Abstract
In Korea, hepatocellular carcinoma is the third frequent cause of cancer death, occupying 17.2% among the whole deaths from cancer, and the rate of death from hepatocellular carcinoma comes to about 21 out of 100,000. This paper proposes an automatic method for the extraction of areas being suspicious as hepatocellular carcinoma from computed tomography (CT) scans and evaluates the availability as an auxiliary tool for the diagnosis of hepatocellular carcinoma. For detecting tumors in the internal of the liver from a CT scan, first, an area of the liver is extracted from about 45–50 CT slices obtained by scanning in 2.5-mm intervals starting from the lower part of the chest. In the extraction of an area of the liver, after the unconcerned areas outside of the bony thorax are removed, areas of the internal organs are segmented by using information on the intensity distribution of each organ, and an area of the liver is extracted among the segmented areas by using information on the position and morphology of the liver. Because hepatocellular carcinoma is a hypervascular tumor, the area corresponding to hepatocellular carcinoma appears more brightly than the surroundings in a CT scan, and also takes a spherical shape if the tumor shows expansile growth pattern. By using these features, areas being brighter than the surroundings and globe-shaped are segmented as candidate areas for hepatocellular carcinoma in the area of the liver, and then, areas appearing at the same position in successive CT slices among the candidates are discriminated as hepatocellular carcinoma. For the performance evaluation of the proposed method, experimental results obtained by applying the proposed method to CT scans were compared with the diagnoses by radiologists. The evaluation results showed that all areas of the liver and hypervascular tumors were extracted exactly and the proposed method has a high availability as an auxiliary diagnosis tool for the discrimination of liver tumors.
Key words: Contrast enhancement computed tomography(CT), hepatocellular carcinoma, hypervascular tumor
INTRODUCTION
In Korea, hepatocellular carcinoma is the third frequent cause of cancer death, recording the highest rate of occurrence in the whole world.1 For the diagnosis of hepatocellular carcinoma, while the histological diagnosis by a liver biopsy shows the highest accuracy, radiological inspections such as ultrasonography, computed tomography (CT), and magnetic resonance imaging and the clinical diagnosis by serologic-alpha-fetoprotein test are generally used.2,3 Because hepatocellular carcinoma is almost asymptomatic, it is detected at the terminal stage in the most cases during the past decades. Recently, examination using multidetector CT (MDCT) scanner is performed for early detection of liver tumors, presurgical planning, and evaluation of treatment. Early detection of hepatocellular carcinoma leads to an operation for the elimination of carcinoma cells. The greatest advantage in using MDCT is that the scan is completed over a wide range in a short time. On the other hand, a large amount of data is generated, which heavily burdens the physician. Thus, there is a need to develop a computer-aided diagnosis system.4
This paper proposed a novel method that automatically provides information on position and morphology of hepatocellular carcinoma through the analysis of CT scans. The proposed method extracts an area of the liver and detects relative positions of hepatocellular carcinoma in the area by applying a sequence of digital image processing techniques to about 45–50 CT slices obtained by scanning by 2.5-mm intervals starting from the lower part of the chest.
Technical Background
In recent years, various approaches have been presented for area segmentation of the liver and hepatocellular carcinoma from CT scans. While the approaches showed good performance in the normal case, the approaches did not generate acceptable results for various types of CT scans with any exceptional conditions.
Intensity thresholding method segments an area of the liver from CT scans only by thresholding intensity values based on a native intensity range of the liver.5–7 The histogram-analyzing method segments areas corresponding to the intensity range of the liver on the histograms of CT slices.8 Model-based contour extraction method segments by extracting contours of the internal organs and using morphological information on the liver.9 The three approaches adopted only conventional image processing techniques and had a common problem as follows: In a CT scan, other internal organs may have a similar distribution of intensity with the liver and border on the liver. In such a case, the approaches have difficulties in measuring the intensity range being able to discriminate between the liver and other organs and determining the boundaries of other organs connecting with the liver.
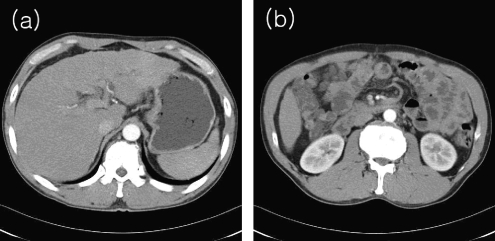
Blob coloring method segments all areas of the internal organs by using automatic threshold intensity (ATI) and morphology methods and lastly extracts an area of the liver by applying Blob coloring method to segmented areas.10 These approaches uses ATI method and morphology method when segmenting interested areas by image binariziation in the initial phase. Therefore, as showed in Figure 1a, if other organs have a similar distribution of intensity with the liver and border on the liver, the approach may not segment interested areas precisely. Also, while the approach extracts an area with the largest extend after applying Blob coloring to the segmented areas as an area of the liver, if, as shown in Figure 1b, there are other areas larger in the extend than the liver, the method may not extract an area of the liver precisely.
