Abstract
Interactions between cells and the extracellular matrix are at the core of tissue engineering and biology. However, most studies of these interactions have used traditional two-dimensional (2D) tissue culture, which is less physiological than three-dimensional (3D) tissue culture. In this study, we compared cell behavior in four types of commonly used extracellular matrix under 2D and 3D conditions. Specifically, we quantified parameters of cell adhesion and migration by human foreskin fibroblasts in cell-derived matrix or hydrogels of collagen type I, fibrin, or basement membrane extract (BME). Fibroblasts in 3D were more spindle shaped with fewer lateral protrusions and substantially reduced actin stress fibers than on 2D matrices; cells failed to spread in 3D BME. Cell–matrix adhesion structures were detected in all matrices. Although the shapes of these cell adhesions differed, the total area per cell occupied by cell–matrix adhesions in 2D and 3D was nearly identical. Fibroblasts migrated most rapidly in cell-derived 3D matrix and collagen and migrated minimally in BME, with highest migration directionality in cell-derived matrix. This identification of quantitative differences in cellular responses to different matrix composition and dimensionality should help guide the development of customized 3D tissue culture and matrix scaffolds for tissue engineering.
Introduction
The development of optimal biocompatible scaffolds for tissue engineering requires an in-depth understanding of the interactions between cells and the extracellular matrix of the tissue of interest. Recently, natural three-dimensional (3D) matrices have been adopted as more physiological models for analyzing cell–matrix interactions than traditional two-dimensional (2D) tissue culture.1–14 Initial studies comparing cell behavior in 2D and 3D matrices have revealed differences in cell morphology,9,11 migration,1,9 adhesions,1 and signaling.4,15–20 These findings have established the importance of matrix dimensionality, that is, 3D versus 2D. However, studies to date have generally focused on a single specific 3D model, even though in vivo tissue environments can vary substantially in matrix composition.
Four of the most commonly used in vitro 3D matrix models are collagen gels,21–26 cell-derived matrix (CDM) from fibroblasts,1,6 fibrin gels,27,28 and basement membrane extract (BME or Matrigel).29 Collagen I is the most widely used matrix protein for in vitro 3D studies.30,31 Collagen I exists in vivo as fibers and is a major component of connective tissue. The primary integrin receptor used by cells to bind to 3D collagen is α2β1.11
CDM is a 3D composite of matrix proteins produced naturally by fibroblasts. After fibroblasts produce a dense 3D matrix in vitro, the matrix is denuded of cells to yield an acellular 3D matrix. A key component of these matrices is fibronectin, but it also contains significant amounts of collagen I and heparan sulfate proteoglycans such as perlecan.32 The integrin α5β1 is the primary receptor for fibronectin and CDM.1
Fibrin forms a network of fibers assembled after thrombin cleavage of fibrinogen. Fibrin serves as a provisional matrix in wound healing33 and is also involved in pathological states, such as tumor invasion.34 Fibrin, like collagen, is a self-polymerizing matrix protein that can form a gel, which permits studying 3D cell interactions with a single matrix protein. The fibers in a fibrin matrix are typically thinner, shorter, and straighter than collagen fibers.35 Polymerized fibrin was first used in 1998 as a scaffold for transplantation of tissue-engineered grafts in burns and chronic wounds.36 Cells adhere to fibrin using a number of integrins, most notably α5β1, αvβ1, and αvβ5.37
BME, also known as Matrigel, is used as a reconstituted basement membrane gel after polymerization of extracts from the Engelbroth-Holm-Swarm mouse sarcoma.38 In vivo, basement membranes separate the epithelium from connective tissue, and they are composed of collagen type IV, laminins, entactins, and proteoglycans, particularly perlecan.39,40 Depending on the cell type or malignant state, cells can proliferate or differentiate when cultured on BME.41 Cells also differentiate to varying extents depending on whether they are plated on top or inside of BME.41 The integrins α1β1, α2β1, α3β1, α4β1, α6β1, α7β1, and α10β1 are used by fibroblasts to bind to BME.42,43
Although 3D cell–matrix adhesions were reported to exist in vivo,1 a recent study on 3D collagen gels reports the absence of any discrete cell adhesions in 3D.44 Because 3D cell–matrix interactions may differ markedly depending on the specific type of ECM, a systematic comparison of the behavior of cells in diverse 3D matrices is needed to identify shared and divergent morphological and biological responses, including the nature of cell–matrix adhesions. This study presents a comparative quantitative investigation of the influence of these four matrices on the morphology, migration, and adhesion structures of primary human fibroblasts.
