Abstract
The radiology community has recognized the need to create a standard terminology to improve the clarity of reports, to reduce radiologist variation, to enable access to imaging information, and to improve the quality of practice. This need has recently led to the development of RadLex, a controlled terminology for radiology. The creation of RadLex has proved challenging in several respects: It has been difficult for users to peruse the large RadLex taxonomies and for curators to navigate the complex terminology structure to check it for errors and omissions. In this work, we demonstrate that the RadLex terminology can be translated into an ontology, a representation of terminologies that is both human-browsable and machine-processable. We also show that creating this ontology permits computational analysis of RadLex and enables its use in a variety of computer applications. We believe that adopting an ontology representation of RadLex will permit more widespread use of the terminology and make it easier to collect feedback from the community that will ultimately lead to improving RadLex.
Key words: Ontologies, terminologies, vocabularies, RadLex, software tools
INTRODUCTION
A key challenge for the Transforming the Radiological Interpretation Process (TRIP) initiative is to enable computer applications to manage the increasing volume and diversity of radiological information.1 Controlled terminologies—lists of standard terms describing a domain—and an architecture for integrating such terminologies into radiological applications are essential components that will be needed by future systems to manage the diversity of radiological information. Controlled terminologies permit computer applications to recognize that multiple synonymous terms refer to the same entity; they permit radiologists to add structure to radiological information to facilitate data mining and query, and they give developers capabilities needed to create a diversity of radiology applications, such as structured reporting, coded radiology teaching files, and decision support.2–6
The radiology community is building RadLex,7 a controlled terminology for radiology reporting, teaching, and research.8 RadLex terms are being acquired by experts to describe the domain of radiology, with the goal of disseminating standardized terminology to researchers, radiologists, and developers to facilitate analysis of radiological information, to permit uniform indexing of image libraries, and to enable new applications for structured capture of image information.
There are significant obstacles to the development and use of terminologies such as RadLex:
Tool support for creating RadLex. RadLex is being acquired using text files and spreadsheets, formats that simplify the task of collecting terms by radiology experts. However, these formats make it difficult to browse, manage, and modify the large RadLex hierarchy as it grows.
Maintenance of RadLex. The flat-file formats of RadLex are cumbersome for analyzing it from a structural perspective—for detecting omissions, duplications, and inconsistency (“curation” of the terminology). As RadLex increases in size, maintaining it will become even more challenging.
Deploying RadLex in applications. For RadLex to be successfully deployed, it must be shared among diverse computer applications. These include applications that display RadLex to users as well as programs that use RadLex terms for tasks such as structured reporting, teaching file coding, and indexing research data. Because RadLex changes over time, it can be cumbersome to update all client applications whenever RadLex changes.
A suitable software architecture is needed for managing and accessing terminologies such as RadLex and for addressing the above challenges. In addition to support for terminology creation and curation, a tool is needed to permit users to browse terminologies and to enable developers to incorporate RadLex in new terminology-driven radiology applications.
We are studying the use of Protégé (http://www.protege.stanford.edu), an open source ontology management suite of tools,9 for maintaining and developing RadLex and other terminologies pertinent to radiology. An ontology describes a domain and comprises terms, attributes of those terms, and relationships among the terms. A terminology can be represented as an ontology, and this representation permits the terminology to be accessed and analyzed with advanced tools such as Protégé.
Our hypothesis is that representing RadLex in an ontology will facilitate RadLex development by (1) providing a shared model for both humans and machines to work with RadLex; (2) helping curators analyze RadLex to identify omissions and structural inconsistencies; (3) eliminating need for separate applications to author, view, and deploy RadLex; and (4) enabling developers to integrate RadLex in computer applications. In this paper, we highlight particular advantages that the ontology format and Protégé provide: (1) accessing source terminologies that are stored in flat file formats, (2) analyzing the terminology to identify omissions and duplications, and (3) deploying RadLex on the Internet using a Web server extension.