Fig. 1.
a CT image with obscure boundaries of the liver. b CT image with the liver being smaller than other internal organs
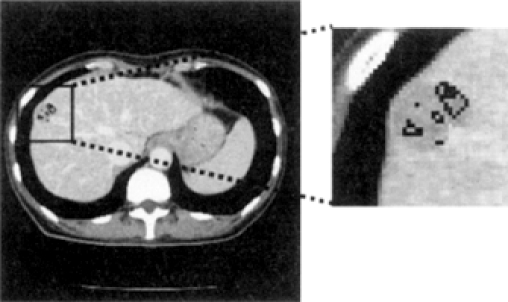
The recently proposed method using 3-dimensional multiphase multislice CT images, first, refines CT slices by using 3-dimensional convergence index filter, finds feature points of liver tumors based on information on the intensity distribution, segments candidate regions by applying region growing processing, and discriminates real regions of liver tumors among the candidates by using Mahalanobis distance method.4 This approach, by using the feature that the central part of tumors appears more brightly than the backgrounds, selects local maxima of intensity values as candidate points and performs region growing processing, segmenting candidate regions of liver tumors. However, as shown in Figure 2, being quoted by Nakagawa et al.,4 because the hepatic portal vein or the vena cava has the similar distribution of intensity with liver tumors and the approach does not support the discrimination between the organs and liver tumors, regions of the organs may be wrongly segmented as candidate regions, thus influencing the region segmentation of liver tumors.
Fig. 2.
Example of underestimated tumor candidates in the method using 3-dimensional multiphase multislice CT images
As stated above, the conventional approaches for the segmentation of an area of the live have used only feature information on intensity distribution and morphology of the liver. Therefore, the approaches have difficulties in extracting an area of the liver from CT slices when some areas have a similar distribution of intensity with the liver and the areas are adjacent to or connect with the liver. To solve this difficulty, this paper extracts every organ individually by using feature information on all organs presented in CT scans and removes unconcerned areas. And, for boundary areas between the liver and neighboring other organs, this paper analyzes degrees of inclusion by the liver and other organs, thus segmenting the boundary of the liver more precisely.
Also, while in the conventional approaches segmentation results differ from each other according to initial values of parameters given subject to a special CT slice, this paper processes synthetically all CT slices, including the liver among successive CT slices photographed from the chest to the abdomen and traces the morphological changes of the liver and liver tumors based on the correlation between successive CT slices, thus being able to extract areas of the liver and liver tumors precisely.
METHODS FOR EXTRACTION OF THE LIVER
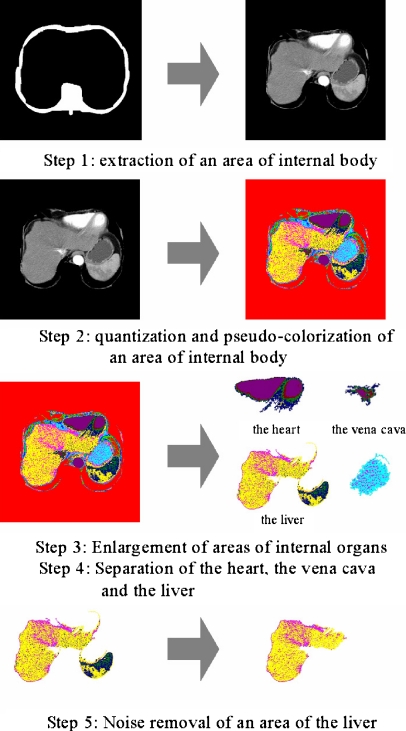
This paper proposed a novel method that extracts an area of the liver from a CT scan by using a feature that CT slices have higher brightness in areas of the blood vessel and the bone. The proposed extraction method consists of five steps seen in Figure 3.
Fig. 3.
