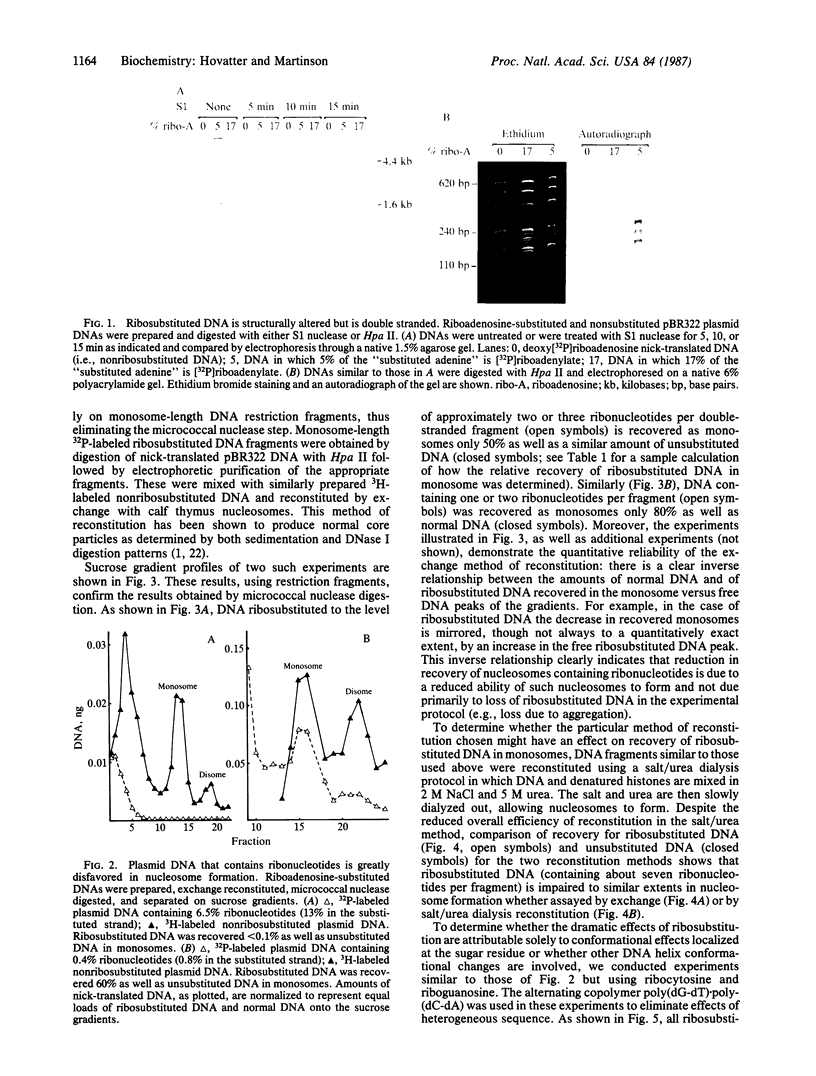
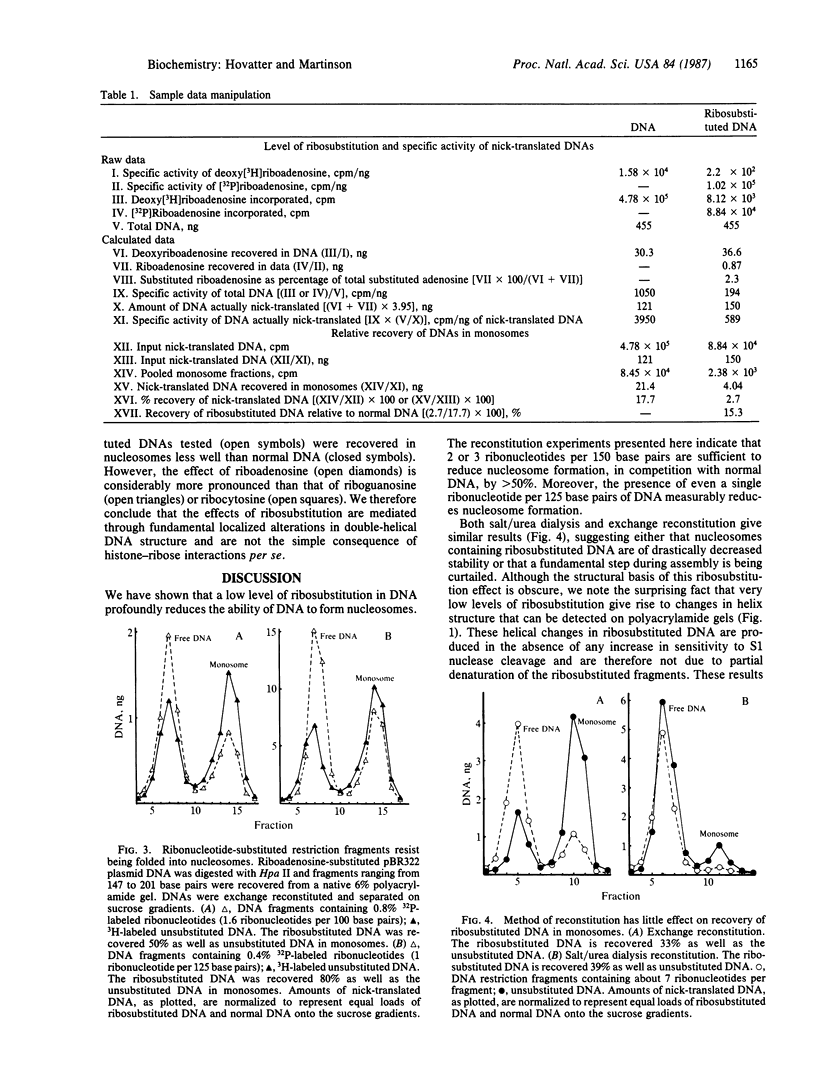
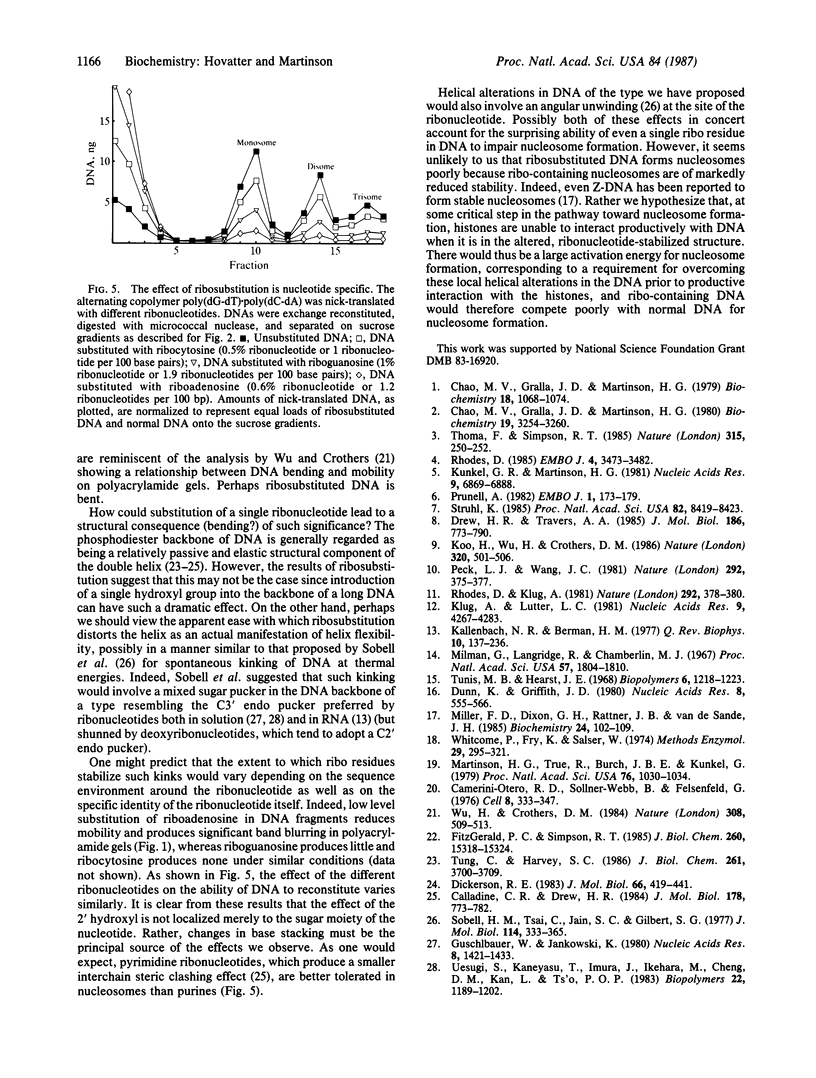
Abstract
Several polynucleotides that assume an A-form helical structure in solution are unable to form nucleosomes. We attempted to establish a relationship between the ease of the A-form----B-form helix transition and ease of nucleosome formation by reconstituting nucleosomes using ribosubstituted DNA containing various levels of ribonucleotides. Instead we discovered that, when riboadenosine is substituted for deoxyriboadenosine, even one ribonucleotide per 125 base pairs of DNA reduces nucleosome formation and that DNA containing greater than 5% ribonucleotide is completely unable to form nucleosomes. Ribosubstituted DNA restriction fragments exhibited altered mobility on native 6% polyacrylamide gels, indicating an altered helical structure (probably bending). The effects on both nucleosome formation and gel mobility are nucleotide specific and are correlated, being greatest for riboadenosine and decreasing in the order riboadenosine greater than riboguanosine greater than ribocytosine. The results are consistent with the hypothesis that the rate of nucleosome formation can be drastically reduced by isolated local perturbations, such as kinking or bending, in the helical structure of DNA.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Calladine C. R., Drew H. R. A base-centred explanation of the B-to-A transition in DNA. J Mol Biol. 1984 Sep 25;178(3):773–782. doi: 10.1016/0022-2836(84)90251-1. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976 Jul;8(3):333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Chao M. V., Gralla J. D., Martinson H. G. lac Operator nucleosomes. 1. Repressor binds specifically to operator within the nucleosome core. Biochemistry. 1980 Jul 8;19(14):3254–3260. doi: 10.1021/bi00555a024. [DOI] [PubMed] [Google Scholar]
