Abstract
Image retrieval at the semantic level mostly depends on image annotation or image classification. Image annotation performance largely depends on three issues: (1) automatic image feature extraction; (2) a semantic image concept modeling; (3) algorithm for semantic image annotation. To address first issue, multilevel features are extracted to construct the feature vector, which represents the contents of the image. To address second issue, domain-dependent concept hierarchy is constructed for interpretation of image semantic concepts. To address third issue, automatic multilevel code generation is proposed for image classification and multilevel image annotation. We make use of the existing image annotation to address second and third issues. Our experiments on a specific domain of X-ray images have given encouraging results.
Key words: Image classification, image annotation, image processing, machine learning
INTRODUCTION
Content-based image retrieval is an active research area in the image processing and computer vision fields.1,2 However, most of the existing content-based image retrieval techniques only support feature-based image retrieval. These low-level features cannot provide much help during the retrieval process, as naïve user may not be familiar with the low-level visual features. High-level semantic information is needed for image retrieval at the semantic level so that the user can specify their query easily by relevant concept. Because of large amount of medical images, there is an urgent need for a mechanism that can classify and search medical images at the different semantic level. Automatic annotation is the process of assigning text to the semantic content of images, and it is an intermediate step to image retrieval. The manual annotation of medical image is expensive, time-consuming, and varies according to different individuals. Therefore, automatic medical image annotation is becoming increasingly important for more effective image classification.3 Three approaches are widely used in automatic image annotation: (a) Translation model, which uses segmentation algorithm to extract blob from image, and then machine translation is applied to translate blobs vocabulary to words vocabulary;4 (b) Relevance model, whereby image is described as small vocabulary of blobs and joint distribution of words and blobs can be learnt from training set of annotated images;5 (c) Classification approaches, whereby image is classified according to predefined classes and class label is considered as image label.6 This can be from more general image categories to specific image category.7 We use similar concept and exploit the hierarchical relation between different classes. Most of the existing system ignores the hierarchical relationships between the different semantic levels. The hierarchy is intended to increase the performance of retrieval. For example, in medical domain, if a user searches for spine images, there are three types of spine images, i.e., cervical, thoracic, and lumbar spine. By exploiting the semantic relation, our system can retrieve all types of spine images, which is not very common in existing medical image retrieval systems. The existing medical image retrieval system only matches low-level features. To retrieve image at semantic level, three issues should be addressed:8 (1) Automatic feature extraction; (2) Concept modeling; (3) Semantic automatic annotation. To address the first issue, multilevel feature extraction approach is used,9 which provides global and local level features with pixel intensity information. To address the second issue, concept hierarchy is constructed by exploiting existing annotation. To address the third issue, support vector machine (SVM)-based approach by support vector machine at different semantic level is used. Support vector machine learns the maximum margin between the positive and negative images.10
AUTOMATIC FEATURE EXTRACTION
There are three levels of feature extraction, namely, global, local, and pixel. The pixel value itself is the simplest visual feature of the image. This pixel intensity value produces pretty good result in medical content-based image retrieval domain.11 Extracting features from the subpart of the image is called local features extraction, and any feature extracted from whole image is a global feature. We combined all three, global, local, and pixel, in one big feature vector.9 Images are partitioned into set of regular grids. Texture and shape features are extracted from whole image and from each subpart of the image. When building feature vector, firstly, 53 dimensions feature vector is obtained from the entire image by applying gray level co-occurrence matrix12 for texture extraction and edge histogram for shape feature extraction. Secondly, 53 dimensions (16 texture + 37 edges) feature was extracted from each grid patch. Finally, by resizing the image to 15 × 15, and pixel vector of size 225 was obtained. Combination of all these features into one big vector gives us a dimension of 490. Due to this resizing of the image, our method can cater for the variations of the original size of the image.
CONCEPT HIERARCHY
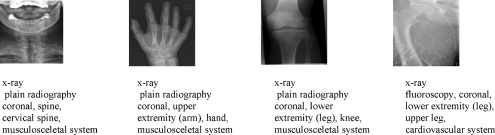
In this work, database used is from imageCLEF 2005.13 It consists of 9,000 training sets and 1,000 test sets of radiograph images. Training set is classified into 57 predefined classes, and each class has been annotated with short description as shown in Figure 1.
