Abstract
Airway neutrophil infiltration is a pathological hallmark observed in multiple lung diseases including pneumonia and cystic fibrosis. Bacterial pathogens such as Pseudomonas aeruginosa instigate neutrophil recruitment to the air space. Excessive accumulation of neutrophils in the lung often contributes to tissue destruction. Previous studies have unveiled hepoxilin A3 as the key molecular signal driving neutrophils across epithelial barriers. The eicosanoid hepoxilin A3 is a potent neutrophil chemoattractant produced by epithelial cells in response to infection with P. aeruginosa. The enzyme phospholipase A2 liberates arachidonic acid from membrane phospholipids, the rate-limiting step in the synthesis of all eicosanoids, including hepoxilin A3. Once generated, aracidonic acid is acted upon by multiple cyclooxygenases and lipoxygenases producing an array of functionally diverse eicosanoids. Although there are numerous phospholipase A2 isoforms capable of generating arachidonic acid, the isoform most often associated with eicosanoid generation is cytoplasmic phospholipase A2α. In the current study, we observed that the cytoplasmic phospholipase A2α isoform is required for mediating P. aeruginosa-induced production of certain eicosanoids such as prostaglandin E2. However, we found that neutrophil transepithelial migration induced by P. aeruginosa does not require cytoplasmic phospholipase A2α. Furthermore, P. aeruginosa-induced hepoxilin A3 production persists despite cytoplasmic phospholipase A2α suppression and generation of the 12-lipoxygenase metabolite 12-HETE is actually enhanced in this context. These results suggest that alterative phospholipase A2 isoforms are utilized to synthesize 12-lipoxygenase metabolites. The therapeutic implications of these findings are significant when considering anti-inflammatory therapies based on targeting eicosanoid synthesis pathways.
Keywords: lung inflammation, neutrophils, arachidonic acid, prostaglandin E2, hepoxilin A3
inflammation as a consequence of Pseudomonas aeruginosa colonization is a complex process that can lead to severe tissue destruction, particularly in the lungs of individuals suffering from cystic fibrosis (20, 28). Host cell infection with Pseudomonas aeruginosa results in production and secretion of numerous inflammatory mediators such as cytokines, chemokines, and eicosanoids that collaborate to mount an immune response (5, 13, 20, 31, 35). Eicosanoids are bioactive lipid mediators that are metabolized from arachidonic acid (AA). Despite their structural similarity, eicosanoids exhibit a wide range of distinct activities in the context of inflammation (7, 10, 19, 27, 33).
Mucosal breach by neutrophils or polymorphonumclear cells (PMNs) is a major event in the inflammatory process that can result in considerable pathology (2, 3, 29, 37). PMNs migrate across airway epithelial barriers in response to mucosal infection and release noxious products in an attempt to prevent pathogenic colonization (2, 3, 29, 37). We have shown that P. aeruginosa infection of lung epithelial barriers results in secretion of the neutrophil chemoattractant eicosanoid hepoxilin A3 (HXA3; Ref. 13). HXA3 release from the apical surface of polarized airway epithelial monolayers results in directed migration of neutrophils across the epithelial barrier from the basolateral to the apical side (13, 14). The migration process is dependant on the actions of the signaling enzyme protein kinase C (PKC) as well as the lipolytic enzyme phospholipase A2 (PLA2: Refs. 13, 15). PLA2 cleaves membrane phospholipids releasing AA, which serves as the precursor to a diverse array of eicosanoids including HXA3 (12, 24). PLA2-mediated liberation of AA is considered to be the rate-limiting step in the synthesis of all eicosanoids (12, 24, 32).
PLA2-specific enzymatic activity is possessed by 20 distinct enzymes in human cells, and these enzymes are categorized into multiple groups based on distinguishable biochemical properties (12, 24, 32). Many of these PLA2 isoforms are expressed by airway epithelial cells (12). We and others (15, 17) have previously demonstrated that P. aeruginosa infection of airway epithelial cells results in the activation of cytoplasmic PLA2α (cPLA2α). This isoform, also referred to as group IVA, is widely studied in the context of eicosanoid generation and has been demonstrated to be critical towards the synthesis of the eicosanoid prostaglandin E2 (PGE2; Refs. 4, 8, 16, 22, 26, 32). Whether cPLA2α is required for HXA3-mediated PMN transepithelial migration in response to P. aeruginosa infection has yet to be explored. Herein, we explore the role of cPLA2α during P. aeruginosa-induced generation of various functionally distinct eicosanoids.
MATERIALS AND METHODS
Bacterial strains.
P. aeruginosa strain PAO1 and nonpathogenic Escherichia coli strain MC1000 were grown aerobically in Luria-Bertani broth overnight at 37°C. For infection of epithelial cells, overnight cultures were washed once in HBSS and resuspended at a concentration of 6 × 107 bacteria/ml of HBSS.
Cell culture.
A549 and H292 lung epithelial cell lines were maintained in Ham's F-12K medium or RPMI-1640, respectively, supplemented with 10% FBS and antibiotics. Polarized monolayers of A549 or H292 were grown on the underside of 0.33 cm2 collagen-coated Transwell filters to study PMN migration in the physiological basolateral to apical direction (13–15).
Inhibitors.
Chelerythrine chloride (CCL), an inhibitor of PKC, was purchased from Biomol (Plymouth Meeting, PA). The ERK kinase inhibitors U0126 and PD98059 were purchased from Cell Signaling Technology (Danvers, MA). General PLA2 inhibitors ONO-RS-082 and ACA were purchased from Biomol (Plymouth Meeting, PA). The cPLA2α-specific inhibitor (cPLA2α inhibitor, product no. 525143) was purchased from Calbiochem. A549 cells were pretreated 1–2 h with each inhibitor before infection and subsequent performance of each assay.
PGE2 enzyme immunoassay.
A549 cells were grown in 24-well plates and used 5–7 days after seeding. After being washed in HBSS, wells were infected with 6 × 107 bacteria/ml for 1 h at 37°C. Each well was washed three times in HBSS followed by incubation at 37°C for 2 h. Supernatents are collected, and the amount of PGE2 in each well was quantified using the Prostaglandin E2 Express enzyme immunoassay (EIA) kit from the Caymen Chemical (Ann Arbor, MI).
Arachidonic acid release assay.
A549 cells were grown in 24-well plates and used 5–7 days after seeding (15). Cells were washed three times with PBS(−), treated with media containing 0.2 μC/ml 3H-arachidonic acid (AA), and incubated for 18–24 h. Cells were then washed three times to remove unincorporated 3H-AA and treated with 0.5 ml of bacteria (6 × 107 bacteria/ml) for 1 h. Following infection, each well was washed three times in HBSS and incubated at 37°C for 2 h. Supernatants (100 μl) were then collected and measured by scintillation counting. After collection of supernatant, cells were solubilized with 500 μl/well of 1% SDS, 1% Triton-X-100 and sampled (250 μl) for measurement by scintillation counting.
Detection of cPLA2α activation.