Materials and Methods
Cell culture
Primary human foreskin fibroblasts were a gift from Susan Yamada and Marinilce dos Santos (National Institute of Dental and Craniofacial Research, NIH), using tissue samples provided by the Cooperative Human Tissue Network funded by the National Cancer Institute. The cells were cultured in Dulbecco's modified Eagle's medium (HyClone) containing 10% fetal bovine serum (HyClone), 100 U/mL penicillin, 100 μg/mL streptomycin, and 100 μg/mL glutamine in tissue culture plastic dishes (BD Falcon). Cells were passaged at a 1:5 ratio throughout the study, and all cells were used between passages 10 and 17.
2D matrices
Twenty-millimeter glass coverslips (Deckgläser Cover Glasses) were coated with a 10 μg/mL solution of BME, collagen, or fibrinogen in phosphate-buffered saline (PBS) at 37°C for 1 h. The fibrinogen-coated coverslips were treated with thrombin (1 NIH U/100 μL) to create a fibrin coating. 2D CDM was created by physically compressing fibroblast-derived 3D matrix as described previously,45 and confocal microscopy was used to confirm that the matrix was confined to the X–Y plane. Fibroblasts were plated onto the 2D matrices at a density of 5 × 103 cells/cm2.
3D matrices
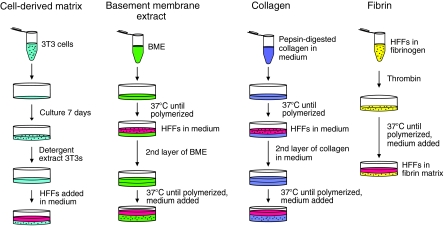
The standard steps for preparation of each of the four types of 3D matrix are indicated diagrammatically in Figure 1.
FIG. 1.
Generation of 3D extracellular matrices. Each matrix was produced as described in the Materials and Methods section at the protein concentration most commonly used in the literature (5 mg/mL CDM, 10 mg/mL BME, 2.5 mg/mL collagen, and 4 mg/mL fibrin). 3D, three-dimensional; BME, basement membrane extract; CDM, cell-derived matrix.
Cell-derived matrix
CDM was produced from high-density 7-day cultures of mouse NIH-3T3 cells as described.45 Ascorbic acid (50 μg/mL; Sigma) was added every 2 days. The total protein concentration of these matrices was determined by solubilizing in 6 M guanidine, measuring using a NanoDrop ND-1000 spectrophotometer (n = 3), and then back-calculating to the original volume of matrix. Primary human fibroblasts were plated on top of the CDM, and they rapidly penetrated the matrix as demonstrated previously by 3D confocal microscopy reconstructions showing the primary human fibroblasts fully embedded within the matrix.1
Basement membrane extract
Growth factor-reduced BME without phenol red (Trevigen; 12–18 mg/mL) was polymerized as described,46 with the following modifications: 60 μL BME was plated onto a 14 mm MatTek dish and polymerized at 37°C for 30 min. Cells were added to 3 mL medium at 1 × 104 cells/mL and incubated at 37°C for 45 min. The medium was aspirated and a second layer of 70 μL BME was added on top of the first layer. This 3D BME system was placed at 37°C to polymerize for 45 min. Alternatively, BME was diluted 1:10 with media containing 5 × 103 cells/mL, and 100 μL of this homogenous BME–cell mixture was plated onto a 14 mm MatTek dish to polymerize at 37°C for 45 min. Then, 2 mL of medium was added, and the cells were cultured for an additional 24 h. In experiments directly comparing cells seeded in a sandwich between two layers of BME with cells embedded in a single layer of BME, the same concentrations of BME were used for all layers.