Overall procedure for the extraction of an area of the liver
Extraction of an Area of the Internal Body
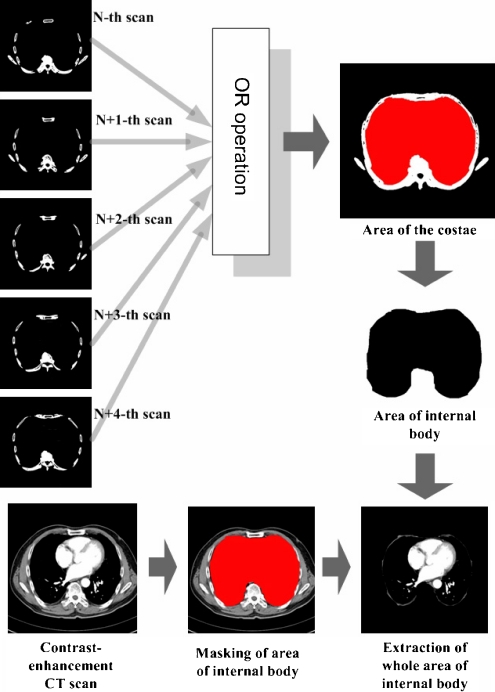
To extract an area of the liver from a CT scan, first, this paper divides the whole area of a CT slice into the area of internal body, including all internal organs and the external area consisting of the subcutaneous fat layer, the muscular layer, and the background, and then removes the unconcerned external area for reducing the computation overhead caused by successive image processing operations. Because the bony thorax exists in the boundaries between the area of internal body and the external areas, it becomes a criterion for the division of the whole area of a CT slice. Figure 4 shows the proposed procedure to extract an area of internal body, including the liver based on an area of the bony thorax.
Fig. 4.
Extraction of an area of internal body using an area of the bony thorax
This paper binarizes CT images simply by using the threshold of a gray value of 180 to segment an area of the bony thorax from CT slices. An area of the bony thorax has higher intensity than other organs in CT slices and various experiments showed that the intensity value discriminating precisely the bony thorax and other organs is comparatively high like 180. Because all types of CT scans show commonly that the difference of intensity between the bony thorax and other organs is distinctly great, the binarization by the threshold of 180 does not generate any exceptions in the segmentation by binarization. A bit of modification on the threshold may generate some noises not influencing the segmentation of an area of the bony thorax, while the threshold being greatly lower than 180 generates parts of the internal organs as noises and does not preserve the boundaries of the liver and the one being greatly higher causes the loss of information on an area of the bony thorax.
However, while an area of the bony thorax with a fully connected elliptical shape is needed for the division process, the area segmented from only a CT slice does not form a fully connected elliptical shape as shown in Figure 4. Based on the feature that the position and shape of the internal organs change slightly in several successive CT slices, this paper conjoins by OR operation areas of the bony thorax segmented from the current CT slices and additional contiguous four CT slices in the up-and-down direction, forming an area with a fully connected elliptical shape. It was shown heuristically through various experiments that five successive CT slices are optimal for forming an area of the bony thorax with a fully connected elliptical shape, preserving well the boundaries of the liver. Using more than five CT slices may cause the loss of information on an area of the liver. If the conjoined area is cut off, a connected area is made by applying smearing operation to broken areas.11 Lastly, by using the area of the bony thorax as a criterion, this paper extracts the interested inner area not including the area of the bony thorax.
Quantization and Pseudocolorization of an Area of the Internal Body
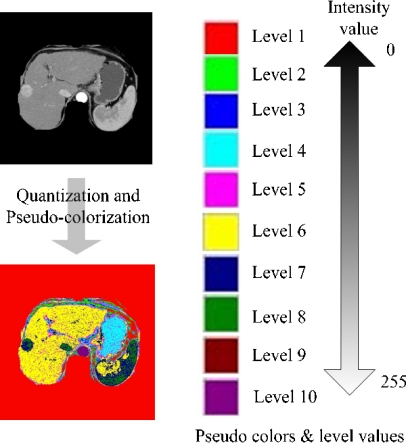
It is difficult to discriminate computationally only the liver in an area of internal body because CT slices are grayscale and other organs may have the similar distribution of intensity with the liver in CT slices. So, all areas of the internal organs need to be encoded into different colors from each other for the precise discrimination of the liver from other organs. This paper classifies intensity values between 0 and 255 into intensity levels of 10 represented by pseudocolors on a CT slice by using quantization and pseudocolorization methods.12
Intensity values on the area of internal body and the background are quantized into values of 10 and, according to quantized values, each pixel is pseudocolorized randomly with 10 colors. And for successive computational processing, the pseudocolors representing quantized intensity values are marked with relative level values from 1 for the lowest intensity to 10 for the highest one (see Fig. 5). These operations support the higher discriminating power between the internal organs in a CT scan, thus making it easy to extract an area of the liver from the CT scan.
Fig. 5.