Fig. 1.
Images from different categories with annotation.
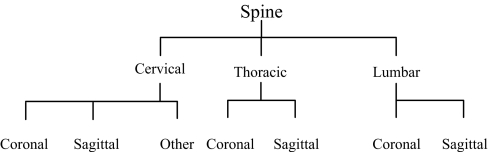
We design hierarchy of 57 different classes using existing annotation. For example, class 3 has annotation “03: x-ray, plain radiography, coronal, spine, cervical spine, musculosceletal system”, and we obtain six different parts from this annotation, and part four for each annotation is selected as main concept for hierarchy. So, we have nine main concepts, which are cranium, spine, arm, chest, abdomen, pelvis, leg, breast, and thorax. We build hierarchy under these nine topics. Figure 2 shows hierarchy of one of the main concepts. As shown in hierarchy, spine is a combination of coronal cervical spine, sagittal cervical spine, other orientation cervical spine, coronal thoracic spine, sagittal thoracic spine, coronal lumbar spine, and sagittal lumbar spine. Our system automatically distributes all 57 classes into nine main classes. In the second level of hierarchy, there are 29 different classes, i.e., cervical spine, thoracic spine, lumbar spine, etc. Therefore, cervical spine is a combination of three different classes, i.e., coronal cervical spine, sagittal cervical spine, and other orientation cervical spine. The last level of hierarchy represents the original 57 classes. This class distribution according to hierarchy is very helpful in semantic image retrieval.
Fig. 2.
Hierarchy of spine.
CLASSIFIER TRAINING
SVM is used as a classifier because of its excellent results in various classification experiments.14,15 Given set of training samples {xi, yi∣i = 1,...,N} where xi is a vector of visual features and yi = ±1 is a class label. During SVM classifier generation, low-level image features are mapped in to new higher dimension feature space f(x) = W × Φ(xi) + b ≥ ±1. Φ(xi) is the function that maps xi into higher dimensional space where W and b are transformation parameters. The optimal separation hyperplane is constructed in higher dimensional space by a kernel function defined as K(xi, xj) =Φ(xi) × Φ(xj). In our implementation, radial basis function (RBF) is selected, K(xi, xj) = exp(−γ∣∣xi − xj∣∣2), γ > 0 with one against all rules to label images. All together, three classifiers are generated according to each level of hierarchy. The first classifier is for 57 classes, second for 29 classes, and third is for 9 classes. The classifier can characterize different visual properties of image more effectively and efficiently if more visual features are given to classifier, but this high-dimensional feature vector needs more memory and computational power. Principal component analysis was used to shrink the dimension of the feature vector, which reduces memory requirement and computational cost.
AUTOMATIC ANNOTATION AND RETRIEVAL
Once extracting multilevel features and obtaining classifiers from training images are done, our system assigns two types of annotation to test image, the class number and code according to hierarchy. There are three steps for multilevel image annotation. (1) Test image is tested for top-level classes of the hierarchy, which is, in our case, nine classes. Third classifier is used to classify, and our system automatically annotates test image with a character. The character helps to recognize the level of hierarchy. (2) The process of classifying image for the second level of hierarchy is by using second classifier and annotated with a character. (3) For more specific classification, use first classifier and annotated with character and class label. With these three steps, we achieve multilevel annotation, which gives great advantage at retrieval process. It is important to note that once a test image is classified into one class, the characters that are used to represent the relevant class become the characters for annotating the multilevel semantic of the corresponding image. Each character interprets one text keyword.
If keyword is used for retrieval, for example, “spine” keyword is given, then our system will display seven different images from classes like cervical spine, thoracic spine, lumbar spine, etc., as shown in Figure 3.
Fig. 3.
Result of spine keyword.
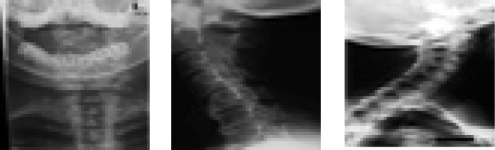
The result of “cervical spine” keywords will be from three different classes as in Figure 4.