A549 monolayers seeded on 6-well plates or 4.5-cm2 permeable filters were treated with HBSS alone, 1 μM PMA, or 6 × 107 bacteria/ml for 45 min (15). Cells were scraped off wells or permeable filters in buffer containing 150 mM Tris pH 8.0, 15 mM EDTA, 6 mM EGTA, 200 mM PMSF, 4 mM Na3VO4, 40 mM NaF, and 1 Complete Mini protease inhibitor cocktail tablet/10 ml buffer. Scraped cells were subjected to sonication followed by centrifugation at 55,000 rpm for 1 h. Cell pellets were resuspended in a lysis buffer containing 0.1% Triton-X-100, 0.2% SDS, 50 mM Tris pH 8.0, 5 mM EDTA, 2 mM EGTA, 200 mM PMSF, 4 mM Na3VO4, 40 mM NaF, and 1 Complete Mini protease inhibitor cocktail tablet/10 ml buffer. Lysates were again subjected to sonication followed by centrifugation at 30,000 rpm for 10 min. The supernatant was collected and concentrated using Centricon filters (30,000 molecular weight cutoff), and lysates were normalized for protein concentration and electrophoresed on an 8–16% gradient polyacrylamide gel (Bio-Rad Laboratories, Hercules, CA) followed by transfer to nitrocellulose. Blots were probed with anti-GAPDH (Santa Cruz Biotechnology), anti-cPLA2 antibody, or anti-phospho cPLA2 antibody (Cell Signaling Technology) followed by incubation with the HRP-conjugated goat anti-rabbit antibody and detection with ECL reagent.
PMN isolation.
PMNs were isolated from blood of healthy consenting human volunteers anti-coagulated with acid citrate/dextrose (Institutional Review Board Protocol No. 1999-P-007782). The buffy coat was resolved by centrifugation. Plasma and mononuclear cells were removed by aspiration, and the majority of the red blood cells were removed using 2% gelatin sedimentation. Residual red blood cells were removed by lysis in cold NH4Cl lysis buffer (13–15).
PMN transmigration assay.
Transwell inserts containing A549 cell monolayers seeded on the underside were exposed to 25 μl of 6 × 107 bacteria/ml for 1 h (13–15). After infection, PMNs (1 × 106) were added to the top (basolateral) chamber and incubated at 37°C for 2 h. PMNs that fully migrated across the cell monolayer reaching the bottom (apical) chamber were quantified by the myeloperoxidase assay. Uninfected A549 monolayers or monolayers infected with nonpathogenic E. coli strain MC1000 served as negative controls for PMN transmigration and establishment of a concentration gradient of fMLP across the epithelial barrier served as a positive control (0.1 μM fMLP added to apical chamber of uninfected A549 monolayers at the same time that PMNs were added to the basolateral chamber).
Cell viability/barrier integrity assays.
The amount of LDH released into the supernatant with and without infection of PAO1 in the presence or absence of each of the inhibitors employed was quantified using the LDH-based In Vitro Toxicology Assay kit (Sigma, St. Louis, MO). Barrier integrity of lung epithelial monolayers grown on Transwells was assayed by the horseradish peroxidase (HRP) flux assay as previously described (13).
Bacterial/epithelial association assay.
Monolayers were infected with 6 × 107 bacteria/ml for 3 h at 37°C (11). After infection, monolayers were washed followed by addition of 1 ml/well 1% Triton X-100. Cells were shaken at 4°C for 1 h, plated on Pseudomonas isolation agar plates, and incubated at 37°C overnight for subsequent colony forming unit determination.
Generation of stable cPLA2α-deficient A549 cells.
Plasmids used to generate small interfereing RNAs were constructed using the pSUPER vector (Oligoengine, Seattle, WA) by the method described by Brummelkamp et al. (1). Oligonucleotides were designed incorporating a 19-nt sequence (in italics) from the targeted human cPLA2α (PLA2 group IVA; Genbank Accession No. NM_024420) transcript, its reverse compliment (in italics) separated by a short spacer region, and complementary restriction sites for annealing into the BglII and HindIII sites of the vector as follows: for cPLA2α: 5′-GATCCCCGAACAGTCGTTAAGAAGTATTCAAGAGATACTTCTTAACGACTGTTCTTTTTGGAAA-3′ and 5′- AGCTTTTCCAAAAAGAACAGTCGTTAAGAAGTATCTCTTGAATACTTCTTAACGACTGTTCGGG-3′; or random control sequences: 5′- GATCCCCAGGATATTAGGACCATAAATTCAAGAGATTTATGGTCCTAATATCCTTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAAGGATATTAGGACCATAAATCTCTTGAATTTATGGTCCTAATATCCTGGG-3′. Oligonucleotides were annealed creating double-stranded DNA that have overhanging restriction sites and were ligated into digested pSUPER. Constructs were transformed into competent E. coli DH5α by standard methods, and transformants were plated on Luria-Bertani with ampicillin (50 μg/ml). Plasmids were extracted, purified (QIAprep Spin Mini-Prep kit; Qiagen, Valencia, CA), and sequenced for confirmation. Once confirmed, bulk plasmid was prepared for transfection using Qiagen Plasmid Midi kit (Qiagen).
A549 cells were transfected with the modified pSUPER using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) per manufacturer's instructions. Briefly, 4 μg of plasmid was diluted into F12K media without serum (Invitrogen). Separately, Lipofectamine 2000 (Invitrogen) was diluted into F12K with FBS and incubated at room temperature for 5 min. Following incubation, the two mixtures were combined and incubated at room temperature for 20 min. This cocktail was added to A549 cells with F12K, and the cells were incubated in F12K with 8% vol/vol FBS without selection. Cells were passaged into fresh media with selection added the next day (neomycin-G418, 1 mg/ml; Sigma-Aldrich, St. Louis, MO). Cells underwent two additional cycles of growth/passage in G418 before use.
Extraction of lipids from supernatants.
A549 cells were seeded on 162 cm2 flasks and grown to confluence. Confluent cell monolayers were treated with or without 6 × 107 bacteria/ml HBSS for 1 h at 37°C, washed three times with HBSS, and incubated an additional 2 hrs at 37°C. Supernatants were collected acidified to pH 4.0 and mixed with lipid extraction standards [LTB4-d4/15(s)-HETE-d8 obtained from Caymen Chemical; Refs. 18, 34]. Acidified supernatants were poured through a Supelco Discovery DSC-18 SPE column and eluted with methanol. The lipid fraction suspended in methanol was dried under a stream of nitrogen to 100 μl and stored at −80°C until further processing.
For assessment of PMN chemoattractant activity, each extracted lipid sample (prepared in triplicate) was resuspended in 1.7 ml HBSS. A volume of 0.6 ml resuspended lipids was diluted in 1.2 ml HBSS (referred to as diluted 1:3). One milliliter of diluted 1:3 resuspended lipids was added to the apical well of a Transwell containing H292 polarized lung epithelial monolayer. The remaining 0.8 ml diluted 1:3 resuspended lipids were further diluted by adding 0.8 ml HBSS (referred to as diluted 1:6). One milliliter of diluted 1:6 resuspended lipids was added to the apical well of a Transwell containing H292 polarized lung epithelial monolayer. The remaining 0.6 ml diluted 1:6 resuspended lipids were further diluted by adding 0.6 ml HBSS (referred to as diluted 1:12). One milliliter of diluted 1:12 resuspended lipids was added to the apical well of a Transwell containing H292 polarized lung epithelial monolayer. PMNs (1 × 106) were added to the top (basolateral) chamber in a volume of 120 μl for all diluted resuspended lipid samples and incubated at 37°C for 2 h. PMNs that fully migrated across the cell monolayer reaching the bottom (apical) chamber were quantified by the myeloperoxidase assay.
Measurement and quantification of eicosanoids.
HXA3, PGE2, 12-HETE, and 15-HETE were quantified by LC/MS/MS-based lipidomics. In brief, extracted samples were analyzed by a triple quadruple linear ion trap LC/MS/MS system (MDS SCIEX 3200 QTRAP) equipped with a LUNA C18–2 mini-bore column using a mobile phase (methanol:water:acetate, 65:35:0.02, vol:vol:vol) with a 0.50 ml/flow rate. MS/MS analyses were carried out in negative ion mode, and prominent fatty acid metabolites were quantified by multiple reaction monitoring (MRM mode) using established transitions for HXA3 (335→27 m/z), PGE2 (351→271, 351→189 m/z), 15-HETE (319→175 m/z) 12-HETE (319→179 m/z), LTB4-d4 (339→197 m/z), and 15-HETE-d8 (327→182 m/z). Calibration curves (1–1000 pg) and specific LC retention times for each compound were established with synthetic standards (Cayman Chemical, Ann Arbor, MI). Structures were confirmed for selected autacoids by MS/MS analyses using enhanced product ion mode with appropriate selection of the parent ion in quadrupole 1.
