Abstract
Bronchiectasis is an airway disease caused by the dilatation of the bronchial tree, and a bronchovascular pair is formed between a bronchus and a vessel. An abnormal bronchovascular pair is one that has a larger bronchus compared to its accompanying vessel. Typically, bronchi and vessels running perpendicular to the plane of section appear as near-circular rings on computed tomography (CT) scans. This paper describes BV_pairs, a system capable of detecting abnormal bronchovascular pairs in high-resolution CT scans of sparse datasets using a three-stage process: (1) detection of potential bronchovascular pairs, (2) detection of discrete pairs, where there exists no ambiguity as to the artery that accompanies a bronchus, and (3) identification of abnormal pairs with severity levels. The system was evaluated at every stage. The automated scoring for the presence and severity of bronchial abnormalities was demonstrated to be comparable to that of an experienced radiologist (i.e., kappa statistics κ > 0.5). In addition, BV_pairs was also evaluated on images containing honeycombing regions, since honeycombing cysts appear very similar to bronchi, and the system could successfully differentiate honeycombing cysts from bronchi.
Key words: Algorithms, classification, computed tomography, computer-aided diagnosis (CAD), feature extraction
Introduction
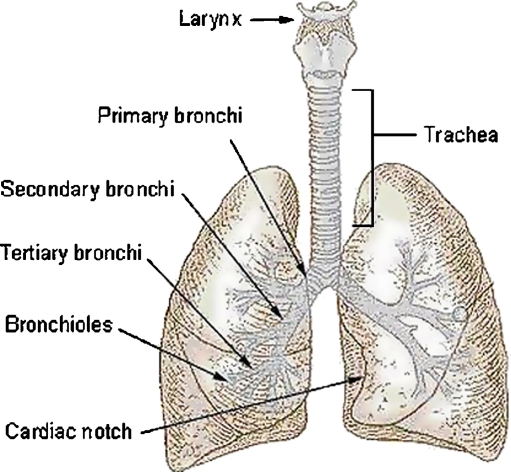
The human airway system comprises the trachea that bifurcates into two main bronchi, one for each lung as shown in Figure 1. Each bronchus further divides into lobar bronchi, which in turn divide into segmental bronchi. Each lung also contains pulmonary arteries that supply blood. The arteries descend along with the main bronchus and further divide into lobar and segmental arteries. Inside the lung, the artery and the bronchi are paired together and branch simultaneously, running on parallel courses. Together, they form the bronchovascular bundle. Bronchiectasis is defined as localized, irreversible dilatation of the bronchial tree.1 On computed tomography (CT) images, near-perpendicular cross-sections of bronchi and the vessels appear as elliptical high-attenuation rings as shown in Figure 2.
Fig 1.
The airway system in the lungs.2
Fig 2.
a Example of a bronchovascular pair. The circular black region corresponds to the bronchus, and the adjacent white region corresponds to the artery. Panel b contains a normal bronchovascular pair seen on a HRCT scan.
One of the most important direct signs for bronchiectasis is the dilatation of the bronchi. A bronchus is enlarged when its size is significantly larger than the size of its adjacent artery, and together, they constitute a signet ring sign. In normal lungs, the size of the adjacent artery is equal to or slightly larger than that of the accompanying bronchus.
Detection of bronchovascular pairs, abnormal or normal, is a challenging problem for many reasons. The deeper airways that are small and thin inside the lungs can appear broken or discontinuous on high-resolution CT (HRCT) scans. Despite improvements in image quality achieved by thin slices and advances in the reconstruction algorithm, if the long axis of the airway is not perpendicular to the imaging plane, its detection is limited by the partial volume effect. Contrast in a CT image is determined by the differential absorption of X-rays by neighboring structures. The airway wall separates the cavity of the airway from the surrounding lung tissue, and thus, we would expect a high contrast border around the cavity, also called the lumen. If the airway orientation tilts toward the image plane, the density values inside the airway cavity increase due to partial volume averaging of air and the airway wall. The apparent airway lumen density increases substantially with decreasing airway diameter due to partial volume effects as well. In addition, cardiac motion that takes place during scanning (i.e., the movement of the heart that shifts the adjacent lung) causes a distorted appearance of bronchovascular pairs. All of these factors make detection of bronchovascular pairs a challenging task.
Very often, a bronchus is accompanied by more than one vessel, or when the bronchus is not completely branched, more than one bronchus may appear together. A bronchovascular pair is said to be discrete if there exists no ambiguity as to the artery that accompanies a bronchus. The clinical challenge involved in signet ring detection lies in the identification of discrete bronchovascular pairs. The identification of grouped pairs on HRCT scans is very complex (i.e., when they are not branched) due to the ambiguity associated with grouped pairs. Examples of discrete bronchovascular pairs and groups of pairs are presented in Figure 3. The task of identifying the abnormality becomes easier once the individual pairs are successfully identified. This work takes this approach and does not address grouped pair detection.
Fig 3.

Panel a contains a discrete bronchovascular pair. Panels b and c show an example where the bronchovascular pairs are grouped (or not branched). The images are zoomed at different magnitudes.
It is important to recognize that while most accurate airway measurements may ultimately be based on three-dimensional (3D) data analysis, in many clinical situations, it is impractical to routinely gather and analyze 3D data. The purpose of this paper is to present a fully automated system that is capable providing accurate detection of bronchial dilatation on sparse datasets containing HRCT lung images. From a clinical perspective, the proposed technique is intended to direct the radiologists’ attention to significant sets of pairs from which diagnostic information can be obtained.
