V, Ruggiero FM. Role of cardiolipin peroxidation and Ca2+ in mitochondrial dysfunction and disease. Cell Calcium 45: 643–650, 2009 [DOI] [PubMed] [Google Scholar]
- 34. Robinson AB, Rudd CJ. Deamidation of glutaminyl and asparaginyl residues in peptides and proteins. Curr Top Cell Regul 8: 247–295, 1974 [DOI] [PubMed] [Google Scholar]
- 35. Rolo AP, Palmeira CM. Diabetes and mitochondrial function: role of hyperglycemia and oxidative stress. Toxicol Appl Pharmacol 212: 167–178, 2006 [DOI] [PubMed] [Google Scholar]
- 36. Rosca MG, Vazquez EJ, Kerner J, Parland W, Chandler MP, Stanley W, Sabbah HN, Hoppel CL. Cardiac mitochondria in heart failure: decrease in respirasomes and oxidative phosphorylation. Cardiovasc Res 80: 30–39, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scharfe C, Lu HH, Neuenburg JK, Allen EA, Li GC, Klopstock T, Cowan TM, Enns GM, Davis RW. Mapping gene associations in human mitochondria using clinical disease phenotypes. PLoS Comput Biol 5: e1000374, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shen X, Zheng S, Thongboonkerd V, Xu M, Pierce WM, Jr, Klein JB, Epstein PN. Cardiac mitochondrial damage and biogenesis in a chronic model of type 1 diabetes. Am J Physiol Endocrinol Metab 287: E896–E905, 2004 [DOI] [PubMed] [Google Scholar]
- 39. Stojanovski D, Pfanner N, Wiedemann N. Import of proteins into mitochondria. Methods Cell Biol 80: 783–806, 2007 [DOI] [PubMed] [Google Scholar]
- 40. Turko IV, Murad F. Quantitative protein profiling in heart mitochondria from diabetic rats. J Biol Chem 278: 35844–35849, 2003 [DOI] [PubMed] [Google Scholar]
- 41. Weytjens C, Cosyns B, D'Hooge J, Droogmans S, Lahoutte T, Garbar C, Roossens B, Van Camp G. Evaluation of contractile function and inotropic reserve with tissue velocity, strain and strain rate imaging in streptozotocin-induced diabetes. Eur J Echocardiogr 11: 622–629, 2010 [DOI] [PubMed] [Google Scholar]
- 42. Williamson CL, Dabkowski ER, Baseler WA, Croston TL, Alway SE, Hollander JM. Enhanced apoptotic propensity in diabetic cardiac mitochondria: influence of subcellular spatial location. Am J Physiol Heart Circ Physiol 298: H633–H642, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Y, Wada J, Hashimoto I, Eguchi J, Yasuhara A, Kanwar YS, Shikata K, Makino H. Therapeutic approach for diabetic nephropathy using gene delivery of translocase of inner mitochondrial membrane 44 by reducing mitochondrial superoxide production. J Am Soc Nephrol 17: 1090–1101, 2006 [DOI] [PubMed] [Google Scholar]