Statistics.
Data displayed in results (see Figs. 1–6) are presented as a representative experiment with a mean (SD) of at least three independent data points/conditions. Each experiment has been repeated multiple times yielding similar results. Statistical analysis was performed by Student's t-test for all comparisons and considered significant when P values were <0.05.
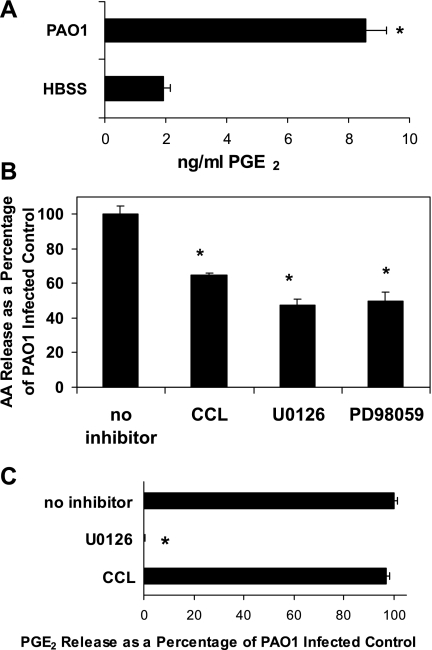
Fig. 1.
Effect of signaling kinase pathway inhibitors on PAO1-induced release of arachidonic acid (AA) and PGE2 from lung epithelial cells. A: amount of PGE2 (ng/ml) secreted from A549 cells measured by enzyme immunoassay in the presence or absence of PAO1 infection. B: relative amount of AA released by PAO1-infected A549 cells following a 1-h pretreatment with 5 μM Chelerythrine chloride (CCL), 40 μM U0126, or 40 μM PD98059 compared with vehicle control. Vehicle control (0.1% DMSO) represents the amount of AA released upon infection with PAO1 in the absence of inhibitors and is set at 100%. C: relative amount of PGE2 secreted by PAO1-infected A549 cells preincubated for 1 h before infection with either 5 μM CCL or 40 μM U0126 compared with vehicle control. Vehicle control (0.1% DMSO) represents the amount of PGE2 secreted upon infection with PAO1 in the absence of inhibitors and is set at 100%. *P < 0.05, statistically significant difference compared with control using the Student's t-test. Each data point represents an average of at least 3 separate wells. Evaluation of each inhibitor was performed in at least 3 internally controlled experiments yielding similar results.
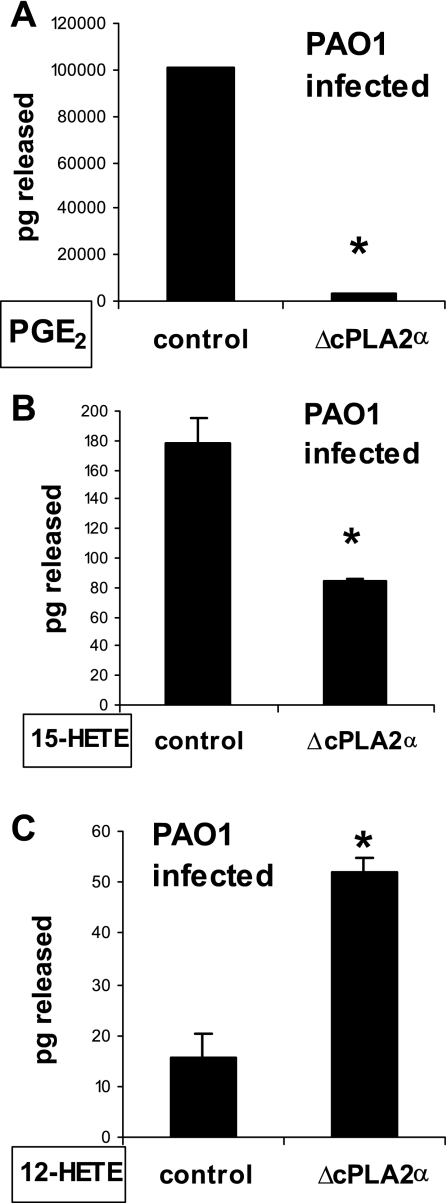
Fig. 6.
Inhibition of cPLA2α expression altered the quantity of eicosanoids released from PAO1-infected A549 cells. Lipids were extracted from the supernatants of PAO1-infected A549 cells transfected with vector containing irrelevant sequences (control) or with vector containing sequence specifically inhibitory towards cPLA2α. The following select eicosanoids were quantified by LC/MS/MS: PGE2 (A), 15-HETE (B), 12-HETE (C). *P < 0.05, statistically significant difference compared with control using the Student's t-test. Each data point represents an average of 2 separate extractions. Each above experiment was performed twice yielding similar results.
RESULTS
We have shown that infection of lung epithelial cells with P. aeruginosa results in increased release of AA accompanied by production of the eicosanoid HXA3 (13, 15). This biochemical process underlies the directed migration of PMNs across infected lung epithelial barriers. The mechanism by which P. aeruginosa induces increased HXA3 production is unclear, as is the extent to which other eicosanoids are also produced in response to P. aeruginosa infection. PGE2 is an eicosanoid generated from AA through the cyclooxygenase pathway (7). Despite the structural similarity to HXA3, PGE2 exhibits very distinct functional properties. For example, PGE2 plays a key role in resolving inflammation in the lung, whereas HXA3 facilitates inflammation by orchestrating neutrophilic mucosal breach (13, 21, 36). Several pathogens, including P. aeruginosa, have been shown to instigate PGE2 production upon infection of lung cells (6, 25, 31, 35). As shown in Fig. 1A, treatment of A549 cells with PAO1 results in an approximately fourfold increase in secretion of PGE2 after a 3-h infection, demonstrating that indeed multiple eicosanoids (PGE2 as well as the previously reported HXA3) are produced by lung epithelial cells in response to infection with P. aeruginosa.
Investigations into the signaling pathways involved in PMN transepithelial migration have revealed that release of HXA3 is highly dependant on PKC but unaffected by inhibitors of the MAP kinase ERK1/2 pathway (13). In contrast, ERK1/2 has been shown to be required for facilitating the synthesis of PGE2 in various cell types responding to different stimuli (16, 26). Since both HXA3 and PGE2 are synthesized from the common precursor AA, we investigated whether inhibitors of either PKC or ERK1/2 were capable of interfering with PAO1-induced AA release. CCL, a potent inhibitor of PKC caused an incomplete, but significant, reduction of PAO1-induced AA release (Fig. 1B). A similar result was observed with inhibitors of ERK1/2 (U0126 and PD98059), as each inhibitor prevented ∼50% of PAO1-induced AA release (Fig. 1B). Next, each inhibitor was evaluated for its ability to prevent enhanced PGE2 production upon infection of A549 cells with PAO1. The ERK1/2 inhibitor U0126 completely blocked PAO1-induced PGE2 release (Fig. 1C). In contrast, the PKC inhibitor CCL had no affect on PGE2 release in response to PAO1 infection (Fig. 1C). Inhibitors of both PKC and ERK1/2 interfere with PAO1-induced AA release, with AA serving as the precursor of both PGE2 and HXA3. ERK1/2 inhibitors interfere exclusively with PGE2 production, while PKC inhibitors interfere exclusively with HXA3 production, suggesting a divergence in the signaling mechanisms utilized to facilitate the production of each functionally distinct eicosanoid upon infection of the lung epithelial cell with P. aeruginosa (13, 21).