- Chao M. V., Gralla J., Martinson H. G. DNA sequence directs placement of histone cores on restriction fragments during nucleosome formation. Biochemistry. 1979 Mar 20;18(6):1068–1074. doi: 10.1021/bi00573a021. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E. Base sequence and helix structure variation in B and A DNA. J Mol Biol. 1983 May 25;166(3):419–441. doi: 10.1016/s0022-2836(83)80093-x. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Dunn K., Griffith J. D. The presence of RNA in a double helix inhibits its interaction with histone protein. Nucleic Acids Res. 1980 Feb 11;8(3):555–566. doi: 10.1093/nar/8.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald P. C., Simpson R. T. Effects of sequence alterations in a DNA segment containing the 5 S RNA gene from Lytechinus variegatus on positioning of a nucleosome core particle in vitro. J Biol Chem. 1985 Dec 5;260(28):15318–15324. [PubMed] [Google Scholar]
- Guschlbauer W., Jankowski K. Nucleoside conformation is determined by the electronegativity of the sugar substituent. Nucleic Acids Res. 1980 Mar 25;8(6):1421–1433. doi: 10.1093/nar/8.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallenbach N. R., Berman H. M. RNA structure. Q Rev Biophys. 1977 May;10(2):138–236. doi: 10.1017/s0033583500000202. [DOI] [PubMed] [Google Scholar]
- Klug A., Lutter L. C. The helical periodicity of DNA on the nucleosome. Nucleic Acids Res. 1981 Sep 11;9(17):4267–4283. doi: 10.1093/nar/9.17.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Kunkel G. R., Martinson H. G. Nucleosomes will not form on double-stranded RNa or over poly(dA).poly(dT) tracts in recombinant DNA. Nucleic Acids Res. 1981 Dec 21;9(24):6869–6888. doi: 10.1093/nar/9.24.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinson H. G., True R., Burch J. B., Kunkel G. Semihistone protein A24 replaces H2A as an integral component of the nucleosome histone core. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1030–1034. doi: 10.1073/pnas.76.3.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller F. D., Dixon G. H., Rattner J. B., van de Sande J. H. Assembly and characterization of nucleosomal cores on B- vs. Z-form DNA. Biochemistry. 1985 Jan 1;24(1):102–109. doi: 10.1021/bi00322a015. [DOI] [PubMed] [Google Scholar]
