Abstract
Medical imaging research and clinical applications usually require combination and integration of various techniques ranging from image processing and analysis to realistic visualization to user-friendly interaction. Researchers with different backgrounds coming from diverse areas have been using numerous types of hardware, software, and environments to obtain their results. We also observe that students often build their tools from scratch resulting in redundant work. A generic and flexible medical imaging and visualization toolkit would be helpful in medical research and educational institutes to reduce redundant development work and hence increase research efficiency. This paper presents our experience in developing a Medical Imaging and Visualization Toolkit (BIL-kit) that is a set of comprehensive libraries as well as a number of interactive tools. The BIL-kit covers a wide range of fundamental functions from image conversion and transformation, image segmentation, and analysis to geometric model generation and manipulation, all the way up to 3D visualization and interactive simulation. The toolkit design and implementation emphasize the reusability and flexibility. BIL-kit is implemented in the Java language so that it works in hybrid and dynamic research and educational environments. This also allows the toolkit to extend its usage for the development of Web-based applications. Several BIL-kit-based tools and applications are presented including image converter, image processor, general anatomy model simulator, vascular modeling environment, and volume viewer. BIL-kit is a suitable platform for researchers and students to develop visualization and simulation prototypes, and it can also be used for the development of clinical applications.
Key words: Medical imaging, visualization, toolkit, Java, anatomy model
Introduction
The capabilities of radiology equipment are increasing dramatically, and three-dimensional (generally n-dimensional) medical images are becoming important and critical in clinical diagnosis and therapy. This situation drives the research interest in medical image analysis and modeling. This also elevates demands for suitable medical imaging research and development platforms and environments that can rapidly turn research ideas into prototypes and clinical applications.
Medical images can be interpreted as n-dimensional digitized signal sequences. Therefore, some of the fundamental signal processing algorithms are frequently being used in preprocessing and advanced analysis. Because of historical reasons and commercial competition as well as of researchers' personal customs and preferences, images are created, communicated, and transferred in numerous formats. The diversity of medical image formats has always been an aggravating problem in our research experience. During analysis of scans, various modeling algorithms may be applied. Finally, the results of analysis and modeling need to be validated or verified by researchers and clinicians, which requires these results to be presented in a user-friendly and quantitative way. To achieve this purpose, modeling and visualization techniques are used frequently to show the investigation, analysis, and modeling results in the most presentable way. Modeling and visualization are also very important communication bridges between researchers and clinicians.
Working in a research institute, we observed that many students have been spending much time doing redundant work because of their different backgrounds and lack of suitable reusable source code. This evokes us that a proper software development and prototyping platform will be useful and important to accelerate our research work. On the other hand, sharing work and data among heterogeneous computer systems is another issue in our research environment. Researchers are reluctant to give up using their favorite computer systems, and they build tools on platforms they are familiar with. Therefore, a cross-platform toolkit certainly benefits the researchers, the students, and the organization.
The goal of this work is to address these issues to increase research efficiency. The emphasis of toolkit design is concentrated on the following:
Portability: The libraries and tools should be platform-independent, so that the toolkit can be used in a heterogeneous computing environment with minimized redevelopment work.
Reusability: The design of the toolkit object classes should follow the object-oriented principle, so that the object classes are well self-encapsulated, with high cohesion and minimum coupling.1 Meanwhile, the object classes shall be well tested and documented so that the users will be able to use them without knowing much about the implementation details.
Flexibility: The toolkit shall be a pool of generalized object classes with the above reusability features, and therefore, researchers and students can select relevant object classes to quickly assemble a tool or a prototype in their research interests.
Extensibility: The new modules developed by researchers and students shall easily be incorporated into the toolkit.
Related Work and Our Approach
Existing Toolkits
There are several medical imaging toolkits available either in the public domain or the commercial market. One of the commercial packages, the de facto gold standard in medical image analysis, is Analyze from Mayo.2 It is an integrated suite of complementary tools for fully interactive display, manipulation, and measurement of multidimensional biomedical images. The system is developed using C++ with Tcl/Tk. VTK3,4 is a public domain software package developed for 3D graphics and visualization. It is developed in C++ and relies on Tcl/Tk or Java as its cross-platform graphical user-interface (GUI) support. VTK provides only basic image-processing functionality. Besides, to the developers, they have to master a second language, either Tcl/Tk or Java. Another packaged under recent active development is the Insight Segmentation and Registration Toolkit (ITK).5 ITK is aiming to implement the commonly used medical image segmentation and registration algorithms. Similarly, it is developed in C++, and therefore, to overcome the cross-platform GUI issues, the application developers also have to master a second language such as Tcl/Tk, Java, etc. The above-mentioned systems require a second language or script to overcome the cross-platform problem. Moreover, they are not as convenient as Java when it comes to developing Web-based applications.
There are also medical imaging packages developed by using Java. One of them is ImageJ—a medical imaging software developed by Rasband6 at NIH. ImageJ provides a considerable number of basic image operations. However, most of the image-processing operations are actually 2D, sometimes with capabilities to process image stacks. In addition, we found that its 3D modeling and visualization functions are not sufficient enough to fully support our research activities. Another Java-based medical imaging package, NeatVision developed by the Vision Systems Group of Dublin City University,7 provides an impressive visual image-processing flow interface, with strong image-processing functionality. However, similar to ImageJ, they are concentrating more on imaging rather than on geometric modeling and manipulation.
We have observed that most of the similar toolkits are more or less focused on either imaging or visualization or at least inclined to either one area. There are recent attempts8,9 to integrate packages from both areas to provide more comprehensive toolkits for medical imaging researchers; however, they are facing problems of various styles. The development of a comprehensive toolkit was also our objective when we were starting our project a few years ago, with imaging and visualization functionalities considered and implemented together consistently.