Quantization and pseudocolorization of an area of internal body
Intensity Simplification of Areas of the Internal Organs
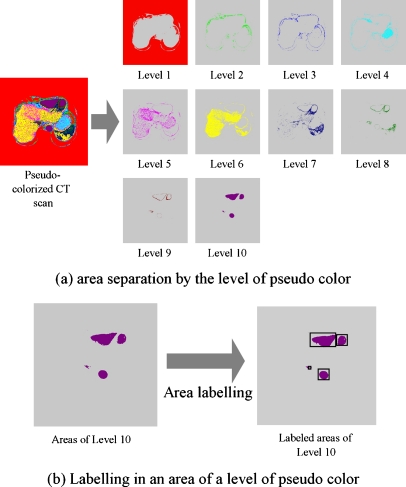
Generally, in any CT scan, an internal organ does not have one unique intensity value but has a narrow distribution of intensity including only several adjacent intensity values. Therefore, as shown in Figure 5, an area of an internal organ is represented by a few pseudocolors with consecutive level values in a pseudocolorized CT slice. For examples, yellow and pink colors appear in an area of the liver and green and blue colors in an area of the heart, as in Figure 5. Based on this feature, this paper simplifies the intensity distribution of an internal organ by merging areas with different pseudocolors covered by the internal organ, thus making the boundaries of the internal organs more distinct. The intensity simplification for an internal organ is achieved by scanning from areas with a pseudocolor of high level to ones with a pseudocolor of low level and merging areas bordered on each other as follows:
-
Step 1
Apply quantization and pseudocolorization to a CT slice, mark all pseudocolors with relative level values based on quantized values of intensity, and segment areas with each pseudocolor separately from the pseudocolorized CT slice.
-
Step 2
Let N be the level value of a pseudocolor currently scanned and processed. After labeling all areas of level N, select an area of interest among labeled areas according to a sequence of labeling, such as in Figure 6.
-
Step 3
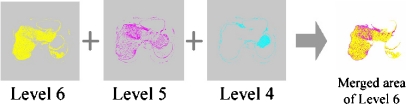
Select target areas being adjacent to the area of interest of level N among areas of level N − 1, inspect the boundaries of target areas, and merge areas bordering on the area of interest among target areas with the area of interest. And repeat these operations as decreasing the level value of target areas. However, as stated above, an internal organ is represented by only a few pseudocolors with consecutive level values, and if restricting the range of levels being searched in this step by using information on a distribution of pseudocolors for each internal organ, search overhead is able to be greatly reduced. For example, an area of the liver is represented by pseudocolors of levels from 6 to 4 and the heart and the vena cava by pseudocolors of levels from 10 to 7. In this paper, for the area mergence for the liver, levels of from 6 to 4 were used, such as those in Figures 7 and 8. And, if merging areas of pseudocolors of levels with a great difference, the merged area of an internal organ would cover other internal organs.
-
Step 4
Repeat steps 2 and 3 for the remaining labeled areas of level N and repeat this operation, decreasing the level of interest from the highest to the lowest.
-
Step 5
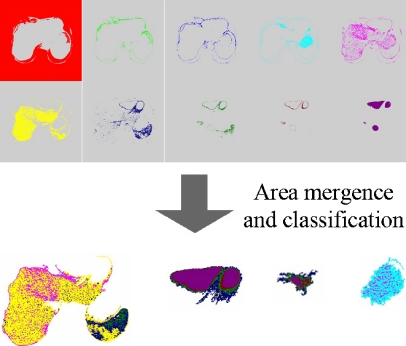
The above operations determine approximately the boundaries of the internal organs, but sometimes an area of an internal organ may contain pseudocolors representing other internal organs. So, lastly, classify areas of the internal organs into only three areas corresponding to the liver, the heart, and the vena cava by using features on position and morphology for each internal organ.
Fig. 6.
Area segmentation by levels of pseudocolors in a pseudocolorization CT slice
Fig. 7.
Area mergence for the liver
Fig. 8.
Example of results of area mergence and classification
Extraction of an Area of the Liver
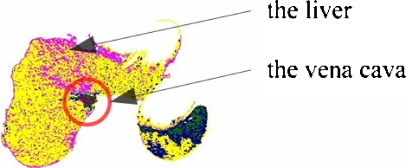
Because in CT scans the intensity distribution of a hypervascular tumor is similar with the heart and the vena cava and sometimes an extracted area of the liver contains the vena cava like in Figure 9, the heart or the vena cava contained in the area of the liver may be mistakenly discriminated as a hypervascular tumor.13 Therefore, this paper extracts an area of the liver not containing the heart or the vena cava by using feature information on the distribution of pseudocolors and the position of each organ.