Collagen matrix
Neutralized solutions of pepsinized collagen (Nutragen) at 2.5 mg/mL were prepared as previously described23 with the following modifications: 10 × Dulbecco's modified Eagle's medium (Invitrogen) was added to the collagen solution in a 1:1 ratio with 0.2 M HEPES. NaOH (0.1 N) was used to adjust the collagen solution to pH 7.4. Collagen solution (60 μL) was spread onto a 14 mm MatTek dish and polymerized at 37°C for 150 min. Cells were added to 3 mL medium at 1 × 104 cells/mL for 30 min. The medium was aspirated and a second layer of 70 μL of collagen was added on top of the first layer. It was then placed at 37°C to polymerize for 180 min. Alternatively, 5 × 103 cells/mL were mixed in with 2.5 mg/mL collagen, and 100 μL of this mixture was plated as a single layer in a 14 mm MatTek dish and polymerized at 37°C. The 3D collagen gels used for this study remained fixed to the substrate, and gels that detached or became free floating were discarded.
Fibrin matrix
Fibrin matrices were produced as previously described37 with the following modifications: 100 μL of solution containing fibrinogen, human fibroblasts, and thrombin was spread onto 14 mm MatTek dishes. Alexa 647-labeled fibrinogen (Molecular Probes) was mixed with unlabeled fibrinogen (American Diagnostica) at a 1:10 ratio.
Light microscopy
After 24 h of culture and fixation in 4% paraformaldehyde (PFA) and 5% sucrose in PBS, images of human fibroblasts in each of the 2D and 3D matrices were acquired using a Photometrics CoolSNAP ES CCD camera on an inverted microscope with a 20 × objective (Zeiss Axiovert S100).
Antibodies and reagents
Labeled activated anti-β1 integrin antibody (mAb 9EG7) was obtained from Pharmingen-BD Biosciences, paxillin antibody (clone 349) from Transduction Laboratories, and monoclonal anti-vinculin-FITC antibody from Sigma. Species-specific secondary antibodies were from Jackson ImmunoResearch. Phalloidin conjugated with rhodamine was obtained from Molecular Probes. Rabbit antifibronectin antibody R5836 was generated by our laboratory using human plasma fibronectin purified by gelatin- and heparin-Sepharose affinity chromatography. Concavalin-A (ConA) was directly labeled with DyLight 649 N-hydroxysuccinimide (Pierce). Briefly, DyLight was first adjusted to 1 mg/mL in DMSO, and ConA to 10 mg/mL in sodium borate buffer (100 mM, pH 8.65). Then, 100 μL of DyLight 649 was added per 1 mL of ConA and incubated at room temperature for 1 h. Excess dye was removed and buffer was exchanged (PBS, pH 7.4) through a desalting column (Pierce).
Immunofluorescence staining
Fibroblasts were plated on 20 mm glass coverslips or added to 3D matrices and cultured overnight. Samples were fixed and permeabilized with 4% PFA and 0.3% Triton X-100 in PBS containing 5% sucrose for 5 min and then were fixed for an additional 30 min in 4% PFA (4% PFA and 0.25% glutaraldehyde for BME) in PBS containing 5% sucrose. Immunostaining was performed with the indicated primary antibodies and rhodamine-conjugated phalloidin at room temperature for 45 min, followed by secondary Cy2- or Cy5-conjugated antibodies for 30 min. The PBS washes contained 0.05% Tween 20. CDM was stained using an antibody to fibronectin, and the fibrin matrix was visualized using Alexa 647-labeled fibrinogen. Stained samples were mounted in Gel/Mount (Biomeda).
Laser scanning confocal microscopy
Immunofluorescence microscopy images were obtained using a Zeiss LSM 510 confocal microscope equipped with an A-Plan Apochromat 63× objective (1.4 N.A.). The 488 nm Argon (∼17% power), 543 nm HeNe1 (∼60% power), and 633 nm HeNe2 (∼30% power) lasers were used to excite 488 Alexa fluor and Cy2, rhodamine–phalloidin, and Cy5, respectively. The pinholes for each laser line were aligned for optimal confocality. The BME and collagen matrices containing cultured cells close to the glass-bottom culture chamber were visualized using reflected light imaging.27,46,47 Z slices were acquired at 1 μm intervals in the 3D matrices. Digital images were obtained using LSM AIM Browser software. Each figure shown is representative of a minimum of three independent experiments analyzing at least 5–10 cells each.
Image processing
Images were processed using ImageJ (NIH) and MetaMorph Offline 7.0 software (Molecular Devices). Z slices of cells in 3D matrices were maximally projected to create a 2D image. A smoothing filter and background subtraction was used (filter details provided upon request). Data from these images were quantified by performing maximal z projections, filtering to reduce background, setting a threshold to create a binary image, and comparing the number of pixels highlighted.