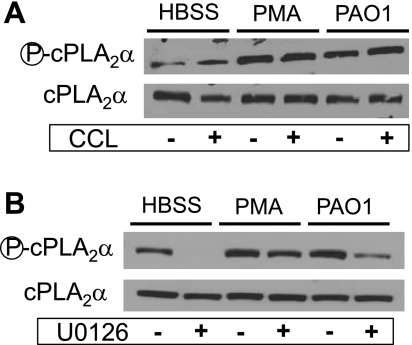
AA liberation for subsequent synthesis of eicosanoids is mainly mediated by PLA2 (24). There are numerous PLA2 isoforms expressed in human tissue that could account for AA release and eicosanoid generation (12, 32). The most widely investigated PLA2 isoform associated with eicosanoid generation is cPLA2α (8, 12, 32). An important step in cPLA2α activation is phosphorylation mediated by the ERK1/2 MAP kinase pathway (8, 16, 26). We and others (15, 17) have shown that infection of A549 cells with PAO1 results in activation of the PLA2 isoform cPLA2α. Treatment of A549 cells with either PMA or PAO1 results in a noticeable increase in expression of the phosphorylated form of cPLA2α, while the overall cPLA2α expression levels remain unchanged (Fig. 2). Increased phosphorylation of cPLA2α is unaffected by pretreatment of epithelial cells with PKC inhibitor CCL (Fig. 2A); however, pretreatment with the ERK1/2 inhibitor U0126 clearly interferes with expression of the phosphorylated form of cPLA2α (Fig. 2B).
Fig. 2.
Inhibitors of ERK1/2, but not PKC, interfered with cytoplasmic PLA2α (cPLA2α) phosphorylation. Stimulation of A549 cells with either PMA or PAO1 resulted in an increase in expression of the phosphorylated form of cPLA2α without any significant change in the expression of total cPLA2α. A: pretreatment of A549 cells with 5 μM CCL had no effect on the expression of the phosphorylated form of cPLA2α. B: pretreatment of A549 cells with 40 μM U0126 substantially reduced expression of phosphorylated cPLA2α both in the presence or absence of stimulation.
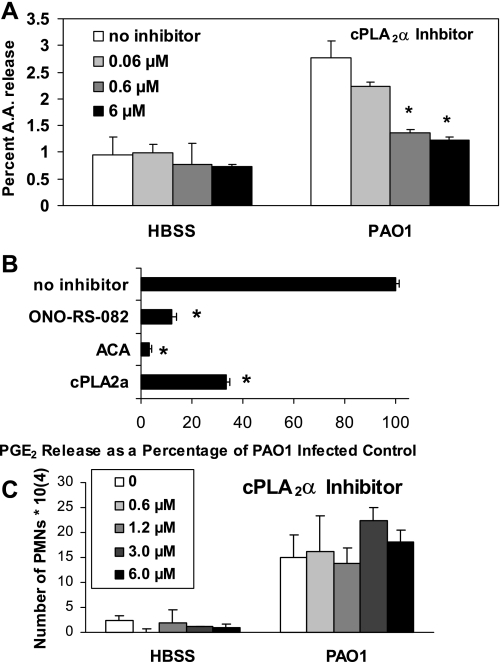
Next, we investigated whether the cPLA2α isoform is involved in the release of AA from lung epithelial cells in response to infection with PAO1. Inhibition of PAO1-induced AA release is observed when A549 cells were pretreated with the cPLA2α-isoform-specific inhibitor in a dose-dependant manner (Fig. 3A). PLA2 inhibitors that interfere with activity exhibited by all isoforms of PLA2 such as ACA and ONO-RS-082 have previously been shown to completely block HXA3-mediated PAO1-induced PMN transepithelial migration (15). These inhibitors also largely prevent PAO1-induced PGE2 release (Fig. 3B). In an attempt to specifically evaluate whether the cPLA2α isoform is involved in the generation of PGE2, we pretreated A549 cells with the cPLA2α-isoform-specific inhibitor before infection with PAO1. The cPLA2α-isoform-specific inhibitor significantly reduced PAO1-induced PGE2 release (Fig. 3B). Interestingly, pretreatment of A549 monolayers with the cPLA2α-isoform-specific inhibitor exhibited no effect on PAO1-induced PMN transepithelial migration (Fig. 3C). This result occurred despite the fact that, as reported previously, both ACA and ONO-RS-082 prevent PAO1-induced PMN transepithelial migration, demonstrating that PLA2 activity is crucial to this process (15). Taken together, it appears that although PLA2 activity is required to facilitate P. aeruginosa-induced HXA3 release culminating in PMN transepithelial migration, the cPLA2α isoform may not be involved in this process.
Fig. 3.
A specific inhibitor of cPLA2α reduced both arachidonic acid (AA) and PGE2 release but did not affect PMN transepithelial migration. A: treatment of A549 cells with PAO1 resulted in a significant increase in the release of AA, which is reduced in a dose-dependant manner by pretreating A549 cells for 2 h with cPLA2α-specific inhibitor. B: response to PAO1 following a 2-h pretreatment of A549 cells with universal PLA2 inhibitors ONO-RS-082 (10 μM) or ACA (50 μM) or with the specific cPLA2α inhibitor (6 μM) is reported as a percentage of the response to PAO1 infection pretreated with vehicle control [0.1% DMSO, which was set at 100% (no inhibitor)]. C: pretreatment of A549 monolayers grown on Transwells with cPLA2α inhibitor (0.6 to 6 μM) for 2 h had no effect on PAO1-induced polymorphonumclear cells (PMNs) transepithelial migration compared with A549 monolayers pretreated with 0.1% DMSO vehicle control (0 μM). *P < 0.05, statistically significant decrease compared with control using the Student's t-test. Each data point represents an average of at least 3 separate wells. Each above experiment was performed on at least 3 separate occasions yielding similar results.
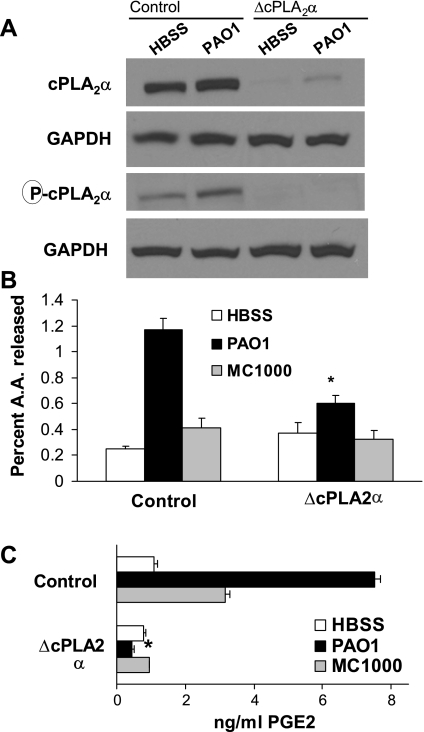
To more specifically target cPLA2α in our model system, we developed stable RNA inhibition (RNAi) of the message encoding the cPLA2α isoform. Short sequences from cpla2gIVa or irrelevant control sequence were cloned into pSUPER, and each plasmid was transfected separately into A549 cells (1). Transfectants resistant to G418 were selected for the purposes of achieving a cell line with a stabilized reduction in cPLA2α expression. Expression of the cPLA2α isoform and its activated phosphorylated form were analyzed by Western blotting for A549 transfected with either pSUPER containing irrelevant RNA sequence (referred to in Figs. 4–6 as control) or pSUPER with RNAi sequence specific for cPLA2α (referred to in Figs. 2–6 as ΔcPLA2α). As shown in Fig. 4A, transfected A549 cells generating cpla2gIVa-specific inhibitory sequence exhibit dramatically reduced expression of both phosphorylated cPLA2α and total cPLA2α in the context of uninfected and PAO1-infected epithelial cells. Demonstration that equal amounts of protein have been loaded into each well was achieved by measuring the expression of the housekeeping protein GAPDH, which was shown to be equivalent between control and ΔcPLA2α A549 cells (Fig. 4A).