Java and Our Strategy
Our medical imaging and visualization toolkit aims to provide a foundation infrastructure for institutes conducting medical imaging researches. Based on years of our medical imaging research experience, both medical analysis ability as well as strong visualization capability are required. Medical image research tasks need generic image-processing algorithms ranging from simple thresholding, contrast enhancement, etc., up to advanced sophisticated image analysis algorithms incorporating anatomical knowledge.10,11
The Java language was developed by Sun Microsystems Inc.12,13 Its key feature is “write once and run everywhere.” Working in a dynamic and fast-pacing biomedical research environment, we have acquired experience to deal with heterogeneous hardware and software platforms and constructing research prototypes within minimum time limits.
Java is designed to meet the challenges of application development in the context of heterogeneous, network-wide distributed environments. Paramount among these challenges is secure delivery of applications that consume the minimum of system resources, can run on any hardware and software platform, and can be extended dynamically.14
Besides the advanced design philosophy of Java language, it has also brought the quality and productivity to developers. In addition, Java provides some optional packages that are very helpful in the development of image processing and visualization functions, namely, the Java Advanced Imaging (JAI) components of Java Media Framework (JMF) and the Java3D component. The advantages of these components will be addressed in The Toolkit Architecture and Modules.
The Java3D15,16 component provides the visualization developers with a set of higher-level APIs compared to most graphics packages which many developers are still using, such as OpenGL.17 This feature will significantly reduce the heavy burden of the toolkit developers and hence the further application developers using our toolkit.
Network-based related medical imaging research and applications are increasingly popular. Java, as a language environment designed for heterogeneous platforms and distributed computing, is an ideal language in producing network-based end-user applications.14 The recent developed technology of Java Web Start has delivered a very convenient deployment solution to deploy Java applications.18
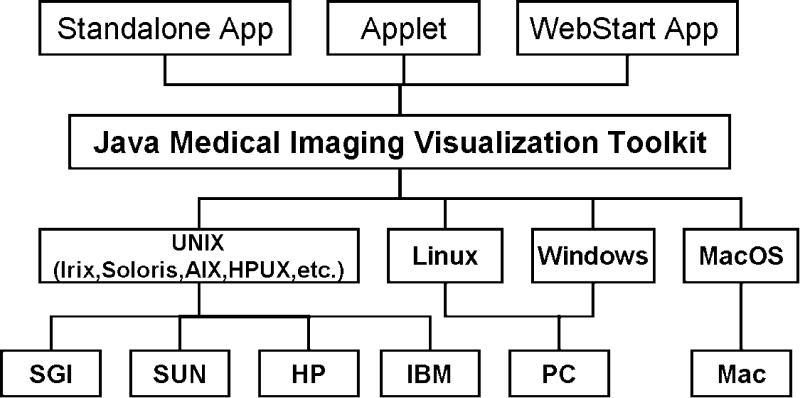
These advanced features, the seamless compatibility of Java optional components and the run-everywhere promise, allow us to build a medical imaging and visualization toolkit adapted to dynamic and heterogeneous hardware and software platforms of our research environment, such as what is shown in Figure 1.
Fig 1.
The BIL-kit development and application paradigm.
One of the questions debated is Java performance. Indeed, the performance of Java is still not as good as some optimized languages, such as C++. However, the speed of Java has been significantly improved in the recent years and is almost comparable to C++.19 On the other hand, the dramatic growth of hardware capacity and the decreasing hardware price have enormously reduced the user concerns about the hardware performance.
In view of software productivity and quality, Java is observably better than C++. A detailed investigation of Java vs. C++ comparison found out Java to be certainly better in both software quality and productivity.20 In addition, a toolkit developed by using pure Java requires the developers to master only one language; those developers who use other packages need to know more than one language, such as C++ with another language or scripts, to overcome the cross-platform problem.
The Toolkit Architecture and Modules
The Java Medical Imaging and Visualization Toolkit called BIL-kit developed in our Biomedical Imaging Lab (BIL) has been built on top of the Java Software Development Kit (SDK), JAI, and Java3D packages. The toolkit can be divided into four major modules: foundation module, medical image input/output (I/O) module, image-processing module, and modeling and visualization module (Figure 2). These modules form the majority of the reusable libraries. On top of these libraries, tools and applications can be built. By using these tools as templates, and making use of the reusable classes, more advanced specific applications or prototypes can be built for researchers and clinical users.
Fig 2.
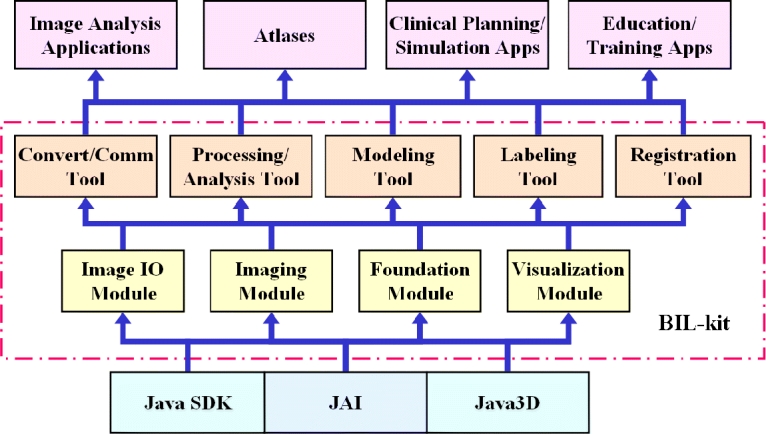
The Java Medical Imaging and Visualization Toolkit (BIL-kit) architecture.