Quantification of cell morphology
Cells in CDM were stained with phalloidin and fibronectin antibody to visualize the cells and the matrix, respectively. Several confocal z slices were acquired to encompass the entire cell. Phalloidin images were projected together and thresholded manually or automatically in MetaMorph to calculate the length, width, and total cell spread area of the fibroblasts in each of the matrix environments. For cells on glass, collagen, fibrin, and BME, the cells were labeled with DyLight 649-concavalin A. Confocal images were captured and the spread area was calculated in MetaMorph as described above.
Time-lapse microscopy
Cells were plated onto matrices at a density of 1 × 104 per mL. After overnight incubation, cell movements were monitored with inverted microscopes equipped for phase-contrast microscopy (Zeiss Axiovert 25) with a 37°C humidified chamber (Precision Plastics) using a 5 × (A Plan, 0.12 N.A.) objective. Images were collected with digital cameras (Infinity2; Luminera) at 10 min intervals for 24 h using Infinity2 software and stored as image stacks using MetaMorph 7.0 software (Universal Imaging/Molecular Devices). Velocity and persistence of migratory directionality were determined by tracking the positions of the cell body using the Track Point function of MetaMorph. GraphPad InStat was used to perform analysis of variance with Tukey posttests. To visualize the interactions of fibroblasts with ECM fibers, imaging of cells was initiated at 3 h after plating and continued for 24 h using phase-contrast microscopy as described above, but using a 20 × (LD A Plan, 0.3 N.A.) phase-contrast objective.
Statistical analysis
Quantitative data were compared by performing analysis of variance with Tukey posttests using InStat software (GraphPad).
Results
Fibroblast morphology in 2D and 3D extracellular matrices
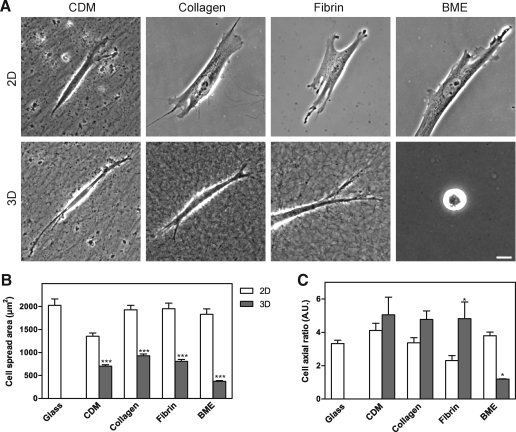
We first characterized morphological differences between fibroblasts migrating in the four extracellular matrices. Human foreskin fibroblasts were cultured on 2D or within 3D matrices overnight before being fixed and imaged using phase-contrast microscopy (Fig. 2A). The cells on 2D collagen, fibrin, and BME/Matrigel substrates were broader and more flattened and had more cell protrusions and lamellae than in the corresponding 3D matrices. Fibroblasts cultured on 2D compressed CDM displayed a more slender appearance with fewer protrusions than on the other 2D matrices. Conversely, the fibroblasts cultured in 3D CDM, collagen, and fibrin all displayed a spindle-shaped, elongated, or cylindrical phenotype with fewer protrusions than on the corresponding 2D matrices and with small or no lamellae. In 3D BME, the large majority of the cells did not spread and remained rounded. A small minority (10%) of the cells in BME could spread partially, but their overall spread area was substantially less than in the other 3D matrices.
FIG. 2.
Morphology of human fibroblasts in 2D and 3D matrices (A) and quantification of cell length, width, and spread area in 2D and 3D matrices. (A) Cells were imaged using phase-contrast microscopy at 24 h after plating on 2D matrix-coated coverslips or into 3D matrices. Scale bar represents 10 μm. (B, C) Cell outlines were determined as described in the Materials and Methods section. The cell spread area (B) was calculated on 2D and 3D substrates after concanavalin A or phalloidin staining. The cells on a 2D surface were nearly twice as large in terms of cell spread area as cells in the comparable 3D matrix. The cell axial ratio (cell length/width) was calculated (C); the cells in 3D BME were unable to spread and thus had an axial ratio close to 1 because their length and width were nearly identical. Scale bar represents 10 μm and error bars indicate standard error. *p < 0.05 and ***p < 0.001. 2D, two-dimensional.