Fig. 9.
Area of the liver containing the vena cava
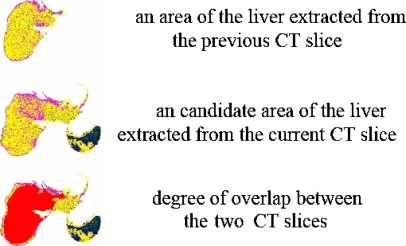
Important features of three organs which are the heart, the vena cava and the liver are summarized in Table 1. The features are used when extracting areas of the three organs only from the first CT slice and in the processing on successive CT slices position information of each organ obtained in the last image is used. For each internal organ, if the degree of overlap between areas extracted from the current and the previous CT slice is above 70% of the area previously extracted, the area currently extracted is newly selected as an area of the internal organ. The threshold value of 70% was determined heuristically through several experiments.
Table 1.
Features on Distribution of Pseudocolors and Position of the Heart, Vena Cava, and Liver
| Organ | Feature | |
|---|---|---|
| Level Distribution of Pseudocolors | Position | |
| Heart | 10–7 | The center of a CT slice including the lung |
| Vena cava | 10–7 | The right neighborhood of the spine |
| Liver | 6–4 | Right below the heart |
Noise Removal Using Rough Edge
If areas of other organs have the same pseudocolor with an area of the liver and these areas border on each other, areas of the liver and other organs are extracted together as an area of the liver like in Figure 10. For a case like in Figure 10, this paper defines an area of the liver as a primary object and areas of other organs as secondary objects. If the primary object and a secondary object are connected with a length of one pixel, the method of noise removal described in Kim and Kim14 is able to separate the two objects. On the other hand, the method of Kim and Kim14 fails in the separation when the length of connection is above 2 pixels. Therefore, this paper proposes a novel method of noise removal, which is able to separate the primary object and a secondary one with a length of connection above 1 pixel.
Fig. 10.
Degree of overlap between areas of the liver extracted from two successive CT slices
Features used to discriminate and remove secondary objects from the primary one are as follows:
While secondary objects are smaller than the primary, the objects have a boundary with a length of above 30 pixels at a minimum.
The length of connection between the primary object and a secondary one is above 1 pixel and below L pixels.
The shape of the primary object is near a circle and does not have sharply protruding parts.
The removal procedure of a secondary object is as follows:
-
Step 1
On a candidate area of the liver, mark edge pixels by an interval of R pixels along with the boundary of the area and connect two neighboring edge pixels by a straight line, generating rough edges on the area.
-
Step 2
Measure the length of a line between two edge pixels and stop the process if the length is greater than L because there are no secondary objects connecting to the primary object. Otherwise, go to step 3.
Because two detected edge pixels are connecting points between the primary object and a secondary one, divide the two objects using a line between two edge pixels.
-
Step 4
Apply the morphological reduction operation for the separation of two objects.
-
Step 5
Extract a pure area of the liver by using 8-diectional contour tracking algorithm.15
METHODS FOR EXTRACTION OF HYPERVASCULAR TUMORS FROM THE LIVER
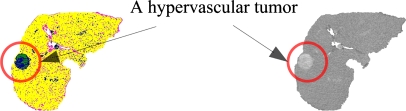
It is shown in Figure 11 that a hypervascular tumor like hepatocellular carcinoma appears more brightly than the surroundings on an area of the liver because neovasculars aggregate densely in the internal of the tumor, and when showing expansile growth pattern it takes a spherical shape.16 This paper discriminates and extracts areas of the tumor from an area of the liver by using features on the intensity distribution and the morphology of a hypervascular tumor.
Fig. 11.
A hypervascular tumor in an area of the liver
Segmentation of Candidate Areas of Hypervascular Tumors
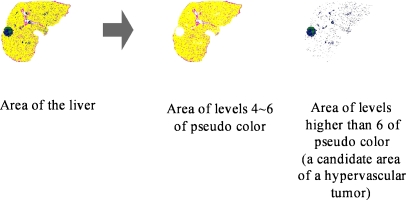
Using the feature that areas of hypervascular tumors are brighter than the surroundings, this paper segments areas having pseudocolors of higher levels than the pseudocolor representing an area of the liver as candidate areas of the tumors. Figure 12 shows an example on the segmentation of candidate areas of hypervascular tumors from an area of the liver.
Fig. 12.