Foundation Classes Library
A comprehensive set of foundation classes has to be built to form the base of any toolkit. Java SDK has created a broad range of foundation classes that can meet the requirements of medical imaging and visualization in most cases. However, as BIL-kit is specially designed for medical imaging research and applications, some of the frequently used foundation classes needed to be extended from the standard Java SDK, such as data structures used for presenting medical information and some GUI widgets frequently used in medical imaging.
Medical Image I/O Module
In our research experience, we have encountered numerous image formats from many sources. Besides the general photograph formats such as JPG, GIF, and TIFF, the image formats used by different computer platforms also vary, such as RGB format for SGI and PNM format for X-Windows. Medical images tend to carry patients and diagnosis information as well as descriptive information of acquisition procedure together. These formats more or less depend on image acquisition systems. We encountered problems in dealing with image formats from different vendors in the early days; for example, image formats from GE scanners were different from Siemens. Even different models from the same vendor had variations.
The American College of Radiologists (ACR) and the National Electrical Manufacturers Association (NEMA) recognized the need for standards to facilitate multivendor connectivity to promote the development of PACS and networking more than 20 years back. Digital Imaging and Communications in Medicine (DICOM)21 has been released as the result of these standardization efforts, and it has become prevalent nowadays.
DICOM is the ubiquitous standard in the radiology for the exchange and management of images and image-related information. This makes the DICOM standard a huge, sophisticated, and hard-to-read standard, which means it is also difficult to interpret. DICOM Structured Reporting has recently become an interesting topic.22 Our toolkit also considered Structured Reporting in its DICOM images encoding and decoding functions. Because various user groups have different expectations and requirements of the Structured Report, the toolkit allows the users to supply their own structured report definition in a text format, which screens the user data and instructs how the reports will be encoded and decoded. Although this is a simple implementation to support structured reporting, it is also practical to keep flexibility and extensibility and to produce a more elegant document with an appropriate structure for specific applications. Currently, this feature is used in our DICOM image converter, where the report structure is simple. It will be enriched and extended to handle real structured reports according to users' requirements when the toolkit is used to develop patient-specific medical image analysis applications.
The Java language not only provides the image readers with generic network image formats but also forms a framework for users to extend their work in handling unusual image formats. Java Image I/O component is part of the Java Standard Edition. It is designed as a pluggable framework into which any developer may add their own “plug-ins.” A plug-in is defined as a set of Java programming language classes that may be loaded into the API at run time and that add functionality to the API. In the context of the Java Image I/O API, a plug-in may provide the ability to read image data from a new file format, to write image data in a new format, to “transcode” nonimage metadata between two formats, or to read or write streaming data from or to a new data source or sink. A plug-in may also provide support for the same format as another plug-in, perhaps providing better performance, with more features, or a different view of the data stored by the format.23
This mechanism provides a flexible and useful interface for the developers to handle specific medical image formats. It also provides a framework for the additions of format-specific plug-ins. The plug-ins are standard Java classes that are loaded into the API at run time, adding functionality. These plug-ins can also be embedded in Java archive files. Some of the standard format plug-ins are available with the SDK itself, whereas the developers add the plug-ins for reading and writing specific image formats.
This operation of loading and instantiating a plug-in may be expensive. To alleviate this difficulty, another inexpensive class known as the Service Provider Interface is made available to provide information about a plug-in. The information provided about the plug-in includes the ability of the plug-in to decode a certain file format. The developer also needs to specify the nonstandard plug-ins developed in the form of a text file. This information is utilized by the registry of the Image I/O architecture to know which image formats can be read or written and selects the appropriate plug-in for each image file.
Another advantage of the introduced architecture is a powerful API available for handling metadata. The metadata are converted into an XML document, which can be handled using standard XML tools. The presence of different image readers and writers and the availability of API for writing transcoder plug-ins make the ability to transcode images available.
This Image I/O module uses JAI framework and extends a number of plug-ins that handle generic image formats, e.g., JPG, GIF, TIFF, BMP, PNG, including the latest compression format JPEG2000. It is also able to decode image formats used in specific computer systems, e.g., RGB format used by SGI workstation, PNM and XBM formats used by X-Windows-based systems, and some other formats used in other platforms. Most importantly, it also supports popular medical image formats, such as DICOM and ACR/NEMA 1.0 and 2.0 (the predecessor of the DICOM formats), as well as some image formats that are available in commercial products, such as Analyze AVW and Analyze 7.5.
By extending the Java Image I/O framework, the BIL-kit offers support to most of medical image formats, and more importantly, it is highly extensible. It can be easily extended by simply upgrading or adding codec (coder/decoder) in the form of plug-ins to handle new medical image formats in the future or keep paced with the standards upgrade. This implementation significantly saves the researchers and developers of doing redundant and tedious work and brings great convenience to the users.
Medical Image Processing and Analysis Module
Typical medical image analysis requires various generic image-processing algorithms; thus, most of them need to be implemented in the toolkit. However, there are some specific requirements to be taken care of. For instance, image transformation algorithms are frequently required to be performed not just in 2D but also in 3D. Meanwhile, some parameters such as pixel size, image acquisition modality, etc., have to be taken as parameters in image transformation. These requirements are also considered in the design of the image-processing component in our toolkit.