Quantification of cell morphology
Cell outlines of human fibroblasts in 2D and 3D matrices were determined by confocal microscopy of cells stained with concanavalin A or phalloidin. The length, width, and total spread area of fibroblasts in each of the matrix environments were calculated using MetaMorph software. Consistent with the observed differences in cell morphology, the most striking quantitative differences in fibroblasts on 2D versus within 3D matrices involved overall cell shape and cell spread area (Fig. 2). In each of the 3D matrices, cells were substantially less spread than in the corresponding 2D matrix (Fig. 2B). The observed cell cross-sectional area determined from the confocal section of the cell at its widest circumference was twofold greater on 2D compressed CDM than in 3D CDM, and it was fivefold greater for cells on 2D BME than in 3D BME. The overall shape of cells as quantified by cell axial ratio (length divided by width) was the same in 3D CDM, 3D collagen, and 3D fibrin, but the ratio was close to 1 in 3D BME because the cells remained rounded without spreading (Fig. 2C). Although the fibroblasts failed to spread in 3D BME, time-lapse movies revealed that they continuously explored and interacted with the BME via small filopodial projections (data not shown). Overall, the cells tended to be less elongated and more spread on all 2D matrices and 2D glass than in 3D matrices (Figs. 2 and 3A). This difference was most profound for cells on 2D collagen, 2D fibrin, and 2D glass substrates. Parenthetically, the length of cells in 3D could be underestimated by our calculations because cells can tilt out of the X–Y plane used for quantification, so the difference is likely to be even larger. These findings are generally consistent with previous descriptions of cells in the four types of 3D matrix, although the excellent spreading on 2D BME (as great or greater than on a glass substrate) compared with the complete failure to spread in 3D BME was not expected. We conclude that fibroblasts are more elongated and spread substantially less laterally within 3D matrices compared with their morphology on 2D substrates.
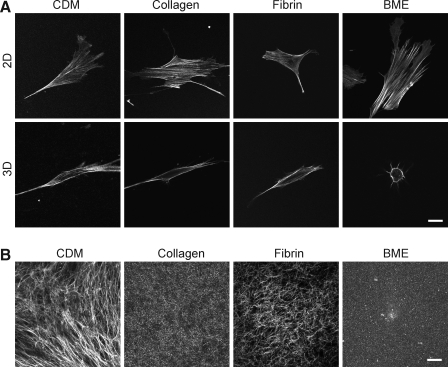
FIG. 3.
Actin distribution in primary human fibroblasts on 2D and within 3D matrices (A) and structure of 3D matrices (B). Images of actin in 3D matrices represent a single z slice through the central region midline of each cell. 3D CDM was stained using an antibody to fibronectin. Collagen and BME were visualized using reflection microscopy. Fluorescently labeled fibrinogen was used to visualize the fibrin matrices. Images were acquired using a confocal microscope. Scale bar represents 10 μm.
Actin phenotype in 2D and 3D extracellular matrices
To explore further the cytoskeletal phenotypes of fibroblasts in each of the 2D and 3D matrices, we compared confocal z slices after staining for F-actin using phalloidin. Figure 3A shows the most central confocal z slice of the fibroblasts on 2D and in 3D matrices. Numerous stress fibers are present in cells on each of the 2D matrices, whereas generally fewer, thinner, and more peripherally located stress fibers are seen in the cells in each of the 3D matrices.
Confocal imaging of 3D matrix structure
Visualizing the arrangement of proteins in a matrix is important to understand the interaction of cells with the matrix. Confocal fluorescence imaging of fibrin and CDM and reflectance/backscatter imaging of collagen and BME were used to view the overall morphology of the 3D matrices (Fig. 3B). These two modes of imaging are known to provide similar results for matrix imaging.27,30 Fibrin and collagen matrices had short fibers arranged randomly in all directions, whereas CDM had longer fibers with local orientation originally produced by the cells that had generated the matrix. BME was the only matrix composed of nonfibrous proteins. The protein concentrations chosen for each matrix were the concentrations most commonly used in studies of cells inside each of these 3D matrices. Parenthetically, it was not possible to prepare the four matrices at the same protein concentration because BME remains a liquid below 9 mg/mL48 and CDM is produced at 5 mg/mL. Fluorescence and reflectance imaging were also used to compare the responses of each type of matrix to living fibroblasts. When fibroblasts were fixed in the matrices and stained for F-actin (phalloidin) or examined by phase-contrast microscopy, accumulation of fibers adjacent to the cells in fibrin was observed (Fig. 2A).