Fig. 4.
Result of cervical spine keyword.
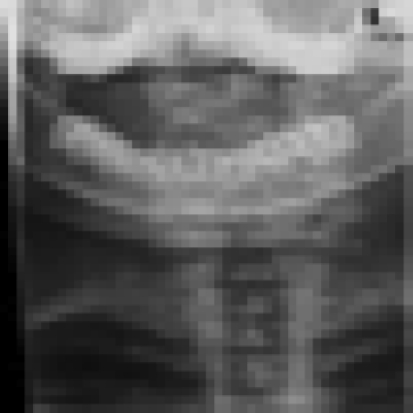
More specific result can be obtained when a user types “coronal cervical spine” as in Figure 5. Same results can be acquired using query image, but user has to specify level of semantic. For example, if coronal cervical spine query image is given and level of semantic is top, then the result is similar as in Figure 3.
Fig. 5.
Result of coronal cervical spine keyword.
EVALUATION
The multilevel annotation by assigning low-level and high-level semantic concept that becomes the semantic label of an image is very attractive to enable semantic image retrieval such that naïve users have more flexibility to specify their query, which is in high demand in medical domain. Eighty percent images were used as training and 20% as test images from each class to ensure that each class has representations in training and testing data.
The benchmark metric for evaluation includes precision ρ and recall τ. They are defined as:
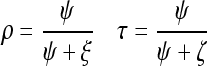 |
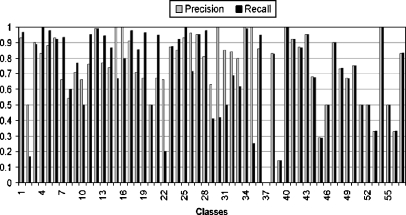
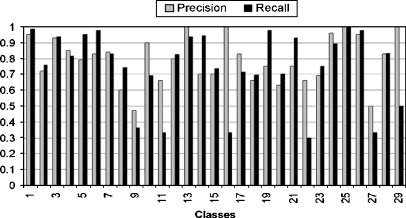
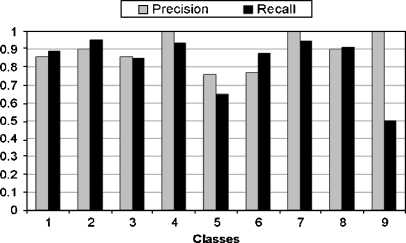
where ψ is the set of true positive images that are related to the corresponding semantic class and are classified correctly, ξ is the set of true negative samples that are irrelevant to the corresponding semantic class and classified incorrectly, ζ is the set of false positive samples that are related to the corresponding semantic class but are misclassified. The kernel selection is a trial-and-error method in SVM. It is impossible to select best kernel in advance. RBF kernel is chosen for our experiment. The classification performance at low, middle, and high semantic level is shown in Figures 6, 7, and 8, respectively.
Fig. 6.
Classification performance (i.e., precision/recall) at low semantic level.
Fig. 7.
Classification performance (i.e., precision/recall) at middle semantic level.
Fig. 8.
Classification performance (i.e., precision/recall) at high semantic level.
The result shows that some classes give a low result, which is due to great imbalance of training set. For example, class 12 has more than 2,000 images, whereas a few other classes have less than 20. We carried out additional experiments in which we examine error rate at different semantic level using two machine learning classifier, SVM and K-nearest neighbor (KNN) as shown in Table 1. One can observe that incorporating class hierarchy in classifier training can improve the classification results at high semantic level.
Table 1.
Error Rate Comparison Between Classifier at Different Levels
| Classifier | Low-level (%) | Middle-level (%) | High-level (%) |
|---|---|---|---|
| SVM | 11 | 9 | 7 |
| KNN | 18 | 17 | 13 |
DISCUSSION
We used existing annotation to create hierarchy of concepts. This hierarchy works as knowledge representation. Currently, most of the medical content-based image retrieval systems focus only on visual similarity. Only few systems exploit semantic information, but they use low-level semantic similarities. For example, similarity between two sagittal view images, called low-level semantic similarity and similarity, i.e., spine image and chest image, is a high-level semantic similarity. Our system can handle high-level semantic queries as well as low-level.