JAI is a set of comprehensive API supplied as an extension package of the standard edition of the Java platform. JAI provides imaging functionality beyond that of the Java Foundation Classes, although it is compatible with those classes in most cases.24,25
JAI is intended to support image processing using the Java programming language as generally as possible. At the same time, JAI presents a simple programming model that can be readily used in applications without a substantial mechanical programming overhead or the requirement that the programmer has to be expert in all phases of the API's design. JAI encapsulates image data formats and remote method invocations within a reusable image data object, allowing an image file, a network image object, or a real-time data stream to be processed identically. Thus, JAI represents a simple programming model while concealing the complexity of the internal mechanisms.
JAI offers several advantages for application developers compared to other imaging solutions, e.g., distributed imaging API, which means it is well suited for client–server imaging by means of the Java platform's networking architecture and remote execution technologies. It is also interoperable, which means that it is integrated with the rest of the JMF APIs, enabling media-rich applications to be deployed on the Java platform.
However, the JAI package only provides 2D image operations, which do not utilize full information contained in 3D medical images. A helpful and practical medical imaging toolkit needs to implement 3D imaging algorithms to take advantage of the latest radiology technology. Image processing in 3D is often considered as a natural extension of 2D image processing, but this is not exactly correct. Implementation of 3D image processing needs to consider the spatial relation of voxels carefully; sometimes, the algorithms need to be redesigned to achieve the optimized result.26 As JAI was designed for general-purpose image processing, it does not support the acquisition of medical images. However, as a medical imaging toolkit, it must be able to capture and keep acquisition information, so it can be used in certain automatic image-processing operations.
The initial image-processing algorithms implemented in our toolkit are the following:
image arithmetic functions, including addition, subtraction, multiplication and scaling, division, and blending;
image algebra transformation, including image AND, OR, XOR, INVERT, and bit-shift operations;
image geometrical transformation, namely, rotate or adjust geometric size and position of image volumes by resampling 3D image voxels with different interpolation functions;
spatial domain transformations, including single- and multiple-range thresholding, region growing (incremental and adaptive), histogram stretch and histogram equalization, mean and median filtering, and distance transformation;
frequency domain transformations, including Fourier transformation, convolution filtering, which includes a cluster of enhance operations, such as Gaussian and Laplacian filtering, sharpening and smoothing, as well as Sobel and zero-crossing edge detection operations;
nonlinear spatial transformations, such as mathematical morphology including dilation, erosion, opening, and closing.
The development of image processing is still in process. We are adding more image-processing functionalities. Some more advanced algorithms are on our roadmap of development, including wavelet transformation, color image processing, and knowledge-based segmentation. All the operations implemented are both in 2D and 3D and are optimized by our best efforts. Most of the algorithmic details are covered by Refs. [26,27].
Modeling and Visualization Module
The modeling and visualization component of our toolkit is built on top of Java3D component. Java3D is a 3D scene graph based on the graphics programming API for the Java language. It is an optional package of the Java standard edition. Java3D API provides routines for the creation of 3D geometries in a scene graph structure that is independent of the underlying hardware implementation for real-time programming. The API provides scene graph compilation and other optimization techniques. It is heavily optimized toward the requirements of real-time 3D rendering.
Java3D is an implementation of the scene graph concepts, which is one level above elementary computer graphics packages, such as OpenGL. The scene graph is used extensively in visualization development platforms.25 More and more programmers choose to use the existing scene graph programming platforms, such as Java3D and Open Inventor,28 to reduce development efforts and costs. Java3D used in our development also uses the scene graph.
The existing visualization languages and packages provide some useful support to build visualization environments for biomedical simulation and education. The popular languages and packages used are Java3D, VRML,29 and Open Inventor. These development environments have their advantages and disadvantages.
We chose Java3D as the foundation of our visualization module because of its several advantages: making use of the scene graph concept, seamless integration with Java SDK, and cross-platform features. Although there are some disputes about the future of Java3D, still, Sun Microsystems announced its plan recently to release the Java3D source code through some form of public source license in the very near future.30 An expert group under the Java Community Process will be formed to define and implement new features for the new version of Java3D API. The open-source strategy is a good sign that it will allow users to contribute bug fixes and utilities, which will increase the availability and accelerate its pace of development.
Java3D has a rich set of APIs, which provides powerful and flexibly interactive functions useful for building educational and simulation systems. It is suitable for building elegantly 3D models and virtual reality environments. However, to achieve the best results of Java3D rendering and interactive effects, visualization professional knowledge is required to make use of the Java3D advanced features. To researchers and students working on medical imaging and without sufficient visualization knowledge and experience, certain simplification and abstraction to the Java3D classes would make their work much easier. Nevertheless, some comprehensive and abstract objects need to be designed and developed to present multimodal anatomy models.
In Java3D, the visualization scenario is created as a virtual universe; objects that impact the visual effect of the virtual universe, such as geometry, lights, location, orientation, appearance, etc., are formed by nodes and arcs. A node is a data element, and an arc is a relationship between nodes. The scene graph is an organization of the nodes and arcs in the form of tree structure. Nodes are categorized into groups and leafs, where groups contain transformation and control information, and leafs contain geometrical information and visual property of visual objects. Groups are further subclassified into transform group, branch group, shared group, ordered group, and switch group. Leafs are further subclassified into shapes that contain geometric information, lights that represent light source, and many others representing the elementary information of visualization scenes. The details of Java3D are beyond the scope of this paper and are too detailed for medical image researchers who are not visualization professionals.