In this paper, a system capable of identifying abnormal bronchovascular pairs called BV_pairs is presented. BV_pairs identifies potential pairs first and then follows it up by detecting discrete pairs. In the last stage, it compares the size of the bronchus and its adjacent artery to determine whether it is abnormal or not. In the next section, related work in the literature is reported. This is followed by a section on materials and methods. The system works in three stages and each of the stages are described in this section. Finally, discussion and conclusions are presented in “Discussion” and “Conclusion,” respectively.
Prior Work
There is very little work reported in the literature on the automated detection of bronchi on CT lung images. Semiautomated techniques require user interaction, based on user dependent grayscale thresholding and manual delineation of bronchial contours on CT images.3 One of the reasons for the paucity of research in the area is the difficulty of matching the subjective assessments made by experienced radiologists. Recently, Chabat et al.4 proposed a fully automated technique called Edge–Radius–Symmetry (ERS) based on the analysis of the distribution of gradient maxima and minima in the neighborhood of each pixel to identify the bronchus. A more detailed description of their approach is provided in the following sections. Heuristics-based and machine-learning techniques have been used to compare the performance of the detection of bronchovascular pairs,5–9 and results reported indicate that machine learning performed comparatively better than heuristic measures.
Sonka et al.10 describe a rule-based method for the segmentation of airway trees from 3D sets of CT images. The method is based on a combination of 3D seeded region growing that is used to identify large airways, rule-based two-dimensional (2D) segmentation of individual CT slices to identify probable locations of smaller diameter airways, and finally a merging of airway regions across the 3D set of slices resulting in a tree-like airway structure. Mori et al.11 used a 3D painting algorithm to extract the bronchus area by gradually increasing a region-growing threshold until the segmented region “leaks” into the lung. Similarly, Wood et al.12 presented a 3D seeded region-growing technique to segment the airway and vascular trees of excised, inflated canine lungs and performed quantitative measurements such as airway tree branch length and cross-sectional area. The area extracted just prior to the leak is identified as the bronchus area. Pisupati13 developed a technique that utilized grayscale mathematical morphology to detect the 3D airway tree, pulmonary artery, and pulmonary trees of isolated canine lungs. Prêteux et al.14 used a combination of morphological filtering, connection cost-based marking, and conditional watershed methods to segment the bronchi of sheep lungs. Aykac et al.15 segmented the airway tree automatically using a stack of 100–120 3-mm section slices to match the candidate airway regions (found on 2D slices) to construct the tree. Their segmentation technique identifies potential airway locations in the 2D cross-sectional images. The identification process involves a preprocessing step to remove noise followed by grayscale reconstruction and thresholding.
The work related to identification of abnormal bronchovascular pairs in the literature does not identify discrete pairs to classify them as being abnormal or normal, considering all bronchovascular pairs would result in ambiguous cases as identification of grouped pairs is very complex on HRCT scans. In addition, the experimentations carried out have been on normal patients or those having only bronchial abnormalities but not other disease findings. Our approach is different as BV_pairs aims to detect abnormal pairs after identifying discrete bronchovascular pairs. BV_pairs is also evaluated on two sets of patients: (1) normal patients or those having only bronchial abnormalities and (2) patients having honeycombing regions. Honeycombing regions were used to evaluate the robustness of the system because the appearance of honeycomb cysts is very similar to bronchi, as honeycombing regions comprise a cluster of circular cysts that appear predominantly along the peripheral regions, where bronchi are not normally expected.
Materials and Methods
Overall System
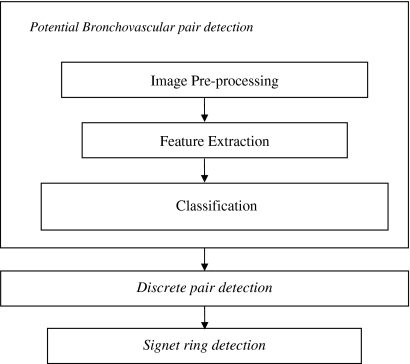
BV_pairs consists of three main modules: potential bronchovascular pair detection, discrete pair detection, and signet ring detection (as shown in Fig. 4). The potential bronchovascular pair detection module is aimed at detecting candidate pairs of bronchi and vessels. It comprises three steps to identify the pairs: image preprocessing to eliminate noise, feature extraction, and classification based on multiview learning and active learning. In a traditional pattern recognition scenario, algorithms have access to the entire set of features (also referred to as a single view) in the domain. By contrast, in the multiview setting, one can partition the domain’s features into subsets (views) that are sufficient to learn the target concept. Such algorithms employ a large number of unlabeled examples to boost the performance of a learning algorithm using a small set of labeled examples. In the multiview setting, one assumes that each view is sufficient to learn the target concept and sufficiently uncorrelated to the other view. Further, Nigam and Ghani16 have shown through extensive experiments that multiview-learning algorithms can outperform single-view algorithms even on tasks where there is no natural split of features.
Fig 4.
Block diagram of BV_pairs.