CONCLUSION AND FUTURE WORK
In this paper, we have presented a novel multilevel automatic medical image annotation and retrieval via keywords method based on concept hierarchy or class hierarchy. Instead of absolute decision for a class, our system is able to decide on its own based on our algorithm that what level of detail the classification is done. This method improves the efficiency of image retrieval and gives more semantic retrieval, which is rare in existing content-based medical image retrieval systems. The experiment results show the strength of our method. We would like to extend this work in the following directions. First, we would like to build more interrelation between concepts besides multilevel hierarchy. Next, we would like to address this kind of hierarchy construction in various other annotated data. Besides medical application, there are other important real-world applications such as management of digital photos, digital scans of important museum art works, and scientific images that can adopt our technique for multilevel image annotation and semantic retrieval. The classification techniques are very helpful in teaching, training, research, diagnostic, and analysis. Our system enables specific retrieval of the images as well as related images.
Acknowledgment
The image data used in this study are courtesy of TM Lehmann, Dept. of Medical Informatics, RWTH Aachen, Germany.
References
- 1.Smeulders AWM, Worring M, Santini S, Gupta A, Jain R. Content-based image retrieval at the end of the early years. IEEE Trans Pattern Anal Mach Intell. 2000;22(12):1349–1380. doi: 10.1109/34.895972. [DOI] [Google Scholar]
- 2.Rui Y, Huang TS, Ortega M, Mehrotra S. Relevance feedback: A power tool for interactive content-based image retrieval. IEEE Trans Circuits Syst Video Technol. 1998;8(5):644–655. doi: 10.1109/76.718510. [DOI] [Google Scholar]
- 3.Carneiro G, Vasconcelos N: Formulating semantic image annotation as a supervised learning problem. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR). San Diego, CA, USA, 2005
- 4.Duygulu P, Barnard K, de Freitas JFG, Forsyth DA: Object recognition as machine translation: Learning a lexicon for a fixed image vocabulary. In: Proceedings of European Conference on Computer Vision, 2002, pp 97–112
- 5.Lavrenko V, Manmatha R, Jeon J: A model for learning the semantics of pictures. In: Proceedings of NIPS’03, 2004
- 6.Li J, Wang JZ. Automatic linguistic indexing of pictures by a statistical modeling approach. IEEE Trans Pattern Anal Mach Intell. 2003;5(19):1075–1088. [Google Scholar]
- 7.Hanbur A: Review of image annotation for the evaluation of computer vision algorithms. Technical Report PRIP-TR-102, PRIP, T.U. Wien, 2006
- 8.Adams WH, Iyengar G, Lin CY, Naphade MR, Neti C, Nock HJ, Smith JR. Semantic indexing of multimedia content using visual, audio and text cues. EURASIP JASP. 2003;2:170–185. [Google Scholar]
- 9.Mueen A, Sapiyan Baba M, Zainuddin R. Multilevel feature extraction and X-ray image classification. J Appl Sci. 2007;7(8):1224–1229. doi: 10.3923/jas.2007.1224.1229. [DOI] [Google Scholar]
- 10.Heisele B, Serre T, Prentice S, Poggio T. Hierarchical classification and feature reduction for fast face detection with SVM. Pattern Recogn. 2003;36:2007–2017. doi: 10.1016/S0031-3203(03)00062-1. [DOI] [Google Scholar]
- 11.Keysers D, Gollan C, Ney H: Classification of medical images using non-linear distortion models. Bildverarbeitung fur die Medizin, Berlin, Germany, March 2004
- 12.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst. 1973;3:610–621. [Google Scholar]
- 13.Lehmann T: IRMA x-ray library. Available at http://irma-project.org, 2005
- 14.Platt JC: Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. In: Smola A, Bartlett P, Schölkopf B, Schuurmans D (Eds.) Advances in Large Margin Classifiers. MIT Press, 1999
- 15.Viola P, Jones M. Robust real-time face detection. Int J Comput Vis. 2004;57(2):137–154. doi: 10.1023/B:VISI.0000013087.49260.fb. [DOI] [Google Scholar]