Candidate areas of hypervascular tumors segmented from an area of the liver
Discrimination of Hypervascular Tumors
Using a morphological feature of hypervascular tumors and the continuity of CT slices, this paper discriminates areas of hypervascular tumors among the candidates. Based on the feature that a protrusive hypervascular tumor has a spherical shape, the degree of circularity is calculated like Eq. 1 for each area with the extent of above 500 pixels among the candidates.
 |
1 |
This paper decides areas having the extent of above 500 pixels and the degree of circularity of above 0.4 among the candidates as hypervascular tumors.
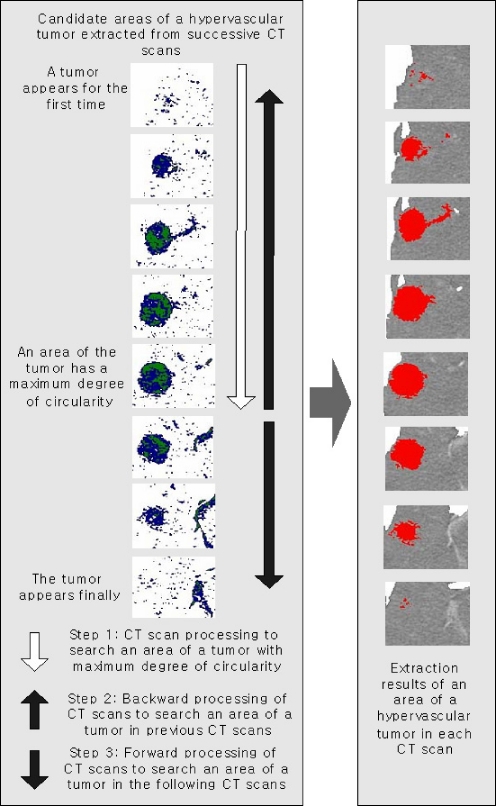
The extraction of hypervascular tumors from a successive CT slice is to extract an area coinciding in position with the area of the tumor currently discriminated and inspects the level of pseudocolor of the extracted area, discriminating the area as a hypervascular tumor when the level is higher than the pseudocolor of an area of the liver. If the area extracted from a successive CT slice does not show pseudocolors of higher levels than an area of the liver, the area of a hypervascular tumor currently discriminated is thrown away. It is because while the blood vessel or the hepatic portal vein has similar features with a hypervascular tumor in CT scans, these physical regions do not appear continuously in several CT slices. Figure 13 shows the extraction procedure of areas of hypervascular tumors from the liver using successive CT slices.
Fig. 13.
Extraction procedure of areas of hypervascular tumors using successive CT slices
PERFORMANCE EVALUATION
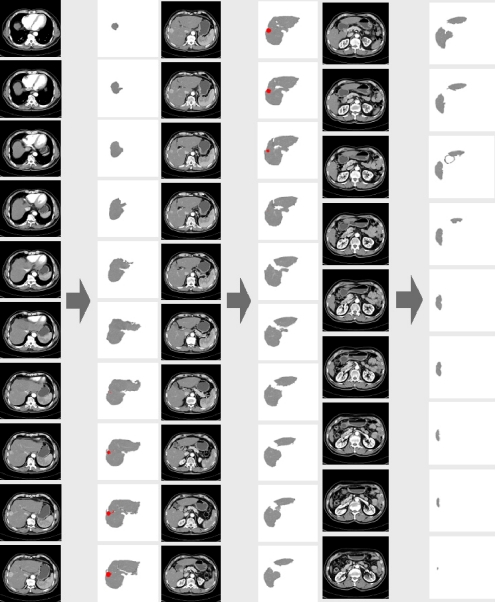
This paper performed experiments using CT scans provided by the specialist to evaluate the availability of the proposed methods. The experimental environment was implemented by using VC++ 6.0 in an IBM-compatible PC equipped with Intel Pentium-IV 2-GHz CPU and 256-MB RAM. Figure 14 shows areas of the liver and hypervascular tumors extracted from a series of CT slices as experimental results and the extracted areas of hypervascular tumors were represented by red color in Figure 14.
Fig 14.
Areas of the liver and hypervascular tumors extracted from a CT scan.