The first formalization of multiview-learning algorithms was provided by Blum and Mitchell17 when they presented a cotraining algorithm. The cotraining approach builds classifiers incrementally over each view by using a large number of unlabeled examples to boost the performance of a learning algorithm built with an initial small set of labeled examples. Specifically, the presence of two distinct views of each example suggests a strategy in which two learning algorithms are trained separately on one view each, and each algorithm’s predictions on new unlabeled examples are used to enlarge the training set of the other.
More recently, extensions to the cotraining algorithm have been provided by many researchers. Co-EM17 uses expectation–maximization (EM)18 for multiview learning. EM can be viewed as a single-view semisupervised learning algorithm by treating the unlabeled examples as having a hidden variable (class). Co-EM integrates cotraining and EM by using the hypothesis learned in one view to probabilistically label the examples in the other view. The primary difference between co-EM and cotraining is that like EM, co-EM assigns a temporary label to each unlabeled example from scratch at each iteration, whereas cotraining selects a subset of the unlabeled examples to permanently label. Additionally, corrected cotraining19 is a similar algorithm in which the informative examples are manually checked and corrected before being added to the training data. The technique is a combination of selective sampling and cotraining motivated by the fact that the quality of the bootstrapped data is the key factor in convergence. Coboost20 is another algorithm for learning in the cotraining setting; it tries to minimize the disagreement on the unlabeled data between classifiers that use different views of the data.
Co-EMT21 uses active learning (i.e., user intervention) when there is a disagreement on the unlabeled data between classifiers that use different views of the data. Krogel and Scheffer22 have explored the effectiveness of using cotraining in functional genomic data that includes relational information. The authors perform an experimental analysis where they show that cotraining fails to improve classification results. A deeper analysis showed that the conditions for performing multiview learning were not met. Research on multiview learning has been largely applied on text analysis. To our knowledge, this work is the first that applies multiview learning to detect local, structural patterns such as bronchovascular pairs on HRCT images.
At the first stage of BV_pairs, the aim is to detect all the visible bronchi on a given slice; it is easier to eliminate some of them with high-level processing in the next stage. In the second stage, discrete pairs of bronchi and vessels are identified through the use of high-level features obtained from the output of stage 1. This is followed by signet ring detection in the third stage where the size of the bronchus and its associated vessel are compared.
Detection of Potential Bronchovascular Pairs
Robust Cotraining with Active Learning
The well-known multiview-learning algorithm, cotraining together with active learning, is used to incrementally perform classification. The goal of active learners is to detect the most informative examples in the instance space and then to ask the user to label them, thus reducing the amount of labeling in the instance space. Active learning is incorporated into the multiview framework as shown in Figure 5. One of the views in the framework is based on relational features. Learning with attribute values has limitations in capturing concise and expressive knowledge structures. To recognize an object in an image, a vision system needs to have knowledge of the object appearance. This knowledge could be about the shape, location, or context in which the object may be found that may be captured by relations. First Order Inductive Learner (FOIL),23 a relational learning program, is used to learn object models after the features are extracted. An attractive feature of FOIL is that it is powerful enough to handle noisy data. FOIL produces output in Horn clauses (a subset of first-order logic) that are concise and comprehensive to the user. For example, the fact that a bronchovascular pair consists of a bronchus, a vessel, and a wall surrounding the bronchus could be expressed by:
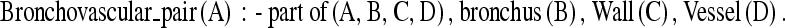 |
Fig 5.
Block diagram of multiview learning to detect potential bronchovascular pairs.
The other view is based on the ERS transform and the learner used is Naive Bayes.24 ERS is a technique used to segment the bronchus and its adjacent vessel on HRCT images. More detailed description of this technique is provided in the following sections. It is based on the analysis of the distribution of gradient maxima and minima in the neighborhood of each pixel. Naive Bayes is a simple probabilistic learner based on Bayes’ theorem24 and has been shown to be very effective at general probabilistic learning using CT images.25–27 To use the Naive Bayes learner, we estimate the prior distribution based on the distribution of the classes (i.e., a “correct” pair or an “incorrect” pair) in the training data. The posterior probability conditioned on the class is calculated using the Bayes theorem. Although the two views appear to be “naturally” partitioned, it has been empirically shown by Muslea et al.21 that active learning in the multiview framework can compensate for view correlation if present. In other words, active learning makes the system robust to the violation of the view correlation assumption.
By using active learning in the multiview learning framework, the user’s burden of labeling data is reduced since only the most informative examples are chosen. After training the two classifiers in each view with an initial training set and applying them to the unlabeled set, the informative examples can be selected by choosing the examples on which the classifiers disagree. Because the target concepts in each view must agree, the use of active learning in the multiview framework can reduce the hypothesis space much more than would otherwise be possible. There are two main advantages of using active learning in the multiview framework: (1) Through the use of two different feature spaces, the most informative examples can be sought; (2) by querying only contention points, the system is tailored to learn from mistakes (i.e., mistakes are more informative than correctly labeled examples).
Image Preprocessing
To extract appropriate features from lung regions, the images must be processed. There are essentially two main steps before features can be extracted: automatic segmentation of the lungs and preprocessing before extracting features. After the lungs are segmented and located, the image is first converted into a binary image by choosing an optimal threshold value as shown in Figure 6. The binary image is then despeckled to remove noise (i.e., regions that have area less than 5 pixels are eliminated). A region-growing algorithm is then applied to connected segments in the binary image. A stopping criterion is set for the region-growing algorithm for nearly closed objects as the deeper airways tend to appear broken or discontinuous on HRCT scans. The resulting image contains potential bronchi regions.