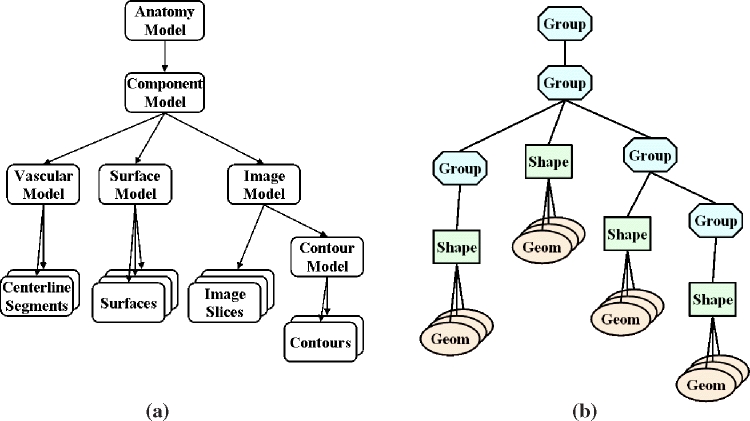
Therefore, simplification and comprehensive wrapping to Java3D is useful and necessary to provide tools with friendly and convenient interactions for medical background users. In our toolkit, the scene graph concept is inherited, although it has been enhanced to adapt to the anatomical model presentation requirements. We call it the anatomy scene graph. A number of comprehensive hierarchical object classes have been designed for building the foundation of virtual anatomy models for biomedical simulation and education. These are enhancement of the Java3D scene graph objects, which give more abstracted and simplified generic medical object models. Some of sophisticated operations and attributes are hidden to the users to have a friendlier user interface, so that users can concentrate on medical research rather than to deal with visualization techniques. Besides the general geometric elements and surface models, the geometrical shape node in the scene graph has been extended to store and interpret more complex models, e.g., tubular models and volumetric mesh models. These models will be very useful in the visualization, medical image-based analysis, and simulation applications. Figure 3 shows an example of an anatomy model that can be used in visualization and simulation and its representation of the anatomy scene graph. The group node is an extension to the Java3D scene graph transform group node, with some features of switch group and links to an anatomy model incorporated, which allows the visibility of submodels to be manipulated easily. The shape nodes are elements of anatomy models composed of the geometry node in Java3D with some functions and attributes extended for presenting more complex anatomy modal data.
Fig 3.
Visualization modeling of the toolkit: (a) anatomy data model; (b) corresponding anatomy scene graph model of (a).
Integration with Anatomical Knowledge
When developing applications or prototypes, anatomical knowledge is often referred, and sometimes, it may also be critically needed, e.g., when building anatomy atlases from various sources of data. Our toolkit contains the official international guide of anatomical terminology—Terminologia Anatomica—released by the Federative Committee of Anatomical Terminology.31 Its hierarchy structure is constructed in the XML format, and a simple built-in database stores the anatomical names. This feature supports researchers and developers to label their models with anatomy terminology.
Anatomy Scene Graph Data Schema
The anatomy scene graph model is used for presenting medical anatomy models being processed or extracted from medical images, so that they can be rendered and manipulated by the toolkit. Somehow there is no straightforward data format for storing this data model because of the complexity of anatomy model presentation and the requirement of catering anatomy information. The more important thing is the data format defined should be also flexible and extensible to handle demands from researchers with different research interests and the variety of applications. We choose XML for this purpose as it is designed to describe data in a simple, flexible, and extensible way.32 We have designed an XML schema33 according to the anatomy scene graph model described above, which incorporates the anatomical terminology along with the XML definitions mentioned above in the Image I/O module. This schema allows us to store the anatomy models and image data in XML files that are understood by the toolkit, so that they are exchangeable in the tools shown in Figure 2.
BIL-Kit-Based Tools and Applications
We have developed several tools and applications built on the toolkit, including DICOM extractor, image converter, image processor, general anatomy model simulator, vascular modeling environment, segmentation validator, and volume viewer.
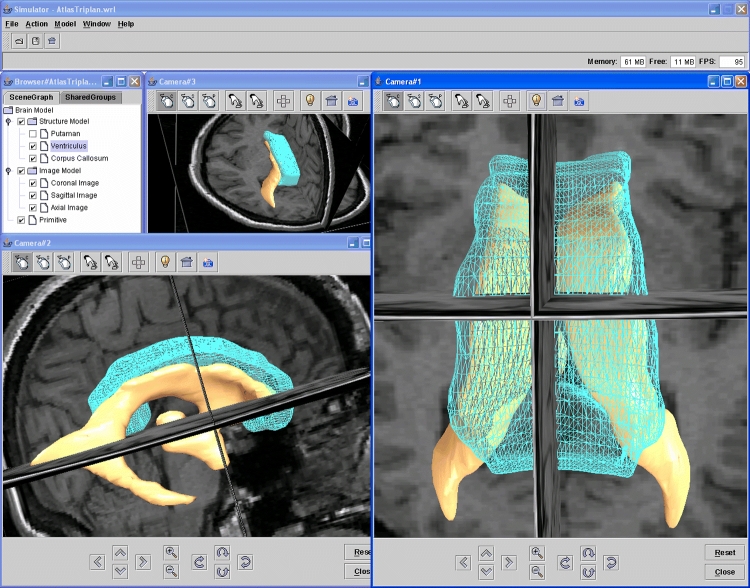
Figures 4–9 present screen snapshots of some of our tools and applications built on the toolkit. Figure 4 shows the image converter supporting a wide range of image formats to be decoded and encoded. It also provides several useful operations to do simple conversion, e.g., changing data type of pixel representation or orientation of images. Figure 5 illustrates the image processor that can be used for image analysis; many common image transformations can be performed in 2D and 3D interactively by using this tool. Figure 6 shows the model simulator where the 3D anatomy scene graph model can be visualized and manipulated. The user can interactively manipulate the hierarchy relationships of the model objects, their spatial positions, and rendering effects. Figure 7 is a vascular centerline model editing environment where the vascular model can be created and manipulated. Figure 8 shows an angiographic image anatomy labeling tool with the Terminologia Anatomica-based index integrated. A 3D cerebrovascular atlas34 is used to label angiography images. Figure 9 is a volumetric view combined with a surface-rendered model; the toolkit provides suitable functionality for hybrid volume and surface rendering. These tools have been developed successfully in a quick manner by using BIL-kit, which demonstrates its capacity and potential.