METHODS
Accessing Source Terminologies
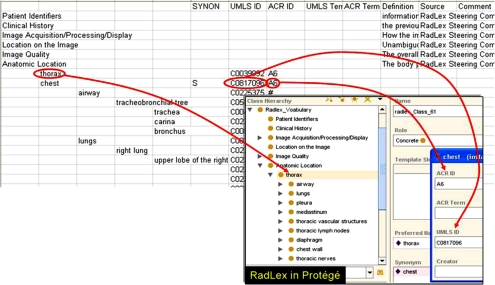
We downloaded an initial release of RadLex in a text file format (2005 release of the chest anatomy subset). This format could be viewed using a spreadsheet application, and the hierarchy was represented in the spreadsheet as an indented list of terms (Fig. 1). RadLex, like other terminologies, consists of terms and attributes of terms. There are two types of RadLex terms—preferred terms and synonyms. Preferred terms are those that experts consider to be the preferred nomenclature, whereas synonyms are alternate ways of naming the preferred terms. The attributes of RadLex terms describe ancillary information, such as the definition of the term, mappings of the term to related terms in other terminologies, and provenance information.
Fig 1.
Constructing an ontology of RadLex from the text format in which it was originally acquired. RadLex was initially acquired in a spreadsheet (upper half), with the terminology structure represented as indented terms (left half of spreadsheet). The attributes for each term is recorded using additional columns, such as synonym, UMLS ID, ACR ID, etc. (column labels shown in right half of spreadsheet). The ontology version of RadLex was produced by creating a class for each term in the RadLex hierarchy and assigning the appropriate attributes for each term (arrows). The resulting ontology can be browsed by expanding and collapsing entire branches of the ontology, and double-clicking on classes to browse the terminology (inset, lower right).
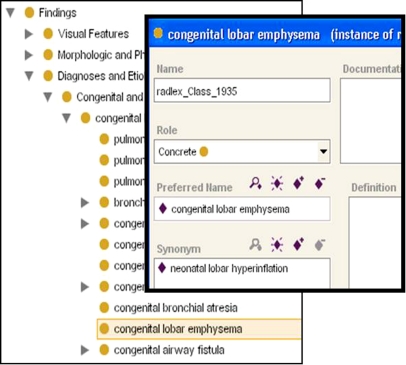
We created an ontology representation of RadLex in Protégé, an ontology development platform.9 Protégé is an open-source, free tool for creating and managing ontologies, as well as for developing applications that use them (http://www.protege.stanford.edu). We built the RadLex ontology by writing a software script that mapped the representational primitives in Protégé to those in the RadLex vocabulary. The Protégé knowledge model consists of classes, instances of those classes, and slots (attributes of classes and instances).10 We created an ontology class in Protégé for each RadLex term (preferred terms as well as synonym terms) and mapped each attribute value for a term in RadLex to the appropriate slot value for the corresponding class in the Protégé ontology (Fig. 1). We assigned the appropriate synonym terms as slot values to each ontology class; in this manner, the RadLex class hierarchy in Protégé contained only preferred terms, and each term referenced all of its synonyms (Fig. 2). These mapping operations were implemented in a Python script that was executed using a scripting component of Protégé.11 This scripting component permits interactive access to ontologies and immediate perusal of the imported ontology in the Protégé GUI, greatly facilitating debugging.
Fig 2.
RadLex in Protégé. RadLex ontology imported into Protégé, showing a hierarchy of classes, representing RadLex preferred terms (left). Each RadLex class contains slots that contain information about the term, such as the preferred name and synonyms. For example, we see that the RadLex term selected in the ontology (left) has a preferred name “congenital lobar emphysema” and a synonym “neonatal lobar hyperinflation” (detail of selected term shown on right).
We provided our ontology and Protégé tool to the RadLex developers to evaluate for qualitative feedback on the usability of the tool and its suitability for browsing and managing RadLex.
Analyzing the RadLex Ontology
We wrote scripts in Python to analyze the RadLex ontology to identify missing term attributes and to find duplicate terms. The scripts performed their actions by visiting all terms in the ontology tree structure and interrogating attribute values. If an attribute value for a term was missing, then the term was recorded. In addition, if a duplicate term name was identified, the term was recorded.