Fig 6.

Panel a corresponds to an original lung scan consisting of bronchovascular pairs and panel b contains the corresponding pre-processed binary image.
Parameter tuning is used to automatically learn the optimum threshold and region-growing stopping criterion through the use of labeled examples. The search space for these parameters was empirically chosen and was between −900 to −520 HU and 1 to 20 pixels, respectively. The parameter-tuning algorithm for finding the optimum parameters is as follows.
Parameter Tuning
For each image
Run the classifier L for the range of threshold values.
Run the model L for the range of region-growing stopping criterion values.
‘+’ve examples are the ones that correspond to the highest f1 measure of completeness and correctness.
Return mean of ‘+’ve examples giving the optimal threshold and region-growing stopping criterion values.
A model L built by the relational learner using an initial training set is used in the parameter-tuning algorithm. The examples corresponding to the highest f1 measure22 of completeness and correctness are used as positive examples. Completeness is also known as recall and sensitivity, while correctness is also known as precision in the pattern recognition literature. They are given by:
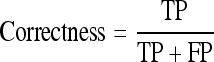 |
1 |
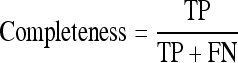 |
2 |
The f1 measure uses the harmonic mean between completeness and correctness and is given by Eq. 3. The mean value of these positive examples will provide the optimum threshold and region-growing stopping criterion.
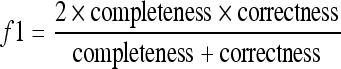 |
3 |
Feature Extraction
Feature extraction is the primary step in creating and recognizing object models in images. In detecting structures such as the bronchi, the features should not be sensitive to translation, rotation, and scale to cover the variability in HRCT images. The two sets of features needed to perform multiview learning are based on relational features and the ERS transform, respectively.
Relational Features. A relational learning system takes a series of relations among local features as input. The relations used are both unary and binary. Unary relations express properties of an object segment, and binary relations describe properties between two segments. The relations that the system can detect are the following:
Proximity between bronchus and vessel: Adjacent_to
Circularity of bronchus and vessel: Circularity
Thickness of wall surrounding the bronchus: Thickness
Location of object in the image: Near_Hilum
Ratio of area between bronchus and vessel: Area_Ratio
The symbolic relations extracted are input to a relational learner, FOIL in the format shown in Figure 7, and the concept to be learned is Bronchovascular_pair. Partof expresses the ownership of a segment by an object. In the case of bronchovascular pairs, partof is used to express that the object owns a bronchus, wall, and a vessel. Thickness corresponds to the wall thickness of the bronchus, which is averaged over eight directions from the center of the bronchus. A bronchus and a vessel are both near-circular structures. Circularity is calculated using the formula:
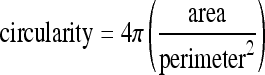 |
4 |
Circularity returns a number between 0 and 1 where 1 indicates a perfect circle. The location of the object in the lung of a pathology-bearing region has significance both clinically and for classification purposes. For instance, according to the medical experts in our team, large bronchovascular bundles predominantly occur near the pulmonary hilum, whereas honeycombing cysts, which look very similar to bronchus, occur predominantly along the periphery of the lung. A bronchovascular pair is said to be near_hilum if it is less than 30 pixels from the hilum. This relation is applied to the bronchovascular object (i.e., the distance to the hilum from the center of the bronchovcasular pair). The user enters a midpoint that separates the two lungs, which is used to construct the region of hilum if trachea bifurcation is present. The region along the lung boundary 100 pixels above and below the midpoint corresponds to the area around the hilum as shown in Figure 8.
Fig 7.
Relational features in FOIL format.
Fig 8.
Lungs segmented along with the hilum region marked in yellow (gray in print).
This is done because the lung segmentation algorithm tends to merge the two lungs or the lungs and the trachea together if they are very close to each other. Area_ratio is used to obtain the ratio between the area of the bronchus and that of the vessel. For a bronchovascular pair to exist, the area of both the bronchus and the vessel must be of a certain value. Adjacent_to is used to denote if two segments are connected. For instance, the bronchus wall and the accompanying vessel are adjacent to each other, and the bronchus and its wall are adjacent to each other. These are binary relations.
Edge–Radius–Symmetry. The second set of features is based on the ERS transform, introduced by Chabat et al.,4 which together with template matching was designed to segment the bronchus and its adjacent vessel on HRCT images. It is based on the analysis of the distribution of gradient maxima and minima in the neighborhood of each pixel. Bronchi that are elliptical or near-circular are symmetrical patterns relative to their centroids, and this property is characterized by the spatial distribution of the edges. Given the densities of air, bronchial wall, and lung tissue, the intensity gradient displays local maxima and minima at the points on the inner border (at the interface between airway and bronchial wall) and outer border (at the interface between bronchial wall and lung regions) of a bronchus, respectively. E, R and S measure edge strength, radial uniformity, and local symmetry, respectively. Features based on ERS constitute the second view.
ERS is computed along eight directions (with increments of 45° starting from the horizontal) from the center of a potential bronchus object. Since the typical size of the elliptical patterns to be identified in CT images is less than 12 pixels, it is possible to derive a precise approximation of them by considering only eight points, rather than considering all points on the ellipse. In addition to the E, R, and S features, the ratio of the maximum radius distance (corresponding to the difference between gradient maxima and minima) to the average radius distance is used as a feature to provide information about the adjacent vessel. The features are input to Naive Bayes to perform classification in the second view.