Fig 4.
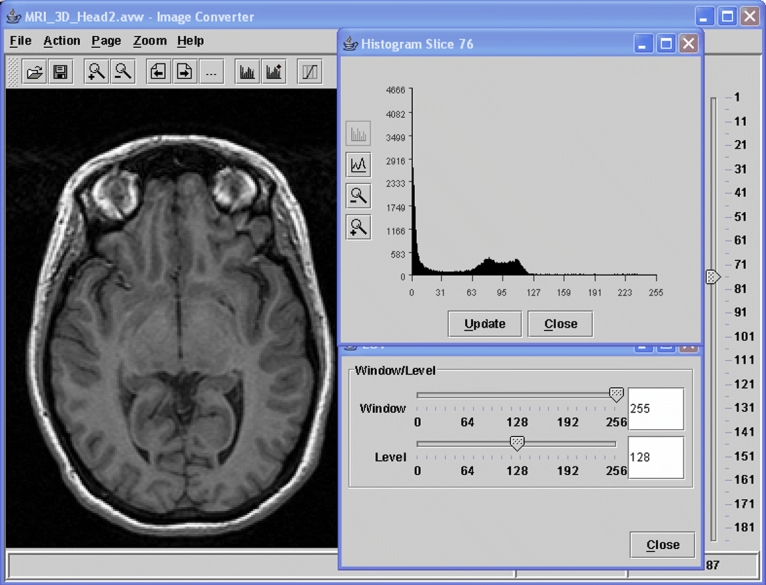
A medical image conversion tool built based on BIL-kit.
Fig 9.
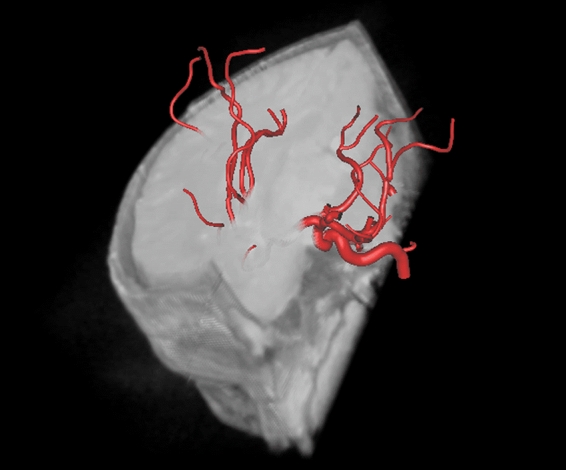
A volumetric viewer for rendering multimodal models, including hybrid volume and surface models.
Fig 5.
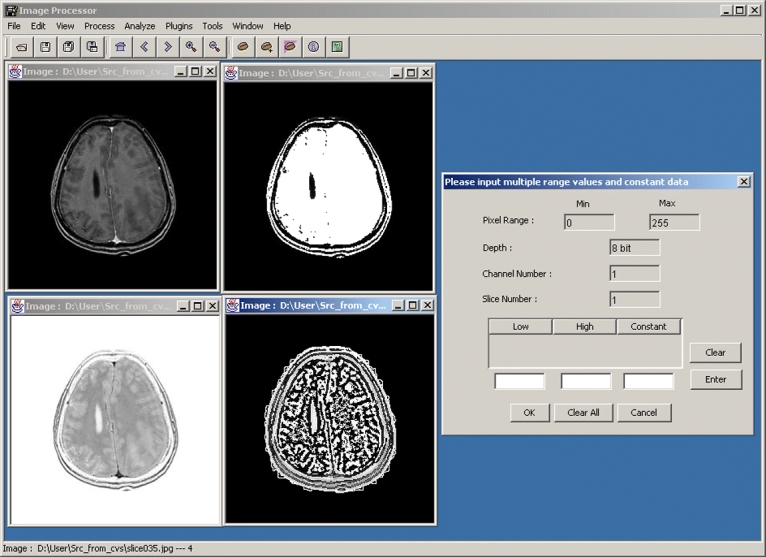
A medical image processor built based on BIL-kit.
Fig 6.
An anatomical modeling tool built based on BIL-kit for manipulating anatomical models.
Fig 7.
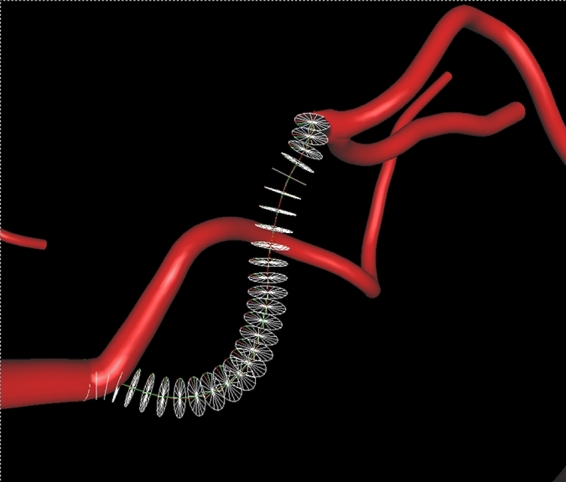
The anatomy simulator and vascular centerline modeling environment.
Fig 8.