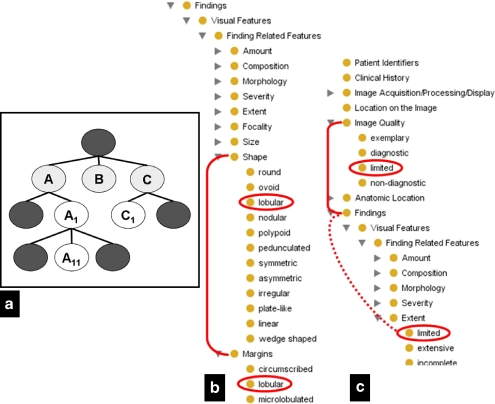
The ontology was also used to measure the distance between duplicate terms in the ontology tree structure to identify semantically similar branches within the RadLex ontology. In ontologies, all classes that are children of a particular class could be similar semantically and overlap in meaning, which require the ontology to be revised to make the distinctions more crisp. If the children of classes at the same level of the ontology are duplicated, this provides evidence that those classes are similar (Fig. 3), and ontology curators may want to scrutinize this portion of the ontology to make sure that there is good semantic separation in those classes (in addition to removing the duplicate classes). We measured the distance between the duplicate terms we found in RadLex by calculating the depth in the RadLex tree one would need to traverse to arrive at the common parent of the duplicate terms. If the duplicate terms had the same depth, this suggested that the parents of those terms were similar semantically.
Fig 3.
Analyzing RadLex ontology structure. The location of duplicate terms in the RadLex ontology structure can suggest whether different branches of the ontology are similar semantically. a In this generic ontology, terms A, B, and C are at the same level of the ontology tree structure and could be similar. If the children of these terms (e.g., A1 and C1) are duplicates, this suggests that the parent terms (A and C) are similar semantically, although not if the duplicate terms lie at different levels below the parents (e.g., A11 and C1). b Fragment of RadLex ontology showing that the duplicate term lobular is a child of terms at the same level of the ontology tree (shape and margins), suggesting that the shape and margins RadLex terms are similar semantically. c In this fragment of the RadLex ontology, the term limited is duplicated but lies at different depths of the tree below the terms Image Quality and Findings, providing no additional information about whether the latter two terms are similar semantically.
Deploying the Ontology on the Internet
The RadLex vocabulary is currently accessible on the Web; however, the RadLex Web application uses its own internal representation of RadLex. When a new version of RadLex is created, the Web application for viewing RadLex must be updated. We sought to explore the ability of Protégé to streamline the distribution of RadLex by providing simultaneous editing and Web browsing of ontologies.
We implemented the WebProtégé extension, a Web application that allows users to access and browse ontologies in Protégé over the Internet. The application runs in a Web server environment, and it provides a Web-based graphical display of ontologies. WebProtégé permits many users in cyberspace to browse ontologies in a similar manner to the desktop Protégé GUI. WebProtégé also permits text-based searches of ontologies for finding concepts and instances. The same ontology file that RadLex curators edit can be simultaneously disseminated and accessed on the Web WebProtégé. We deployed the RadLex ontology on the Web using WebProtégé and compared qualitatively the maintenance process to that required with current RadLex Web application.
RESULTS
RadLex could be browsed easily in the graphical editing environment of Protégé, in which the terminology is represented as an ontology and shown to the user as an expandable tree, with child terms shown indented below parent terms (Fig. 2). When a term is selected, all attributes of a term could be displayed and edited. The RadLex developers reported that this format and display made the terminology easier for curators to manage than using the spreadsheet format, although content authors may still prefer a spreadsheet for acquiring new terms. Following an introductory session with Protégé and some time working with RadLex in the Protégé environment, the RadLex developers decided to adopt Protégé for managing RadLex.
The scripts that analyzed the RadLex ontology revealed that of the 1,326 RadLex terms, 23 were synonymous terms. There were 1,274/1,326 (96%) terms with no definition, but 595/1,326 (45%) of the terms had a source. No terms were missing a RadLex unique identifier. There were 93 terms with an ACR identifier, whereas only 16 terms had an ACR term name. Only 16% (216 out of 1,326) of RadLex terms were associated with a UMLS concept.
Structural analysis of the RadLex ontology tree structure revealed a total of 17 duplicate terms, each duplicate term appearing twice in the RadLex tree structure. For example, the terms “lobular” and “limited” occurred in two different places in RadLex. In 15 out of 17 of the cases, the duplicate terms lay within separate subtrees at a depth of 1–3 terms in the ontology tree (for example, “limited”; see Fig. 3). In the other two cases, the duplicate terms appeared in the same subtree, suggesting that the subtrees have similar semantics (for example, “lobular”; see Fig. 3).