Classification using FOIL and Naive Bayes. FOIL is capable of producing Horn clause definitions of target concepts. A series of positive and negative examples are presented to FOIL for the target concept to be learned. FOIL is a general to specific learning system that searches the hypothesis space from the most general description of a concept to a specific one. The Naive Bayes classifier uses the Bayes theorem where the posterior probability of each class is computed using the feature values present in the instance. The Bayes theorem is expressed by:
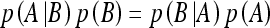 |
5.0 |
The instance is assigned to the class with the highest probability. Naive Bayes classifiers assume that the effect of a variable value on a given class is independent of the values of other variables. This assumption is called class conditional independence and is made to simplify the computation, hence the term “Naïve.”
Detection of Discrete Pairs
The second stage of BV_pairs is aimed at detecting discrete bronchovascular pairs. High-level features are extracted from the detected bronchovascular pairs in the first stage and are further processed. These were designed in consultation with medical experts based on clues that are normally used in a clinical setting. The features include proximity between the centers of nearest bronchi, nearest/common vessels, area, and circularity of the bronchus/vessel (see Fig. 9).
Fig 9.
Illustration of high-level measurements. The arrow corresponds to the distance between the two nearest bronchi. The region in red corresponds to the common vessel for the two bronchi.
The measurements are all performed at pixel level. The proximity between objects along with their area in the lung of a pathology-bearing region has significance both clinically and for classification purposes. For example, according to the medical experts, large bronchovascular pairs are predominantly grouped, whereas discrete pairs tend to be comparatively smaller in size. Classification was performed using the well-known decision tree algorithm, C4.5.24 Decision tree algorithms have several advantages over other classification algorithms. They are computationally not intensive, relevant attributes are selected automatically, and the classifier output is easy to interpret. The size of the bronchi included for evaluation ranged from 2.3 (3 pixels) to approximately 12 mm (18 pixels) on average.
Signet Ring Detection
The third stage of BV_pairs is aimed at identifying signet ring patterns. Because bronchiectasis is defined by the presence and/or absence of bronchial dilatation, recognition of increased bronchial size relative to that of the artery is the key to the CT diagnosis of this abnormality. The ratio of the diameters of the bronchus and its associated vessel along the minor axis and the ratio of the area of the bronchus and its associated vessel are used as features. The diameter is computed along the minor axis of the cross-sections of the bronchus and its accompanying vessel because these best represent the true size of the pairs in oblique situations. It has been observed in many cases that when measurement is performed at the pixel level, the bronchus and the accompanying vessels do not appear as near circles due to the plane of scanning. In addition, rounding effects from averaging the diameter across different directions can make the task of detecting bronchial dilatation a challenging task. The ratio of the area of the bronchus and its accompanying vessel was also used besides the diameter ratio as features to detect bronchial dilatation. Classification was performed using the decision tree algorithm, C4.5, to detect the presence and severity of the abnormality. Severity of bronchial dilatation is defined on a four-point scale (normal, mild, moderate, and severe) adapted from Webb et al.1 and illustrated in Table 1. These scales can be simplified to two-point scales scoring for the presence and or absence of bronchial dilatation.
Table 1.
Severity of Bronchial Dilatation adapted from Webb et al.1
| d ≤ 1 | 1 < d ≤ 2 | 2 < d ≤ 3 | d < 3 | |
|---|---|---|---|---|
| Dilatation | 0 (normal) | 1 (mild) | 2 (moderate) | 3 (severe) |
d is defined as the ratio of the bronchovascular diameter.
Experimental Test
Image Acquisition
Experiments were performed using 75 subjects randomly chosen from a research database.28 Studies were approved by the Human Research Ethics Committee at the University of New South Wales and Ethics Committee at Medical Imaging Australasia. The subjects were scanned at a thickness of 1 mm, and the images were spaced at 10 mm apart each. The images were obtained on a SIEMENS (Volume Zoom) scanner at 140 kVP, ~280 mAs, with a standard filter reconstruction algorithm. Manual labeling of bronchovascular pairs was performed by a radiologist specialized in diagnostic radiology with more than 30 years of experience. When labeling the bronchovascular pairs, the radiologist was shielded from viewing the results to avoid bias. Once the bronchovascular pairs were labeled, the dataset was split randomly into the different categories for experimental analysis as shown in Table 2.
Table 2.
Experimental Setup for Bronchovascular Pair Detection
| Category | Setup |
|---|---|
| Initial training set | 15 patients (24 images) |
| Parameter tuning set | 5 patients (9 images) |
| Cotraining set (unlabeled) | 23 patients (32 images) |
| Evaluation set—seven patients had honeycombing | 32 patients (42 images) |
Note that the sets of images are nonoverlapping.
BV_pairs was cotrained on 32 unlabeled HRCT images selected from 23 patients. It was observed that some images consisted of up to 12 bronchovascular pairs, depending on the position of the scan. Some images that do not contain any bronchovascular pairs were included in the experimental analysis to ensure minimum false positives. Our approach was to add to the initial training set, the most informative pairs, labeled actively, from a group of five HRCT images at each iteration. This was done to ensure variability in the cotraining process. The initial classifier was built using 24 labeled images from 15 patients. Nine labeled images from five different patients were used for the parameter-tuning algorithm. The output of the system was evaluated on a labeled test set (separate from the training and parameter tuning set) comprising 42 HRCT images selected from 32 patients. Out of these, seven patients consisted of honeycombing findings.