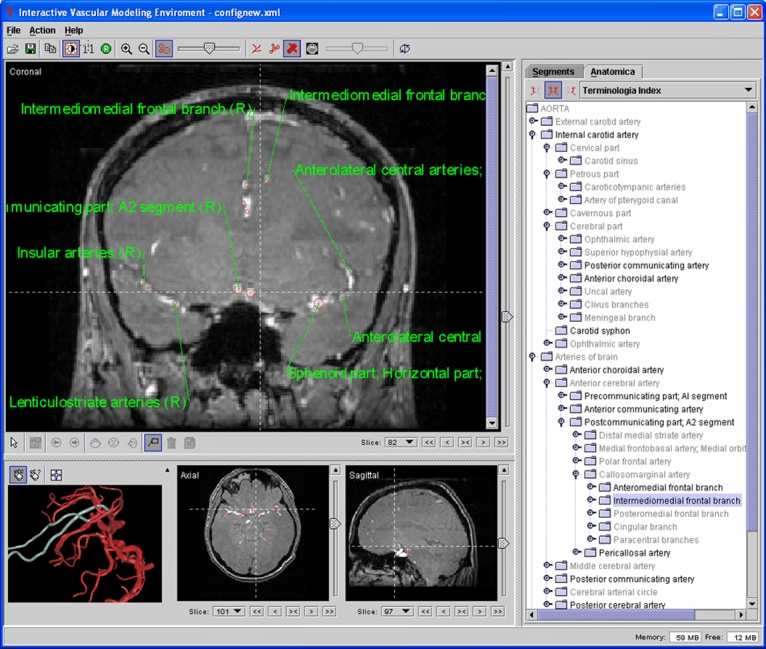
An angiographic image labeling tool with the Terminologia Anatomica index integrated.
Utilizing the Toolkit
Similar to other medical imaging and modeling toolkits, BIL-kit supports two levels of user interfaces: the developer level and the end-user level. For professionals with software engineering background, they can make use of the APIs of the reusable Java class libraries, with their preferred Java development environments, to prototype new algorithms and develop advanced tools and applications. For the users with medical background and no programming skills, they can make use of the tools built on top of the BIL-kit libraries to manipulate medical imaging data as well as to build 3D models, visualize them, and interact with them.
Summary and Future Work
This paper shares our experience and features our work in progress on building a medical imaging and visualization toolkit, which is a useful development and prototyping platform in our lab. It helps researchers and students to conduct their research work much more efficiently by providing a set of reusable libraries to reduce their redundant and tedious development efforts.
Comparing BIL-kit to the similar packages mentioned in Related Work and Our Approach, their scopes of functionality are slightly different; therefore, it is not easy to make a quantitative comparison. Our toolkit design has considered the imaging and visualization requirement from the beginning. This ensures that the two areas are combined seamlessly, which is an explicit advantage for developing medical imaging and modeling tools. This consistent design enriched the reusability and flexibility compared to other packages. The portability is achieved by using the Java language. This is much better compared to the packages that required a multiple language combination; similarly, the extensibility is also much better. In view of about 2-year development history (and a long-term experience in tool development), BIL-kit is still immature. However, the affluent functionality and several useful tools we developed in such a short period illustrate the benefits contributed by the reusability and flexibility of BIL-kit.
The major objective of researchers and students is to conduct research on their favorite domain; therefore, requesting them to learn several different languages is not cost effective. Comparing to those packages requiring various languages to take care of either algorithm codes or GUIs on different platforms, the use of a single language (Java) for both GUI and algorithms on all platforms is a more efficient approach. The advantage of portability is obvious: our tools are able to work on various platforms, such as MS Windows, Linux, Sun OS, Mac OS, and others, without any modification and even without recompiling.
The toolkit is being used in our institute from image formats conversion to image segmentation and analysis, and further to model presentation and visualization. A number of tools for image processing and 3D modeling have been constructed rapidly using the same group of classes, which demonstrated the toolkit's reusability and flexibility. Researchers and students use the reusable object classes to accelerate their research and development. More applications to integrate the latest medical imaging and modeling research results are under development. This demonstrates its potential as a useful infrastructure in medical imaging research and its extensibility. The object-oriented feature of the Java language also plays an important part in promoting its reusability.
More work will be performed in implementing advanced image-processing algorithms, such as wavelet transformation and color image processing, complicated image analysis and recognition functions, and so on, as well as advanced algorithms that are closely related to medical domain knowledge, e.g., segmentation of anatomical structures.10,11 Modeling behaviors and properties of anatomical structures are also a potential area to be explored because the toolkit is extensible to support the data models for physical deformation modeling. Combination of the multimodal virtual anatomy models with knowledge-based and semantic ontology will be an interesting idea to produce genuine and useful virtual models for medical education and simulation. There is still much space to enhance the functionality to make BIL-kit a fully useful medical imaging and visualization toolkit, acceptable by the research and clinical communities.
Acknowledgments
We gratefully acknowledge the support of the Biomedical Research Council, Agency for Science, Technology and Research, Singapore, for this research.