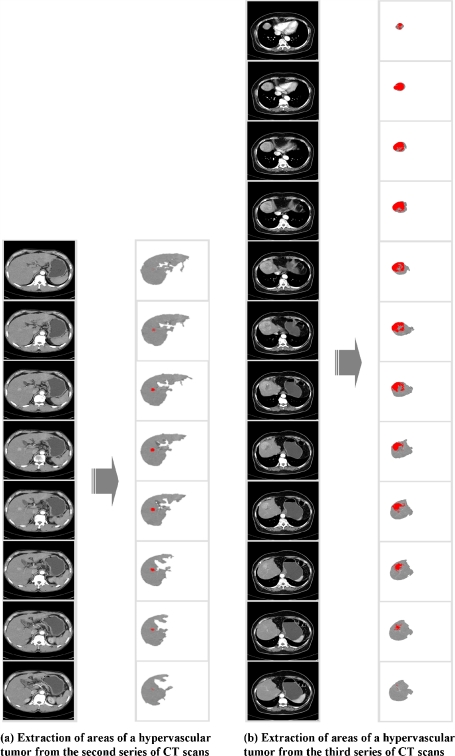
Figure 14 shows all results extracted from a total of 46 CT slices photographed in a CT scan and Figure 15 shows extraction results generated by experiments using two additional CT scans.
Fig. 15.
Extraction of areas of hypervascular tumors from two additional CT scans
Table 2 shows the comparison between the diagnoses of the specialists and extraction results from three CT scans by the proposed method being depicted in Figures 14 and 15.
Table 2.
Analysis of Hypervascular Tumors Extracted from Three CT Scans
| CT Scan | Number of Extracted Tumors | Sensitivity (%) | Specificity (%) | False Positive(%) | False Negative(%) |
|---|---|---|---|---|---|
| (+)→(+) | (−)→(−) | ||||
| 1 | 8 | 100 | 0 | 0 | 0 |
| 2 | 8 | 100 | 0 | 0 | 0 |
| 3 | 12 | 100 | 0 | 0 | 0 |
In Table 2, the number of extracted hypervascular tumors means the number of CT slices in which the area of the liver contains discriminated areas of a hypervascular tumor between the CT slice where an area of the liver begins to appear and the one where the area appears finally.
In the diagnostic radiology, the diagnosis of negative (−) means the normal case being not a tumor, and the diagnosis of positive (+) means the case being a tumor.17 Table 2 classifies extraction results into diagnoses of three types, such as sensitivity, false-positive, and false-negative. Sensitivity (+)→(+) means the diagnosis that hypervascular tumors are correctly extracted in a positive case, and sensitivity (−)→(−) means the diagnosis that no tumors are extracted in a negative case. The diagnosis of false-positive is the case that hypervascular tumors are wrongly extracted from the negative liver. On the other hand, the diagnosis of false-negative is a misjudgment that no tumors are extracted from the positive liver, being the most serious error in the medical image processing. As shown in Table 2, the proposed method generated correct outputs in diagnoses of all types.
This paper is a preliminary study to evaluate the feasibility of the proposed computer-aided diagnosis technique that extracts the liver and hepatocellular carcinoma automatically from CT scans. So we did not enroll many CT scans like a large clinical study, and at present, we have a plan to study clinical efficacy of this technique in our next clinical study with many cases.
DISCUSSION
With the increasing number of medical images, the use of computers in facilitating their processing and analysis has become a prerequisite. In particular, computer algorithms for delineating anatomical structures and detecting lesions are key components in assisting and automating specific radiological tasks.18 This paper proposed a novel image processing method extracting and discriminating areas of the liver and hypervascular tumors like hepatocellular carcinoma from a CT scan, which is able to support information on the position and the morphology of these areas to assist the specialists in detection and diagnosis.
The proposed method first extracts an area of the liver from a CT scan by using a feature that the distributions of intensity of the internal organs are different from each other. In the extraction of an area of the liver, after unconcerned areas outside of the bony thorax are removed, areas of the internal organs are segmented by using information on distributions of intensity of the internal organs, being adaptable to all types of CT scans, and the area of the liver is extracted among segmented areas by using information on position and morphology of the liver. Because hepatocellular carcinoma is a hypervascular tumor, an area of hepatocellular carcinoma appears more brightly than the surroundings in a CT slice, and when the tumor is growing expansively, the area has a spherical shape. Therefore, areas being brighter than the surroundings and globe-shaped are selected as candidate areas of hypervascular tumors in the extracted area of the liver, and areas appearing at the same position with a similar intensity in successive CT slices among the candidates are discriminated as hypervascular tumors. For the performance evaluation of the proposed method, experimental results obtained by applying the proposed method to three CT scans were compared with the diagnoses by the radiologists. The evaluation results showed that all areas of the liver and hypervascular tumors were extracted exactly and the proposed method has a high availability as an auxiliary diagnosis tool for the discrimination of hepatocellular carcinoma.