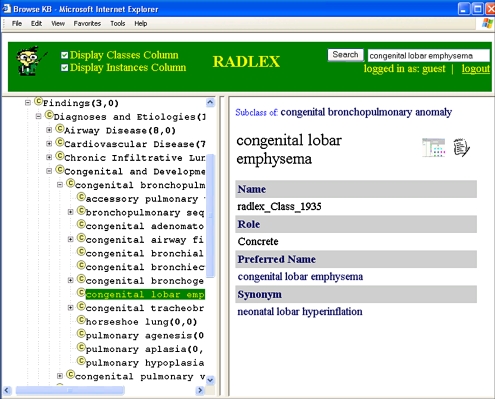
The RadLex ontology was successfully deployed on the Web using the WebProtege extension, accessing the same RadLex file that was being managed by the content authors. Multiple concurrent users could browse RadLex in a Web browser, presenting a GUI similar to that of Protégé itself (Fig. 4). Modifications that were made to the RadLex ontology locally were simultaneously reflected in the Web display once the server cache was refreshed, eliminating the need to update a separate Web application as the terminology evolves. WebProtégé appeared to simplify the process of deploying RadLex on the Web compared with the current RadLex Web application because it was not necessary to modify WebProtege whenever RadLex changed.
Fig 4.
RadLex ontology accessed over the Internet using WebProtege. This application provides Web access to ontologies; a single ontology file can be edited by curators (Fig 2) and immediately deployed on the Web without needing to update any software applications. The ontology is shown on the left and details about selected terms on the right, similar to that provided by the Protégé editing tool (Fig 2).
DISCUSSION
The RadLex project is an effort to develop a standard terminology for radiology. It began on a limited scale, focusing on a subset of the radiology domain (chest radiology). The vision of RadLex is to be comprehensive, it has since expanded to all of radiology, and the first version of RadLex has been released recently. As RadLex has grown, several key challenges have emerged:
Terminology management. It has been cumbersome managing RadLex. Text files and spreadsheets simplify the task of collecting terms, but it is difficult to browse, manage, and modify large terminology hierarchies in these formats.
Terminology quality control. Terminologies need to be curated to detect omissions, duplications, and other problems. It is necessary to browse large-term taxonomies to make sure terms are in the correct place. These tasks are difficult using flat file formats.
Terminology dissemination. For a terminology to be ultimately successful, it must be made available to the community in ways they can easily access and use it. Terminologies change over time, and it is cumbersome to update the applications that disseminate the terminologies when new versions are released.
Our work addresses these challenges to creating and deploying terminologies by converting RadLex from the current flat file representation into an ontology format and by adopting the Protégé suite of ontology management tools. RadLex is a terminology, comprising a lexicon of terms. Ontologies are similar to terminologies, although they provide a general representation formalism for specifying terms and the relationships among terms that is not committed to a specific storage syntax. The ontology formalism is advantageous for several reasons. First, RadLex contains much rich information about terms (the term attributes), and an ontology represents this information in an intuitive manner compared with the flat file formats of RadLex (Fig. 1). Second, users can use existing ontology tools to display, query, and navigate this information. In our work, we used Protégé to manage the RadLex ontology, although other ontology editing tools could be used.12, See footnote1 The particular advantages of Protégé are its rich set of plugins that provide ontology visualization and ontology access through the Protégé scripting tab. In addition, the WebProtégé extension enables widespread dissemination of RadLex on the Web (Fig. 4).
A third reason the ontology representation for RadLex is advantageous is the ability to unify terminologies across file formats. The existing medical terminologies are stored in a diversity of file formats.13 As ontologies provide an abstract model of terms and relationships, we will be able to unify RadLex with other terminologies currently stored in many different terminology formats. Thus, the ontology representation of RadLex will permit it to be enhanced by incorporating references to a variety of existing and related terminologies. In fact, there has already been preliminary work undertaken to harmonize RadLex with Systematized Nomenclature of Medicine. Recently, the Web Ontology Language (OWL)14 has been recommended by W3C as a standard for knowledge interchange. Protégé provides rich support for the OWL language,15 and it could permit RadLex to be translated into OWL in the future, enabling RadLex to play a role in radiology applications on the “Semantic Web.”16
A fourth advantage of ontologies is that they are human-readable and machine-processable. The original flat file format for RadLex was friendly to people, but not to machines that need to visualize and manage RadLex. The ontology format of RadLex will enable software developers to more readily use RadLex in new applications. For example, we showed in this work that the ontology format of RadLex could be used in the WebProtege application to permit the community to browse the terminology on the Web.