Fig. 4.
Inhibition of cPLA2α expression resulted in decreased PAO1-induced AA and PGE2 release. A: expression of cPLA2α, both the phosphorylated form as well as total, was significantly reduced when A549 cells were engineered to express inhibitory RNA sequence specific to the cPLA2α gene. B: A549 cells exhibiting reduced cPLA2α expression were significantly less responsive to PAO1 in terms of the percentage of AA release. C: A549 cells exhibiting reduced cPLA2α expression were unable to secrete PGE2 in response to PAO1 infection. *P < 0.05, statistically significant decrease compared with control using the Student's t-test. Each data point represents an average of at least 3 separate wells. Each above experiment was performed on at least 3 separate occasions yielding similar results.
Upon developing a lung epithelial cell line with suppressed cPLA2α expression, we next investigated the involvement of cPLA2α in various cellular processes. Evaluation of the possibility that reduction of cPLA2α expression might affect A549 cellular viability or the integrity of the A549 barrier when grown on Transwells was considered. We performed LDH release and HRP flux assays, respectively, to investigate each issue independently, both in the presence and absence of PAO1 infection (13). No significant differences were observed between nontransfected A549 cells, transfected A549 ΔcPLA2α, and transfected A549 vector control cells in terms of cell viability as well as barrier integrity (data not shown). Furthermore, the degree of association between PAO1 and A549 cells is not altered to any significant degree in the context of reduced cPLA2α expression (data not shown).
The cPLA2α-isoform-specific inhibitor significantly reduced the ability of PAO1 to mediate AA release from A549 cells (Fig. 3A). Consistent with that observation, AA release in response to PAO1 infection is significantly reduced in A549 cells with a targeted reduction in cPLA2α expression (Fig. 4B). A sixfold induction in AA release is observed with control cells in response to infection with PAO1. The nonpathogenic E. coli strain MC1000 results in only a minor increase in AA release above that observed in uninfected control cells (Fig. 4B). In the context of cPLA2α suppression, PAO1-induced AA release is diminished from sixfold to approximately twofold; however, it is important to note that PAO1 is still capable of inducing AA release in cPLA2α suppressed cells, albeit to a significantly lesser extent (Fig. 4B). When A549 cells were pretreated with the cPLA2α-isoform-specific inhibitor, PAO1-induced PGE2 release was significantly diminished (Fig. 3B). PAO1-induced PGE2 release from A549 cells was completely ablated in the context of reduced cPLA2α expression, clearly suggesting that cPLA2α is critical for PAO1 to stimulate PGE2 production from A549 cells (Fig. 4C). Assays performed using either A549 or A549 transfected with a control vector did not exhibit any significant differences in terms of PAO1-induced AA release or PGE2 production (data not shown).
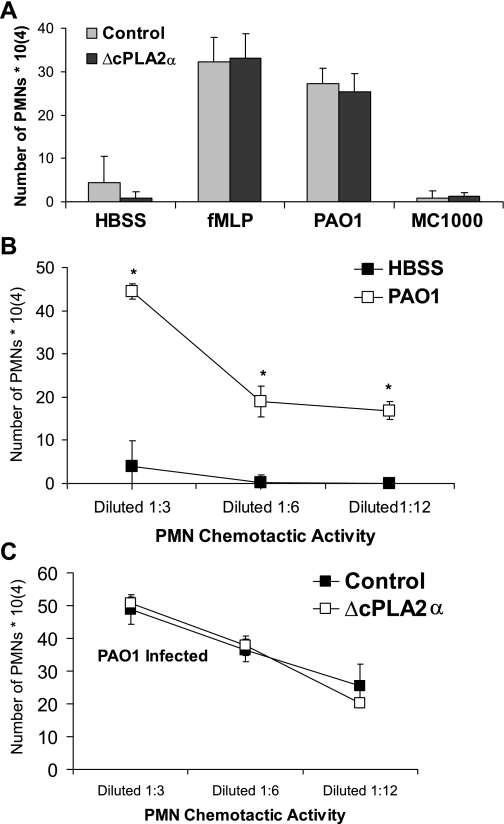
As mentioned above, PAO1-induced PMN transepithelial migration requires production of HXA3 (13). Inhibitors of PLA2 activity potently block PMN migration likely due to interference with HXA3 synthesis (15). Inhibitors specific to the cPLA2α isoform, however, exert no effect on PAO1-induced PMN transepithelial migration (Fig. 3C), deviating from the inhibitor's potent effect on PAO1-induced generation of PGE2 (Fig. 3B). Using the paired set of transfected A549 cells, containing a plasmid expressing either inhibitory cPLA2α sequence or irrelevant control sequence, we established epithelial barriers on Transwells. Certain epithelial monolayers were infected with either pathogenic PAO1 or nonpathogenic E. coli (MC1000) on the apical surface for 1 h, after which PMNs were added to the basolateral surface. The extent of PMN transepithelial migration was quantified after 2 h. Migration of PMNs across each uninfected monolayer in response to an imposed artificial chemoattractant gradient of fMLP resulted in equivalent numbers of PMNs migrating across transfected control and ΔcPLA2α A549 monolayers (Fig. 5A). Migration across uninfected (HBSS) or nonpathogenic infected (MC1000) monolayers was minimal for both cell monolayer groups. Notably, PAO1-induced PMN transmigration was similar between transfected control and ΔcPLA2α A549 monolayers (Fig. 5A), consistent with results obtained from studies employing the cPLA2α-isoform-specific inhibitor (Fig. 3C). This further suggests that PAO1-induced HXA3 production leading to PMN transmigration does not involve the cPLA2α isoform, which is clearly critical for PAO1-induced production of the functionally distinct eicosanoid, PGE2.
Fig. 5.
Inhibition of cPLA2α expression had no effect on PAO1-induced PMN transepithelial migration or PAO1-induced secretion of lipid-based PMN chemoattractants. A: A549 monolayers transfected with vector containing irrelevant sequences (control) or with vector containing sequence specifically inhibitory towards cPLA2α (ΔcPLA2α) demonstrated no significant difference when comparing quantity of PMNs migrating across. B: lipids extracted from the supernatants of A549 cells with PAO1 infection (□) possessed significantly more PMN chemoactivity than A549 cell supernatants in the absence of infection (■). C: lipids extracted from the supernatants of PAO1-infected A549 cells transfected with vector containing irrelevant sequences (■) possessed an equivalent amount of PMN chemoactivity when compared with PAO1-infected A549 cells transfected with vector containing sequence specifically inhibitory towards cPLA2α (□). *P < 0.05, statistically significant increase compared with HBSS control using the Student's t-test. Each data point represents an average of at least 3 separate wells. Each above experiment was performed on at least 3 separate occasions yielding similar results.