This paper is just the beginning of a new technique for analyzing routine CT scans. This technique can possibly be developed to cover several types of tumors with different texture patterns and it could be very useful for surgical and radiation treatment planning if 3-dimensional visualization of the segmented liver and detected tumors were applied. In future works, we refine the proposed method to analyze in detail the intensity level of a hypervascular tumor by using a fuzzy method for the extraction of a hypervascular tumor not being discriminated by the naked eye. Also, we will develop an auxiliary diagnosis system for the discrimination and analysis of hepatocellular carcinoma through clinical testing by the specialists.
In conclusion, the computer-aided diagnosis of hepatocellular carcinoma by the proposed method is feasible on abdominal CT scans.
References
- 1.Jeong YY, Yim NY, Kang HK, et al. Hepatocellular carcinoma in the cirrhotic liver with helical CT and MRI: imaging spectrum and pitfalls of cirrhosis-related nodules. AJR Am J Roentgenol. 2005;185(4):1024–1032. doi: 10.2214/AJR.04.1096. [DOI] [PubMed] [Google Scholar]
- 2.Kamel IR, Liapi E, Fishman EK, et al. Multidetector CT of hepatocellular carcinoma. Best Pract Res Clin Gastroenterol. 2005;19(1):63–89. doi: 10.1016/j.bpg.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Baron RL, Brancatelli G. Computed tomographic imaging of hepatocellular carcinoma. Gastroenterology. 2004;127(5):S133–S143. doi: 10.1053/j.gastro.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 4.Nakagawa J, Shimizu A, Kobatake H. Development of an automated extraction method for liver tumors in three-dimensional multiphase multislice images. Syst Comput Jpn. 2005;36(9):43–54. doi: 10.1002/scj.20179. [DOI] [Google Scholar]
- 5.Henkei RD: Segmentation in scale space. Proceedings of Computer Analysis of Images and Pattern, CAIP, Prague, 1995
- 6.Burdick HE. Digital Imaging, Theory and Application. London: McGraw Hill Inc; 1997. [Google Scholar]
- 7.Umbaugh SE. Computer Vision and Image Processing : A Practical Approach Using CVIPtools. Englewood Cliffs, NJ: Prentice Hall; 1988. [Google Scholar]
- 8.Gao L, Health D, Kuszyk B, Fishman E. Automatic Liver Segmentation Technique for Three-dimensional Visualization of CT Data. Radiology. 1996;201:359–364. doi: 10.1148/radiology.201.2.8888223. [DOI] [PubMed] [Google Scholar]
- 9.Haralick RM, Shapiro LG. Image segmentation techniques. Comput Vis Graph Image Process. 1985;29:100–132. doi: 10.1016/S0734-189X(85)90153-7. [DOI] [Google Scholar]
- 10.Lim O, Kim J, Park S, Lee B: Automatic segmentation of liver region in CT images using Blob coloring. Proceedings of the Korea Information Science Society Fall Conference, 2004, vol. 31, no. 2, pp 760–762
- 11.Kim KB, Kim S: Hierarchical recognition of English calling card by using multiresolution images and enhanced neural network. Lecture Notes in Artificial Intelligence, Springer, 2005, vol. 3801, pp 785–792
- 12.Sonka M, Fitzpatrick JM. Handbook of Medical Imaging, Vol. 2, Medical Image Processing and Analysis. Bellingham, WA: SPIE Press; 2000. [Google Scholar]
- 13.Atasoy C, Akyar S. Multidetector CT: contributions in liver imaging. Eur J Radiol. 2004;52(1):2–17. doi: 10.1016/j.ejrad.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 14.Kim KB, Kim S: A passport recognition and face verification using enhanced fuzzy neural network and PCA algorithm. Lecture Notes in Computer Science, Springer, 2006, vol. 4233, pp 167–176
- 15.Kim KB, Kim YJ. Recognition of English calling cards by using enhanced fuzzy radial basis function neural networks. IEICE Trans Fundam Electron Commun Comput Sci. 2004;E87-A(6):1355–1362. [Google Scholar]
- 16.Murakami T, Hori M, Kim T, Kawata S, Abe H, Nakamura H. Multidetector row CT and MR imaging in diagnosing hepatocellular carcinoma. Intervirology. 2004;47(3–5):209–226. doi: 10.1159/000078474. [DOI] [PubMed] [Google Scholar]
- 17.Kim KB, Kim S, Sim KB: Nucleus classification and recognition of uterine cervical pap-smears using fuzzy ART algorithm. Lecture Notes in Computer Science, Springer, 2006, vol. 4247, pp 560–567
- 18.Pham DL, Xu C, Prince JL. Current method in medical image segmentation. Annu Rev Biomed Eng. 2000;2:315–337. doi: 10.1146/annurev.bioeng.2.1.315. [DOI] [PubMed] [Google Scholar]