- Milman G., Langridge R., Chamberlin M. J. The structure of a DNA-RNA hybrid. Proc Natl Acad Sci U S A. 1967 Jun;57(6):1804–1810. doi: 10.1073/pnas.57.6.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Sequence dependence of the helical repeat of DNA in solution. Nature. 1981 Jul 23;292(5821):375–378. doi: 10.1038/292375a0. [DOI] [PubMed] [Google Scholar]
- Prunell A. Nucleosome reconstitution on plasmid-inserted poly(dA) . poly(dT). EMBO J. 1982;1(2):173–179. doi: 10.1002/j.1460-2075.1982.tb01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Sequence-dependent helical periodicity of DNA. Nature. 1981 Jul 23;292(5821):378–380. doi: 10.1038/292378a0. [DOI] [PubMed] [Google Scholar]
- Rhodes D. Structural analysis of a triple complex between the histone octamer, a Xenopus gene for 5S RNA and transcription factor IIIA. EMBO J. 1985 Dec 16;4(13A):3473–3482. doi: 10.1002/j.1460-2075.1985.tb04106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobell H. M., Tsai C. C., Jain S. C., Gilbert S. G. Visualization of drug-nucleic acid interactions at atomic resolution. III. Unifying structural concepts in understanding drug-DNA interactions and their broader implications in understanding protein-DNA interactions. J Mol Biol. 1977 Aug 15;114(3):333–365. doi: 10.1016/0022-2836(77)90254-6. [DOI] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Simpson R. T. Local protein-DNA interactions may determine nucleosome positions on yeast plasmids. Nature. 1985 May 16;315(6016):250–252. doi: 10.1038/315250a0. [DOI] [PubMed] [Google Scholar]
- Tung C. S., Harvey S. C. Base sequence, local helix structure, and macroscopic curvature of A-DNA and B-DNA. J Biol Chem. 1986 Mar 15;261(8):3700–3709. [PubMed] [Google Scholar]
- Tunis M. J., Hearst J. E. Optical rotatory dispersion of DNA in concentrated salt solutions. Biopolymers. 1968;6(8):1218–1223. doi: 10.1002/bip.1968.360060816. [DOI] [PubMed] [Google Scholar]
- Whitcome P., Fry K., Salser W. The use of ribosubstitution techniques for determining DNA sequences. Methods Enzymol. 1974;29:295–321. [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]