We have previously demonstrated that the lipid-based eicosanoid PMN chemoattractant HXA3 is produced by lung epithelial cells in response to infection with P. aeruginosa (13). Thus significant chemoattractant activity would be predicted to be observed in the lipid fraction of conditioned supernatant isolated from PAO1-infected A549 cell monolayers. Lipid fractions were extracted from the conditioned supernatant of both uninfected and PAO1-infected A549 cells and dried under nitrogen. Lipid extracted conditioned supernatants were subsequently resuspended in buffer (HBSS), and the amount of chemotactic activity was assessed by placing serial dilutions of the resuspended conditioned supernatant in the apical well of the Transwell insert containing H292 lung epithelial barriers. PMNs were placed on the basolateral side, and the number of PMNs fully migrating across the monolayer was quantified after 2 h. The amount of PMN chemtactic activity present in the lipid extracts of conditioned supernatants from PAO1-infected A549 cells is significantly greater than the amount found in the lipid extracts of conditioned supernatants from uninfected A549 cells (Fig. 5B).
Neither the cPLA2α-isoform-specific inhibitor, nor the A549-transfected cell line with reduced cPLA2α expression, displayed any effects on PAO1-induced PMN transepithelial migration, suggesting that the cPLA2α isoform is dispensable for PAO1-induced generation of HXA3. We therefore investigated whether there were any quantitative differences in the amount of lipid-based PMN chemoactivity produced by PAO1-infected A549-transfected control epithelial cells vs. PAO1-infected A549-transfected cells with reduced cPLA2α expression. The amount of PMN chemoattractant activity produced in response to PAO1 infection was in fact strikingly similar between the transfected A549 cells, indicating that cPLA2α has no impact on the amount of lipid-based chemoattractant that is produced by PAO1-infected A549 cells (Fig. 5C). Thus, although PLA2 activity is required for synthesis of HXA3 and subsequent facilitation of PMN transmigration, evidence strongly suggests that PLA2 isoform(s) distinct from cPLA2α are responsible for the generation of HXA3. Despite this finding with regard to HXA3 synthesis, it remains clear that cPLA2α is required for the synthesis of PGE2.
Next, we investigated whether cPLA2α-suppressed lung epithelial cells remain capable of producing increased amounts of HXA3 in response to infection with PAO1. As demonstrated previously, PAO1 induces the release of HXA3 from A549 cells (13). Based on our observations with regard to a lack of an affect on PMN movement in response to pretreatment with the cPLA2α-specific inhibitor or employment of cPLA2α-suppressed cells, we hypothesize that PAO1-induced HXA3 release in the context of cPLA2α suppression will not be affected. The ΔcPLA2α A549 cells were treated with and without PAO1 for 1 h. Cells were then washed and replaced with buffer (HBSS) for 2 h. Conditioned supernatant from ΔcPLA2α A549 cells was collected, and the lipid fraction was extracted for LC/MS/MS-based analysis to quantify HXA3. We measured (2.8 pg ± 3.1) in the lipid extracted supernatants of uninfected ΔcPLA2α A549 cells, whereas (15.5 pg ± 5.1) was recovered in PAO1-infected ΔcPLA2α A549 cells. Data represent a compilation of two separate experiments, and each data point was performed in duplicate (P < 0.05). Thus PAO1 retains the ability to induce a significant increase (>5-fold) in production of the eicosanoid HXA3, despite lack of cPLA2α expression. Consistent with our experiments quantifying PGE2 by EIA displayed in Fig. 4C, PAO1 is unable to cause a significant increase in PGE2 production in the context of cPLA2α suppression when assessed by LC/MS/MS-based quantification (data not shown).
PLA2 activity liberates AA for conversion into many distinct eicosanoids via multiple enzymatic pathways (12). These include the cyclooxygenase pathway, which generates prostaglandins such as PGE2, the 12-lipoxygenase pathway, which generates hepoxilins such as HXA3, and the 15-lipoxygenase pathway that produces anti-inflammatory eicosanoids known as lipoxins (12). Although each of these distinct classes of eicosanoid is synthesized from the same precursor, AA, recent reports (9, 23, 30) have suggested that the fate of free AA within a cell is not merely dependant on the availability of downstream cyclooxygenase/lipoxygenase enzymes but also depends on the particular PLA2 isoform responsible for its generation. Our data presented herein are consistent with this notion whereby blockade of cPLA2α prevents PGE2 production but has no effect on HXA3 production, a 12-lipoxygenase metabolite (27). In an effort to further probe this concept, we explored the fate of representatives of multiple eicosanoid-generating pathways in the context of either the presence or absence of cPLA2α. Transfected A549 cells harboring a plasmid producing either irrelevant sequence or cPLA2α inhibitory sequence were each infected with PAO1 for 1 h. Excess PAO1 was washed away and replaced with buffer (HBSS) for 2 h. Conditioned supernatant was collected, and the lipid fraction was extracted for LC/MS/MS based analysis for quantification of PGE2, 12-HETE, and 15-HETE. HETEs are enzymatic products of lipoxygenases: 12-lipoxygenase and 15-lipoxygenase (10, 33). Consistent with data obtained by EIA (Fig. 4C), the amount of PGE2 produced by A549 cells infected with PAO1 is drastically reduced in the context of reduced cPLA2α expression (Fig. 6A). A similar result is observed with 15-HETE, as reduced release of 15-HETE occurs when cells have reduced cPLA2α expression (Fig. 6B). In contrast, however, production of 12-HETE (representative metabolite of the HXA3-generating pathway) is actually increased in the context of reduced cPLA2α expression (Fig. 6C). These studies indicate that blockade of cPLA2α does not lead to generalized reduction in eicosanoid generation but rather exhibits discrete effects on different eicosanoid-generating pathways.
DISCUSSION
Eicosanoids are important and often overlooked regulatory components of the inflammatory process (10, 33). Leukotrienes and hepoxilins synthesized by 5-lipoxygenase and 12-lipoxygenase, respectively, assist in the orchestration of inflammation by serving as neutrophil chemoattractants, driving PMNs to their site of action (10, 21, 33), whereas, prostaglandins and lipoxins produced by cyclooxygenases and 15-lipoxygenases, respectively, can serve to dampen inflammation as well as promote the resolution of an inflammatory event (7, 10, 33). Although eicosanoids exhibit a diversity of function, they all share structural similarities owing to their common precursor, AA.
We have previously demonstrated that epithelial cell release of HXA3 in response to infection with P. aeruginosa requires PKC but does not involve the ERK1/2 pathway (13, 21). The ERK 1/2 kinase pathway is responsible for the generation of PGE2 in multiple cell types through the activation of cPLA2α by phosphoryation (8, 16, 25, 26). In the current study, we demonstrate that P. aeruginosa-induced AA release, known to precede production of both HXA3 and PGE2, is significantly reduced by inhibitors of either the PKC pathway or the ERK 1/2 pathway. Consistent with previous studies (8, 16, 25, 26), inhibition of the ERK1/2 pathway prevented the phosphorylation of cPLA2α and the production of PGE2 occurring in response to P. aeruginosa infection of the epithelium. Inhibitors of PKC, however, exerted no effect on P. aeruginosa-induced cPLA2α phosphoryation or PGE2 production in our studies. These results suggested a divergence in the mechanism underlying the production of each of these eicosanoids despite the fact that they share a common AA precursor.
PLA2 is generally believed to be the rate-limiting step in the generation of eicosanoids (12, 24, 32). PLA2 activity has been attributed to >20 distinct enzymes; however, the cPLA2α isoform has most often been linked to the generation of eicosanoids, particularly prostaglandins (12, 24, 32). Since our model of P. aeruginosa infection of lung epithelial cells results in the production of multiple eicosanoids, we investigated the role of the cPLA2α isoform in their generation. Pharmacological inhibition of PLA2 activity interferes with the release of AA, prevents the production of PGE2, and potently blocks HXA3-mediated PMN transepithelial migration (15). We observed herein that inhibitors specifically targeting the cPLA2α isoform also interfere with AA release and PGE2 generation but do not impact P. aeruginosa-induced HXA3-mediated PMN transepithelial migration.