While in this work we have argued the advantages of ontology for RadLex and other radiology terminologies, the use of ontology is already widespread in biomedical domains outside of radiology. Biomedical ontologies are now a common tool in the basic sciences; the Gene Ontology17 is providing the foundation for creating a computable description of biological knowledge related to genes, and similar ontologies are being developed to describe microarray experiments, proteomics, and clinical guidelines.18 A key benefit of these biomedical ontologies is reuse: They are separate from application code that uses them, they can be maintained and extended separately from that code, and they can be shared among many different applications. This separation of ontologies from applications permits both the commercial and public sectors to cooperate in developing these ontologies as a public effort, improving them for the benefit of the entire scientific community. We believe that similar benefits will accrue for radiology ontologies such as RadLex. In fact, ontologies could become a critical technology for the TRIP initiative because they can enable developers to create many different data-intensive radiology applications.
A particular advantage of the ontology formalism is that it enables automated routines to be created that identify potential problems in the terminology and bring them to the attention of curators. Our analysis of the RadLex ontology identified several issues that have been subsequently corrected. Analyses such as finding the distance between duplicate terms and identifying semantically similar branches could be readily performed with the ontology form of RadLex (Fig. 3); such analyses would have been difficult in original flat file format of RadLex.
In this work, we have not exhaustively explored all the types of terminological analyses that might be possible. It would certainly be possible to evaluate term consistency across other relationships. Our objective is to demonstrate the value of the ontology representation of the terminology in enabling error checking routines to traverse the ontology components in carrying out the terminology checks.
In detecting duplicate terms, our approach was to find two or more occurrences of an identically named term. However, as some terms can have different meanings in different contexts, not all cases of duplicate terms represent mistakes in the lexicon. For example, in breast imaging, the term “spiculated” may be used to describe the type of margin on a mammogram and correspond the particular descriptor in the Breast Imaging Reporting and Data System (BI-RADS) vocabulary.19 The same term may also be used in lung imaging, and in this context, it would not have the associated BI-RADS code. Accordingly, two occurrences of “spiculated” in the lexicon would be appropriate. Linguistically, this is referred to as “polysemy”—a term that has multiple, related meanings. It would still be useful to RadLex developers to know about the duplicate terms even in cases of polysemy, as they may choose to rename the terms to make their meaning clear to users of the lexicon; for example, the two occurrences of “spiculated” may be renamed to “spiculated breast lesion” and “spiculated lung lesion.”
Our work addresses the problem of terminology concurrency, an issue that arises when multiple copies of a terminology diverge. RadLex is in constant evolution, yet applications such as the RadLex Web browser access a single particular version of the terminology; when new versions of RadLex are released, such applications need to be updated. We have shown that an alternative is to maintain a single central version of RadLex and disseminate it via the WebProtégé extension, which accesses the current version of RadLex at all times. In the Protégé client–server architecture, users and applications read a single central version of ontologies, simplifying their dissemination and ensuring applications can always access the current version. The WebProtege extension gives users access to the same version of RadLex that developers curate, so the need to synchronize versions of the terminology is avoided.
A potential limitation of using ontologies for terminology management is that some users may not find Protégé intuitive to use, or they may experience a learning curve. In fact, RadLex was originally built using flat files to keep the term acquisition process as simple as possible. We are not advocating Protégé or other ontology tools for all users, although we anticipate that terminology curators and developers will find it a useful tool. Other users could continue to work with terminologies in simpler formats and import their work into Protégé; in fact, we created a script to import RadLex flat files into Protégé precisely for this purpose.
While the current work focuses on creating the ontology, many in the radiology community are interested in applications. Future work will be to create ontology-driven applications that use RadLex as a knowledge resource. Such applications could integrate information in the Picture Archiving and Communications Systems (PACS) workstation20 or enable information search and teaching file coding. Tools that enable users to leverage the rich knowledge in RadLex will be helpful in creating the next generation of applications that will assist radiology researchers, practitioners, and educators.
CONCLUSION
Our results suggest that vocabularies such as RadLex can be translated into an ontology and that this representation can permit computational analysis that can help curation efforts to identify omissions, inconsistencies, and redundancies in the terminology. The ontology is a representation that permits users and applications to access the terminology content from a single centralized source. We have demonstrated particular benefits of using Protégé for this purpose, it may help curators improve the quality of their ontologies and vocabularies, and it may also enable developers to exploit controlled terms in new ontology-driven applications.