Analysis
The evaluation of BV_pairs was also performed on subjects having honeycombing disease findings to test the robustness of the algorithms. Completeness and correctness (described in “Image Preprocessing”) were the standard measures used for performance evaluation of BV_pairs in each stage. In short, completeness is the ratio of true positives to the combined value of true positives and false negatives. Correctness is the proportion of positives correctly identified.
Agreement between the radiologist and BV_pairs for scoring the abnormal pairs was measured with the kappa statistic.29 This measure uses a nominal scale where zero denotes random agreement and one denotes perfect agreement. The kappa statistic is an appropriate statistical analysis technique to measure agreement between the radiologist and the automated system as it compares the agreement against that which might be expected by chance. The kappa statistic is standardized to lie on a −1 to 1 scale, where 1 is perfect agreement, 0 is what is expected by chance, and negative values indicate agreement less than chance. However, a commonly cited range within the scale is given by Landis and Koch30 where 0.41–0.6 and 0.61–0.8 are considered to indicate moderate and substantial agreement, respectively. When scoring the severity of bronchial dilatation, weighted kappa was used due to the categorical nature of the data. Weighted kappa is a generalization of the kappa statistic to situations in which the categories are not equal.30 Kappa was computed on the two datasets (i.e., one that included honeycombing and one without the honeycombing findings) for scoring the presence of abnormal bronchial dilatation.
Results
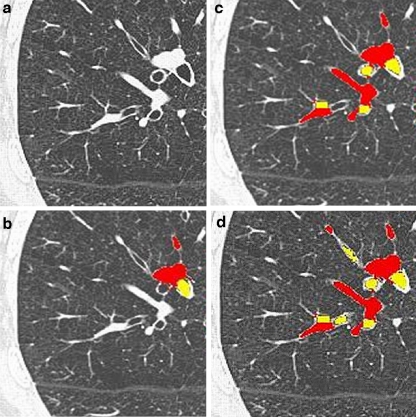
Through the use of clinical and high-level knowledge obtained from the potential bronchovascular pairs (identified in stage 1), the system was able to eliminate the cysts successfully in the second stage of BV_pairs. Some visual results of the algorithm are displayed in Figure 10.
Fig 10.
Panel a contains the original image. Panel b shows the output from iteration 0. Panel c contains the output from iteration 7. Panel d represents the output from iteration 5 using the optimal parameters learned for preprocessing. The regions in yellow and red correspond to the bronchus and its accompanying vessel, respectively.
The graphs in Figure 11 show the average improvement in completeness and correctness over the iterations, measured as the difference between the performance measures relative to iteration 0. The graphs show that the average improvement in completeness increases iteratively, while that in correctness is marginal. The improvement in completeness reaches approximately 29% at iteration 7, compared to iteration 0. This shows that completeness is improved, whereas correctness does not improve a lot.
Fig 11.
Graphs of iterative improvement in completeness and correctness. Panel a corresponds to completeness vs iterations, and panel b shows correctness vs iterations. The improvement is relative to iteration 0.
Results from the last iteration showed that the completeness value was slightly higher when honeycombing images were not included as opposed to including them. The correctness value of the last iteration for the dataset that did not include honeycombing images was higher by approximately 10% compared to the dataset that included them as shown in Table 3. This is because some of the honeycombing cysts were incorrectly identified as bronchi as shown in Figure 12.
Table 3.
Performance Comparison of the Last Iteration for the Two Evaluation Datasets
| Completeness | Correctness | |
|---|---|---|
| Evaluation set without honeycombing images | 82.00 | 72.63 |
| Evaluation set with honeycombing images | 79.39 | 63.73 |
Fig 12.
Output of BV_pairs on images containing honeycombing regions. Panel a contains the original image. Panel b contains the output where the detected regions are misclassified. The arrow points to the peripheral regions where honeycomb cysts predominantly occur. BV_pairs does not identify these as potential bronchovascular pairs because the near_hilum relation serves to eliminate them.
Results indicated that BV_pairs can detect discrete pairs containing larger bronchus with higher completeness and correctness as compared to those containing smaller bronchi as shown in Figure 13. When the bronchi sizes vary between 15 and 18 pixels, the completeness and correctness values were close to 90%.
Fig 13.
Panels a and b contains the plots of completeness and correctness percentages for increasing bronchi size (pixel count). The dashed bars correspond to the dataset augmented with images containing honeycombing regions, and the dark bars (in print) correspond to the dataset not including honeycombing regions.
When images containing honeycombing regions were included, the completeness and correctness values were lower than those when the honeycombing regions were not included by an average of approximately 6%. The training examples that were used to perform the classification of signet rings belonged to one of the four severities (where classes correspond to severity). The decision tree constructed by C4.5 contained both diameter and area as attributes. The visual results of different severities of bronchial dilatation are shown in Figure 14.
Fig 14.
Different severity levels. Each row contains the original image along with the output of third stage of BV_pairs. The bronchus and its adjacent vessel are colored in yellow and red, respectively. Panels a, b, c, and d correspond to normal, mild, moderate, and severe pairs of bronchial dilatation, respectively, outlined in circles.