Although pharmacological inhibitors are useful tools to lend insight into biological processes, their specificity is not always assured. To more specifically target the cPLA2α isoform, we constructed a plasmid capable of constitutively synthesizing an inhibitory RNA sequence designed to target cpla2α message for degradation when expressed in lung epithelial cells (1). We successfully produced a stable lung epithelial cell line that displayed substantially reduced cPLA2α isoform expression. P. aeruginosa-induced release of AA was reduced but not eliminated, suggesting that other PLA2s may be activated within epithelial cells upon infection with P. aeruginosa. However, lack of cPLA2α isoform expression completely ablated PGE2 release by epithelial cells infected with P. aeruginosa, demonstrating a strict requirement for the cPLA2α isoform in generating PGE2.
Experiments whereby the cPLA2α isoform was pharmacologically inhibited indicated that the cPLA2α isoform is not involved in P. aeruginosa-induced HXA3-mediated PMN transepithelial migration. Consistent with this observation, we found that the number of PMNs that migrated across lung epithelial monolayers in response to P. aeruginosa infection was similar between epithelial monolayers expressing the cPLA2α isoform and monolayers with substantially reduced cPLA2α expression. This result suggests that HXA3 production upon infection with P. aeruginosa does not require the presence of cPLA2α. Since HXA3 is a lipid metabolite, we expected to observe increased PMN chemoactivity in lipid extracted supernatants from lung epithelial cells infected with P. aeruginosa. This was indeed the case as P. aeruginosa-infected lung epithelial cells released substantial lipid-based chemotactic activity, whereas uninfected lung epithelial cells produced very little chemotactic activity. Loss of cPLA2α did not appear to affect P. aeruginosa-induced PMN transepithelial migration. In line with this observation, the amount of lipid-based PMN chemotactic activity produced by P. aeruginosa-infected epithelial cells was unaffected by the loss of cPLA2α expression, further suggesting that cPLA2α is not involved in the synthesis of HXA3. Consistent with this hypothesis, we also demonstrated that, in the context of cPLA2α suppression, P. aeruginosa induces a significant increase in HXA3 production, whereas P. aeruginosa-induced PGE2 production is completely ablated.
In aggregate, our observations above suggest that the cPLA2α isoform may only participate in the generation of a subset of eicosanoids when activated in response to inflammatory stimuli such as P. aeruginosa infection. While absolutely required for PGE2 production, the cPLA2α isoform appears completely dispensable during induced synthesis of HXA3. Furthermore, kinase signaling pathways leading to the induced synthesis of either PGE2 or HXA3 are divergent and these distinct pathways may underlie the selective activation of different PLA2 isoforms.
We quantified representative eicosanoids from multiple eicosanoid groups in the context of P. aeruginosa infection of lung epithelial cells with or without cPLA2α isoform expression. Inhibited expression of the cPLA2α isoform had the greatest impact on products of the cyclooxygenase pathway, as represented by substantially reduced PGE2 production. The 15-lipoxygenase product 15-HETE was also reduced in the context of inhibited cPLA2α isoform expression. Interestingly, the quantity of lipid metabolite 12-HETE produced was actually increased in the context of inhibited cPLA2α isoform expression. Therefore, production of 12-HETE and HXA3, both 12-lipoxygenase products, does not appear to require cPLA2α isoform expression. Such results imply that not only is an alternative PLA2 isoform responsible for P. aeruginosa-induced HXA3 production by lung epithelial cells but also the important concept that eicosanoid generation at the rate-limiting PLA2 activity step is a fairly complex process. Our results suggest that different PLA2 isoforms are likely required for the generation of distinct eicosanoid subsets (9, 23, 30). It is interesting to note that in our system cPLA2α appears to contribute to the generation of eicosanoids participating in the anti-inflammatory/resolution pathways (i.e., cyclooxygenase and 15-lipoxygenase) but does not play a role in generation of eicosanoids involved in neutrophil recruitment. In a previous study (9) investigating a model of acute inflammation in rats, calcium-independent PLA2 isoforms (iPLA2s) were demonstrated to act initially in producing proinflammatory eicosanoids, while the cPLA2α isoforms were associated with the generation of proresolving eicosanoids as the inflammatory process progressed. Further research using assorted inflammatory models representing different tissue will be necessary to better understand the apparent complexity of PLA2 isoform utilization during the generation of the eicosanoid profile characteristic of a particular disease process.
In summary, we have demonstrated that P aeruginosa infection results in the production of multiple eicosanoids by distinct mechanistic processes in terms of both signaling and the PLA2 activity. P aeruginosa activates ERK 1/2, which phosphorylates cPLA2α leading to AA release and PGE2 generation. In parallel, P. aeruginosa, likely via a distinct bacterial factor, activates PKC leading to the activation of an as yet to be identified PLA2 isoform culminating in HXA3 release and directed PMN transepithelial migration. Current studies are underway to identify the specific PLA2 isoform that is responsible for HXA3 production and subsequent PMN transepithelial migration. Unraveling the mechanistic distinction between these parallel eicosanoid-generating processes will greatly facilitate the design of therapeutics that specifically target proinflammatory PMN chemtactic eicosanoids but minimize the effect on potentially important anti-inflammatory or proresolving eicosanoids.
GRANTS
This work was supported by Research Scholarship Development Award K22 AI065425 (to B. P. Hurley) and a Cystic Fibrosis Foundation Research Grant (to B. P. Hurley). B. A. McCormick is supported by National Institute of Diabetes and Digestive and Kidney Diseases DK-56754. K. Gronert is supported by National Eye Institute Grant EY-016136.
DISCLOSURES
No conflicts of interest, financial or otherwise are declared by the author(s).
ACKNOWLEDGMENTS
We thank Mike Pazos for expert technical assistance and Aaron Sullivan for the LC/MS/MS analyses. We also thank Dr. Ronald E. Kleinman and Dr. W. Allan Walker for continued support. We thank David L. Tamang for critical review of this manuscript.