References
- 1.Budd T. Understanding Object-Oriented Programming with Java. Reading, MA: Addison-Wesley; 1998. [Google Scholar]
- 2.Analyze Software, Biomedical Imaging Resource, Mayo Foundation: http://www.mayo.edu/bir/Software/Analyze/Analyze.html, accessed March 17 2005
- 3.Schroeder WJ, Martin KM, Lorensen WE: The design and implementation of an object-oriented toolkit for 3d graphics and visualization. IEEE Visualization '96, pp 93–100, 1996
- 4.Schroeder WJ, Martin KM, Lorensen B. The Visualization Toolkit: An Object-Oriented Approach to 3D Graphics. Upper Saddle River, NJ: Prentice-Hall Inc.; 1998. [Google Scholar]
- 5.NLM Insight Segmentation and Registration Toolkit, http://www.itk.org/, accessed March 17 2005
- 6.Rasband W: ImageJ Introduction, http://rsb.info.nih.gov/ij/docs/intro.html, accessed March 17 2005
- 7.Whelan PF, Sadleir RJT, Ghita O. NeatVision: visual programming for computer-aided diagnostic applications. Radiographics. 2004;24:1779–1789. doi: 10.1148/rg.246045021. [DOI] [PubMed] [Google Scholar]
- 8.Zhao M, Tian J, Zhu X, Xue J, Cheng Z, Zhao H: The design and implementation of a C++ toolkit for integrated medical image processing and analyzing. In: Galloway RL Jr (Ed). Proc. SPIE Vol. 5367, Medical Imaging 2004, SPIE The International Society for Optical Engineering, Bellingham, May 2004, pp 39–47
- 9.Wolf I, Vetter M, Wegner I, Nolden M, Bottger T, Hastenteufel M, Schobinger M, Kunert T, Meinzer HP: Themedical imaging interaction toolkit (MITK): a toolkit facilitating the creation of interactive software by extending VTK and ITK. In: Galloway RL Jr (Ed). Proc. SPIE Vol. 5367, Medical Imaging 2004, SPIE The International Society for Optical Engineering, Bellingham, May 2004, pp 16–27
- 10.Xia Y, Hu Q, Aziz A, Nowinski WL. A knowledge-driven algorithm for a rapid and automatic extraction of the human cerebral ventricular system from MR neuroimages. NeuroImage. 2004;21(1):269–282. doi: 10.1016/j.neuroimage.2003.09.029. [DOI] [PubMed] [Google Scholar]
- 11.Hu Q, Nowinski WL. A rapid algorithm for robust and automatic extraction of the midsagittal plane of the human cerebrum from neuroimages based on local symmetry and outlier removal. NeuroImage. 2003;20(4):2154–2166. doi: 10.1016/j.neuroimage.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Lindholm T, Yellin F. The Java Virtual Machine Specification. Reading, MA: Addison-Wesley; 1996. [Google Scholar]
- 13.Gosling J, Joy B, Steele G: The Java Language Specification
- 14.Gosling J, McGilton H: The Java language environment. White paper, May 1996. Sun Microsystems, Inc., http://java.sun.com/docs/white/langenv/
- 15.Bouvier DJ: Getting Started with the Java3D API, Sun Microsystems, 1999, http://java.sun.com/products/java-media/3D/collateral/
- 16.Selman D. Java3D Programming. Greenwich, CT: Manning Publications Co; 2000. [Google Scholar]
- 17.Wood M, et al. OpenGL Programming Guide: The Official Guide to Learning OpenGL. Reading, MA: Addison-Wesley; 1999. [Google Scholar]
- 18.Marinilli M. Java Deployment. Indianapolis, IN: Sams; 2002. [Google Scholar]
- 19.Mangione C: Performance Tests Show Java as Fast as C++. JavaWorld, February 1998
- 20.Phipps G. Comparing observed bug and productivity rates for Java and C++ Softw Pract Exp. 1999;29(4):345–358. [Google Scholar]
- 21.The Dicom Standard. National Electrical Manufactures Association, http://medical.nema.org
- 22.Clunie DA. DICOM Structured Reporting. Bangor, PA: PixelMed Publishing; 2001. [Google Scholar]
- 23.Race P, Rice D, Vera R. Java Image I/O API Guide. Santa Clara, CA: Sun Microsystems Inc.; April 2001. [Google Scholar]
- 24.Programming in Java Advanced Imaging. SUN Microsystems, Inc. 1999, http://java.sun.com/products/java-media/jai/forDevelopers/jai1_0_1guide-unc/JAITOC.fm.html, accessed March 17, 2005
- 25.Rodrigues LH. Building Imaging Applications with Java(TM) Technology: Using AWT Imaging, Java 2D(TM), and Java(TM) Advanced Imaging (JAI) Reading, MA: Addison-Wesley; 2001. [Google Scholar]
- 26.Nikolaidis N, Pitas I. 3-D Image Processing Algorithms. New York, NY: John Wiley & Sons, Inc.; 2001. [Google Scholar]
- 27.Gonzalez RC, Woods RE. Digital Image Processing. Upper Saddle River, NJ: Prentice-Hall Inc.; 2002. [Google Scholar]
- 28.Wernecke J. The Inventor Mentor: Programming object-oriented 3D Graphics with Open Inventor, Release 2. Reading, MA: Addison-Wesley Publishing; 1993. pp. 79–93. [Google Scholar]
- 29.Hartman J, Wernecke J. The VRML 2.0 Handbook : Building Moving Worlds on the Web. Reading, MA: Addison-Wesley; 1996. [Google Scholar]
- 30.Twilleager D: http://archives.java.sun.com/cgi-bin/wa?A2=ind0403&L=java3d-interest&F=&S=&P=21949, accessed March 17 2005
- 31.International Anatomical Terminology (FCAT): Terminologia Anatomica. Stuttgart: Thieme, 1999
- 32.Extensible Markup Language (XML), http://www.w3.org/XML/, accessed March 17 2005
- 33.XML Schema, http://www.w3.org/XML/Schema, accessed March 17 2005
- 34.Nowinski WL, Thirunavuukarasuu A, Volkau I, Baimuratov R, Hu Q, Aziz A, Huang S. Three-dimensional brain atlas ofanatomy and vasculature. Radiographics. 2005;25(1):263–271. doi: 10.1148/rg.251045051. [DOI] [PubMed] [Google Scholar]