Results indicate that the kappa values are higher for images without honeycoming regions as opposed to including them as shown in Table 4. Agreement for the automated scoring of the presence and severity of the signet ring was demonstrated to be comparable to that of an experienced radiologist (i.e., κ > 0.5) as shown in Table 4.
Table 4.
Agreement between the Radiologist and the Automated Technique using Kappa Statistic
| Images with honeycombing regions | Images without honeycombing regions | ||
|---|---|---|---|
| Severity (κd,w) | Presence (κd) | Severity (κd,w) | Presence (κd) |
| 0.51 | 0.59 | 0.56 | 0.638 |
Discussion
Some cases of disagreement between the automated system and the radiologist were traced to cardiac motion effects during scanning. It was also visually observed (Fig. 15a and b) that the pairs that were often missed by the system were very small and faint in appearance. It is also worth noting that the majority of incorrectly identified bronchovascular pairs from the first stage (Fig. 15c) are filtered out by the automated discrete pair detection technique. This is because honeycombing cysts that are incorrectly identified as bronchi appear too close to one another and are removed by BV_pairs through the use of high-level features, as shown by an example in Figure 15d.
Fig 15.
Panel a contains the outputs of the first and second stages. The arrows point to the smaller pairs that have not been identified in the first stage. The distinct larger ones are successfully identified by the automated technique as shown by the boxes. Panel b contains images with honeycombing. The output of the first stage shows honeycombing cysts being incorrectly identified as bronchovascular pairs. The automated technique from the second stage filters out the false positives.
The slightly low completeness and correctness values for very small sized bronchi (ranging between 3 and 6 pixels) can be attributed to the challenges associated in identifying them. These small-sized bronchi appear faint and discontinuous, making the task of segmentation difficult. For the third stage of BV_pairs, it is important to note that when a radiologist selects the pairs, it is based on high-level clinical knowledge, and in many cases, the radiologist may have discarded visible bronchi that may be regarded as diagnostically irrelevant. This results in some cases of disagreement between the radiologist and the automated technique. However, even with the limitations and challenges posed by the dataset, detection of bronchial dilatation on HRCT lung images can be achieved robustly.
Conclusion
This work presents a novel approach for the detection of bronchial dilatation on HRCT lung images. The system introduced, called BV_pairs, works in three stages: (1) detection of potential bronchovascular pairs, (2) detection of discrete pairs, and (3) detection of signet ring. In the implementation of the system, multiview learning in combination with active learning is used to identify potential bronchovascular pairs. The two views are based on relational features and the ERS transform. In the second stage, discrete bronchovascular pairs are identified. Features built on top of the output of potential bronchovascular pairs detected in the first stage are used to perform classification using a simple decision tree algorithm. In the final stage, the presence and severity of bronchial dilatation are detected using the area ratio and diameters of the discrete pairs. The novelty of the work lies in the overall approach taken in designing the different stages of BV_pairs. In particular, the application of multiview learning in identifying local structural patterns is new. The evaluation of the BV_pairs on patients having honeycombing findings ensures robustness of the system. It has been confirmed by medical experts in our team that the significant pairs identified by BV_pairs are useful in diagnosing bronchiectasis. An agreement with an experienced radiologist when automatically scoring for the presence and severity of bronchial abnormalities was demonstrated to be acceptable. Although the work presented in the paper was limited by sparse datasets, the major lesson learned is that detection of bronchial dilatation can be achieved with acceptable accuracy even in two dimensions. With the rapid improvements in scanner technology and data acquisition techniques, a natural improvement to this work would be to extend this work by incorporating information from adjacent slices.
Acknowledgment
This research was partially supported by the Australian Research Council through a Linkage grant (2002–2004), with Medical Imaging Australasia as clinical and Philips Medical Systems as industrial partners.
Contributor Information
Mithun Prasad, Phone: +1-310-9855014, Email: mithunp@cse.unsw.edu.au.
Arcot Sowmya, Email: sowmya@cse.unsw.edu.au.
Peter Wilson, Email: pcwilson2@bigpond.com.au.