REFERENCES
- 1. Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553, 2002 [DOI] [PubMed] [Google Scholar]
- 2. Burns AR, Smith CW, Walker DC. Unique structural features that influence neutrophil emigration into the lung. Physiol Rev 83: 309–336, 2003 [DOI] [PubMed] [Google Scholar]
- 3. Craig A, Mai J, Cai S, Jeyaseelan S. Neutrophil recruitment to the lungs during bacterial pneumonia. Infect Immun 77: 568–575, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Croxtal JD, Newman SP, Choudhury Q, Flower RJ. The concerted regulation of cPLA2, COX2, and lipocortin 1 expression by IL-1beta in A549 cells. Biochem Biophys Res Commun 220: 491–495, 1996 [DOI] [PubMed] [Google Scholar]
- 5. DiMango E, Zar HJ, Bryan R, Prince A. Diverse Pseudomonas aeruginosa gene products stimulate respiratory epithelial cells to produce interleukin-8. J Clin Invest 96: 2204–2210, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fink J, Steer JH, Joyce DA, McWilliam AS, Stewart GA. Pro-inflammatory effects of Burkholderia cepacia on cystic fibrosis respiratory epithelium. FEMS Immunol Med Microbiol 38: 273–282, 2003 [DOI] [PubMed] [Google Scholar]
- 7. Funk CD. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science 294: 1871–1875, 2001 [DOI] [PubMed] [Google Scholar]
- 8. Ghosh M, Tucker DE, Burchett SA, Leslie CC. Properties of the group IV phospholipase A2 family. Prog Lipid Res 45: 487–510, 2006 [DOI] [PubMed] [Google Scholar]
- 9. Gilroy DW, Newson J, Sawmynaden P, Willoughby DA, Croxtall JD. A novel role for phospholipase A2 isoforms in the checkpoint control of acute inflammation. FASEB J 18: 489–498, 2004 [DOI] [PubMed] [Google Scholar]
- 10. Gronert K. Lipid autacoids in inflammation and injury responses: a matter of privilege. Mol Interv 8: 28–35, 2008 [DOI] [PubMed] [Google Scholar]
- 11. Hurley BP, Goodman AL, Mumy KL, Murphy P, Lory S, McCormick BA. The two-component sensor response regulator RoxS/RoxR plays a role in Pseudomonas aeruginosa interactions with airway epithelial cells. Microbes Infect 12: 190–198, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hurley BP, McCormick BA. Multiple roles of phospholipase A2 during lung infection and inflammation. Infect Immun 76: 2259–2272, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hurley BP, Siccardi D, Mrsny RJ, McCormick BA. Polymorphonuclear cell transmigration induced by Pseudomonas aeruginosa requires the eicosanoid hepoxilin A3. J Immunol 173: 5712–5720, 2004 [DOI] [PubMed] [Google Scholar]
- 14. Hurley BP, Sin A, McCormick BA. Adhesion molecules involved in hepoxilin A3-mediated neutrophil transepithelial migration. Clin Exp Immunol 151: 297–305, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hurley BP, Williams NL, McCormick BA. Involvement of phospholipase A2 in Pseudomonas aeruginosa-mediated PMN transepithelial migration. Am J Physiol Lung Cell Mol Physiol 290: L703–L709, 2006 [DOI] [PubMed] [Google Scholar]
- 16. Kawao N, Nagataki M, Nagasawa K, Kubo S, Cushing K, Wada T, Sekiguchi F, Ichida S, Hollenberg MD, MacNaughton WK, Nishikawa H, Kawabata A. Signal transduction for proteinase-activated receptor-2-triggered prostaglandin E2 formation in human lung epithelial cells. J Pharmacol Exp Ther 315: 576–589, 2005 [DOI] [PubMed] [Google Scholar]
- 17. Kirschnek S, Gulbins E. Phospholipase A2 functions in Pseudomonas aeruginosa-induced apoptosis. Infect Immun 74: 850–860, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leedom AJ, Sullivan AB, Dong B, Lau D, Gronert K. Endogenous LXA4 circuits are determinants of pathological angiogenesis in response to chronic injury. Am J Pathol 176: 74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Levy BD, Clish CB, Schmidt B, Gronert K, Serhan CN. Lipid mediator class switching during acute inflammation: signals in resolution. Nat Immunol 2: 612–619, 2001 [DOI] [PubMed] [Google Scholar]
- 20. Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15: 194–222, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McCormick BA. Bacterial-induced hepoxilin A3 secretion as a pro-inflammatory mediator. FEBS J 274: 3513–3518, 2007 [DOI] [PubMed] [Google Scholar]
- 22. Moreira LS, Piva B, Gentile LB, Mesquita-Santos FP, D'Avila H, Maya-Monteiro CM, Bozza PT, Bandeira-Melo C, Diaz BL. Cytosolic phospholipase A2-driven PGE2 synthesis within unsaturated fatty acids-induced lipid bodies of epithelial cells. Biochim Biophys Acta 1791: 156–165, 2009 [DOI] [PubMed] [Google Scholar]
- 23. Munoz NM, Kim YJ, Meliton AY, Kim KP, Han SK, Boetticher E, O'Leary E, Myou S, Zhu X, Bonventre JV, Leff AR, Cho W. Human group V phospholipase A2 induces group IVA phospholipase A2-independent cysteinyl leukotriene synthesis in human eosinophils. J Biol Chem 278: 38813–38820, 2003 [DOI] [PubMed] [Google Scholar]
- 24. Murakami M, Kudo I. Phospholipase A2. J Biochem 131: 285–292, 2002 [DOI] [PubMed] [Google Scholar]
- 25. N'Guessan PD, Etouem MO, Schmeck B, Hocke AC, Scharf S, Vardarova K, Opitz B, Flieger A, Suttorp N, Hippenstiel S. Legionella pneumophila-induced PKCα-, MAPK-, and NF-κB-dependent COX-2 expression in human lung epithelium. Am J Physiol Lung Cell Mol Physiol 292: L267–L277, 2007 [DOI] [PubMed] [Google Scholar]
- 26. Nagataki M, Moriyuki K, Sekiguchi F, Kawabata A. Evidence that PAR2-triggered prostaglandin E2 (PGE2) formation involves the ERK-cytosolic phospholipase A2-COX-1-microsomal PGE synthase-1 cascade in human lung epithelial cells. Cell Biochem Funct 26: 279–282, 2008 [DOI] [PubMed] [Google Scholar]
- 27. Pace-Asciak CR, Reynaud D, Demin P, Nigam S. The hepoxilins. A review. Adv Exp Med Biol 447: 123–132, 1999 [PubMed] [Google Scholar]
- 28. Prince AS, Mizgerd JP, Wiener-Kronish J, Bhattacharya J. Cell signaling underlying the pathophysiology of pneumonia. Am J Physiol Lung Cell Mol Physiol 291: L297–L300, 2006 [DOI] [PubMed] [Google Scholar]
- 29. Qin L, Quinlan WM, Doyle NA, Graham L, Sligh JE, Takei F, Beaudet AL, Doerschuk CM. The roles of CD11/CD18 and ICAM-1 in acute Pseudomonas aeruginosa-induced pneumonia in mice. J Immunol 157: 5016–5021, 1996 [PubMed] [Google Scholar]
- 30. Saiga A, Uozumi N, Ono T, Seno K, Ishimoto Y, Arita H, Shimizu T, Hanasaki K. Group X secretory phospholipase A2 can induce arachidonic acid release and eicosanoid production without activation of cytosolic phospholipase A2 alpha. Prostaglandins Other Lipid Mediat 75: 79–89, 2005 [DOI] [PubMed] [Google Scholar]
- 31. Saliba AM, Nascimento DO, Silva MC, Assis MC, Gayer CR, Raymond B, Coelho MG, Marques EA, Touqui L, Albano RM, Lopes UG, Paiva DD, Bozza PT, Plotkowski MC. Eicosanoid-mediated proinflammatory activity of Pseudomonas aeruginosa ExoU. Cell Microbiol 7: 1811–1822, 2005 [DOI] [PubMed] [Google Scholar]
- 32. Schaloske RH, Dennis EA. The phospholipase A2 superfamily and its group numbering system. Biochim Biophys Acta 1761: 1246–1259, 2006 [DOI] [PubMed] [Google Scholar]
- 33. Serhan CN, Chiang N, Van Dyke TE. Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat Rev Immunol 8: 349–361, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shahabi S, Hassan ZM, Jazani NH. Post heat shock tolerance: a neuroimmunological anti-inflammatory phenomenon. J Inflamm 6: 7, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith RS, Kelly R, Iglewski BH, Phipps RP. The Pseudomonas autoinducer N-(3-oxododecanoyl) homoserine lactone induces cyclooxygenase-2 and prostaglandin E2 production in human lung fibroblasts: implications for inflammation. J Immunol 169: 2636–2642, 2002 [DOI] [PubMed] [Google Scholar]
- 36. Vancheri C, Mastruzzo C, Sortino MA, Crimi N. The lung as a privileged site for the beneficial actions of PGE2. Trends Immunol 25: 40–46, 2004 [DOI] [PubMed] [Google Scholar]
- 37. Weiss SJ. Tissue destruction by neutrophils. N Engl J Med 320: 365–376, 1989 [DOI] [PubMed] [Google Scholar]