Acknowledgment
We wish to thank Curtis P. Langlotz, MD, PhD, and Beverly Collins, PhD, for their assistance and support with RadLex and for their outstanding efforts in creating this rich resource.
Footnotes
See http://www.xml.com/2002/11/06/Ontology_Editor_Survey.html for a recent large survey of ontology editing tools.
References
- 1.Andriole KP, et al. Addressing the coming radiology crisis-the Society for Computer Applications in Radiology transforming the radiological interpretation process (TRIP) initiative. J Digit Imaging. 2004;17:235–243. doi: 10.1007/s10278-004-1027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langlotz CP, Caldwell SA. The completeness of existing lexicons for representing radiology report information. J Digit Imaging. 2002;15(Suppl 1):201–205. doi: 10.1007/s10278-002-5046-5. [DOI] [PubMed] [Google Scholar]
- 3.Sinha U, et al.: Evaluation of SNOMED3.5 in representing concepts in chest radiology reports: integration of a SNOMED mapper with a radiology reporting workstation. Proc AMIA Symp:799–803, 2000 [PMC free article] [PubMed]
- 4.Starren J, Johnson SM: Expressiveness of the Breast Imaging Reporting and Database System (BI-RADS). Proc AMIA Annu Fall Symp:655–659, 1997 [PMC free article] [PubMed]
- 5.Reiner BI, Knight N, Siegel EL. Radiology reporting, past, present, and future: the radiologist’s perspective. J Am Coll Radiol. 2007;4:313–319. doi: 10.1016/j.jacr.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 6.Alberdi E, Taylor P, Lee R, Fox J, Sordo M, Todd-Pokropek A: Cadmium II: acquisition and representation of radiological knowledge for computerized decision support in mammography. Proc AMIA Symp:7–11, 2000 [PMC free article] [PubMed]
- 7.Langlotz CP. RadLex: a new method for indexing online educational materials. Radiographics. 2006;26:1595–1597. doi: 10.1148/rg.266065168. [DOI] [PubMed] [Google Scholar]
- 8.RadLex Steering Subcommittee: RadLex: A lexicon for uniform indexing and retrieval of radiology information resources. http://www.rsna.org/radlex/
- 9.Musen MA, Fergerson RW, Noy NF, Crubezy M: Protege-2000: A plug-in architecture to support knowledge acquisition, knowledge visualization, and the semantic Web. J Am Med Inform Assoc:1079–1079, 2001
- 10.Noy NF, Fergerson RW, Musen MA. The knowledge model of Protege-2000: combining interoperability and flexibility. Lect Notes Artif Int. 2000;1937:17–32. [Google Scholar]
- 11.Dameron O: JOT: a scripting environment for creating and managing ontologies. Proc. 7th International Protégé Conference, Stanford, CA
- 12.Lambrix P, Habbouche M, Perez M. Evaluation of ontology development tools for bioinformatics. Bioinformatics. 2003;19:1564–1571. doi: 10.1093/bioinformatics/btg194. [DOI] [PubMed] [Google Scholar]
- 13.Cimino JJ. Review paper: coding systems in health care. Methods Inf Med. 1996;35:273–284. [PubMed] [Google Scholar]
- 14.Web ontology language (OWL) reference version 1.0. http://www.w3.org/tr/owl-guide/
- 15.Knublauch H, Fergerson RW, Noy NF, Musen MA: The Protege OWL Plugin: an open development environment for Semantic Web applications. Semantic Web—Iswc 2004 Proceedings 3298:229–243, 2004
- 16.Berners-Lee T, Hendler J, Lassila O. The Semantic Web: Scientific American. New York: Scientific American; 2001. [Google Scholar]
- 17.Harris MA, et al. The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.OBO: Open Biological Ontologies. http://obo.sourceforge.net
- 19.Breast imaging atlas. Reston, VA: American College of Radiology; 2003. [Google Scholar]
- 20.Kahn CE, Jr, Channin DS, Rubin DL. An ontology for PACS integration. J Digit Imaging. 2006;19:316–327. doi: 10.1007/s10278-006-0627-3. [DOI] [PMC free article] [PubMed] [Google Scholar]