References
- 1.Webb WR, Müller NL, Neidich DP. High-Resolution CT of the Lung. 3. Philadelphia, PA: Lippincott, Williams & Wilkins; 2000. [Google Scholar]
- 2.Young JL: Seer’s training web site. Available at: http://training.seer.cancer.gov/module_anatomy/unit9_4_resp_passages4_bronchi.htm, 2002
- 3.Seneterre E, Paganin F, Bruel JM, Michel FB, Bousquet J. Measurement of the internal size of bronchi using high resolution computed tomography. Eur Respir J. 1994;7:596–600. doi: 10.1183/09031936.94.07030596. [DOI] [PubMed] [Google Scholar]
- 4.Chabat F, Hu X, Hansell DM, Yang GZ. ERS Transform for automated detection of bronchial abnormalities on CT. IEEE Trans Medical Imaging. 2001;20(9):942–952. doi: 10.1109/42.952731. [DOI] [PubMed] [Google Scholar]
- 5.Zrimec T, Busayarat S, Wilson P: A knowledge based approach for automatic detection and measurement of bronchial dilatation and wall thickening on HRCT images of the lungs. In: World Congress on Medical Physics and Biomedical Engineering. Sydney, 2003
- 6.Saba O, Hoffman EA, Reinhardt JM. Maximizing quantitative accuracy of lung airway lumen and wall measures obtained from X-ray CT imaging. Appl Physiol. 2003;95:1063–1095. doi: 10.1152/japplphysiol.00962.2002. [DOI] [PubMed] [Google Scholar]
- 7.Chiplunkar R, Reinhardt JM, Hoffman EA. Segmentation and quantification of the primary human airway tree. SPIE. 1997;3003:403–414. doi: 10.1117/12.274066. [DOI] [Google Scholar]
- 8.Prasad M, Sowmya A: Detection of bronchovascular pairs on HRCT lung images through relational learning. In: Proceedings of IEEE International Symposium on Biomedical Imaging: Nano to Macro, USA, 2004, pp 1135–1138
- 9.Prasad M, Sowmya A: Multi-view learning for bronchovascular pair detection. In: Proceedings of the 2004 Intelligent Sensors, Sensor Networks and Information Processing Conference, Australia, 2004, pp 587–592
- 10.Sonka M, Park W, Hoffman EA. Rule-based detection of intrathoracic airway trees. IEEE Trans Medical Imaging. 1996;15:314–326. doi: 10.1109/42.500140. [DOI] [PubMed] [Google Scholar]
- 11.Mori K, Hasegawa J, Toriwaki J, Anno H and Katada K: Automated extraction of bronchus area from three dimensional X-ray CT images. IEICE Tech Rep 142, 1994
- 12.Wood SA, Hoford JD, Hoffman EA, Zerhouni EA, Mitzner W. A method for measurement of cross sectional area, segment length, and branching angle of airway tree structures in situ. Comput Med Imaging Graph. 1995;19(1):145–152. doi: 10.1016/0895-6111(94)00034-4. [DOI] [PubMed] [Google Scholar]
- 13.Pisupati C: Geometric analysis of dynamic three-dimensional tree structures. Ph.D. dissertation, Johns Hopkins University, Baltimore, MD, 1996
- 14.Prêteux CI, Fetita A, Capderou, Genier P. Modeling, segmentation, and caliber estimation of bronchi in high resolution computerized tomography. J Electron Imaging. 1999;8:36–45. doi: 10.1117/1.482682. [DOI] [Google Scholar]
- 15.Aykac D, Hoffman EA, McLennan G, Reinhardt JM. Segmentation and analysis of the human airway tree from 3D X-ray CT images. IEEE Trans Med Imaging. 2003;22(8):940–950. doi: 10.1109/TMI.2003.815905. [DOI] [PubMed] [Google Scholar]
- 16.Nigam K, Ghani R: Analyzing the effectiveness and applicability of co-training. In: Proceedings of Information and Knowledge Management, USA, 2000, pp 86–93
- 17.Blum A, Mitchell T: Combining labelled and unlabelled data with co-training. In: Proceedings of the 1998 Conference on Computational Learning Theory, 1998
- 18.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc. 1977;39:1–38. [Google Scholar]
- 19.Pierce D, Cardie C: Limitations of co-training for natural language learning from large datasets. In: Proceedings of the 2001 Conference on Empirical Methods in Natural Language Processing, CMU, Pittsburgh, PA, USA, 2001, pp 1–9
- 20.Collins M and Singer Y: Unsupervised models for named entity classification. In: Proceedings of the 1999 Joint SIGDAT Conference on Empirical Methods in Natural Language Processing and Very Large Corpora, USA, 1999, pp 100–110
- 21.Muslea I, Minton S, Knoblock C: Active + semi-supervised learning = robust multi-view learning. In: Proceedings of the 19th International Conference on Machine Learning, Australia, 2002, pp 435–442
- 22.Krogel MA, Scheffer T. Multi-relational learning, text mining, and semi-supervised learning for functional genomics. Mach Learn. 2004;57(1–2):61–81. doi: 10.1023/B:MACH.0000035472.73496.0c. [DOI] [Google Scholar]
- 23.Quinlan JR. Learning logical definitions from relations. Mach Learn. 1990;5:239–266. [Google Scholar]
- 24.Mitchell T. Machine Learning. Maidenhead, UK: McGraw Hill; 1997. [Google Scholar]
- 25.Prasad MN, Sowmya A, Koch I: Feature subset selection using ICA for classifying emphysema in HRCT images. In: Proceedings of the 17th International Conference on Pattern Recognition, 2004, pp 515–518
- 26.Prasad MN, Sowmya A. Multilevel emphysema diagnosis of HRCT lung images in an incremental framework. Proc SPIE. 2004;5370:42. doi: 10.1117/12.533943. [DOI] [Google Scholar]
- 27.Xu Y, Beek E, Hwanjo Y, Guo J, McLennan G, Hoffman E. Computer-aided classification of interstitial lung diseases via MDCT: 3D adaptive multiple feature method (3D AMFM) Acad Radiol. 2006;13(8):969–978. doi: 10.1016/j.acra.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 28.Rudrapatna M, Sowmya A, Zrimec T, Wilson P, Kossoff G, Lucas P, Wong J, Misra A, Busayarat S. LMIK—learning medical image knowledge: an Internet-based medical image knowledge acquisition framework. Proc SPIE. 2004;5304:307–318. doi: 10.1117/12.526290. [DOI] [Google Scholar]
- 29.Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas. 1960;20:37–46. doi: 10.1177/001316446002000104. [DOI] [Google Scholar]
- 30.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]