Cell migration in 2D and 3D matrices
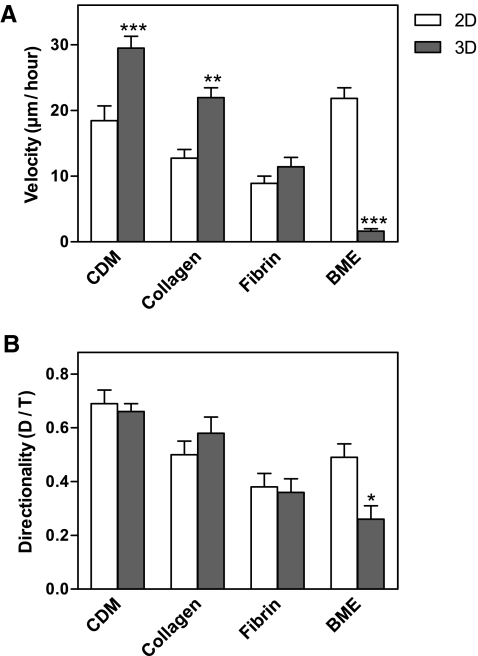
Because of the importance of cell migration in wound healing and tissue remodeling, we quantified human fibroblast migration within each of the four 3D matrices and on their 2D counterparts. Images of the cells were collected every 10 min for 24 h. At least 30 cells were tracked in each of the 2D and 3D matrices. The human fibroblasts displayed significantly different velocities in each of the 2D matrices (Fig. 4A). Interestingly, they migrated most rapidly on 2D BME-coated substrates, followed by 2D CDM, collagen, and fibrin. Among the 3D matrices, the cells migrated with the highest velocities in 3D CDM (29.5 ± 11.7 μm/h), followed by collagen and fibrin; in marked contrast to 2D conditions, they failed to migrate in 3D BME. Overall, the cells translocated 1.6 times more rapidly in 3D CDM than on 2D CDM, 1.7 times faster in 3D collagen than on 2D collagen, and 1.3 times faster in 3D fibrin than on 2D fibrin.
FIG. 4.
Velocity and directionality of fibroblasts migrating in 2D and 3D matrices. Imaging was initiated at 24 h after adding human fibroblasts to matrices. The data were collected by tracking movements of 20–60 cells under each condition in at least three separate experiments. Velocity (A) and directionality (B) were calculated using MetaMorph and Excel. The concentrations of 3T3 CDM, BME, collagen, and fibrin in the matrices were 5.2, 10, 2.5, and 4 mg/mL, respectively. Bars represent standard error. *p < 0.05, **p < 0.01, and ***p < 0.001.
Among the 3D matrices, directionality of cell migration was highest in 3D CDM (0.66 ± 0.2), followed by collagen, fibrin, and BME (Fig. 4B). There were no significant differences in directionality between 2D and 3D for CDM and fibrin substrates. The fibroblasts appeared to use a mesenchymal mode of migration47,49,50 in all of the 2D and 3D matrices, with the exception of 3D BME, in which there was virtually no migration and minimal calculated directionality. These results underscore the markedly different migratory responses of cells to the same matrix material in a planar distribution in 2D versus arrangement as a 3D hydrogel. Although three of the 3D matrices promoted more rapid migration of fibroblasts than on a 2D substrate, BME was the opposite in promoting rapid migration as a flat substrate versus as a hydrogel.
Adhesions to 3D matrices differ from adhesions to 2D matrix
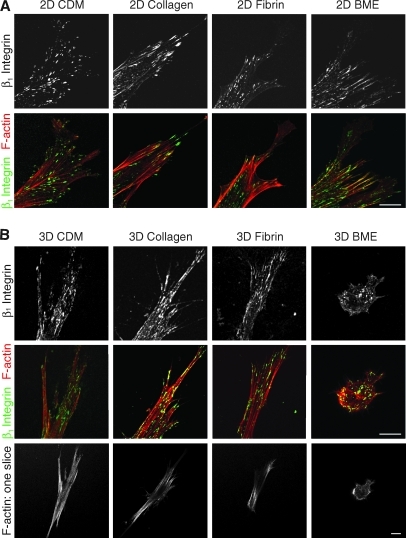
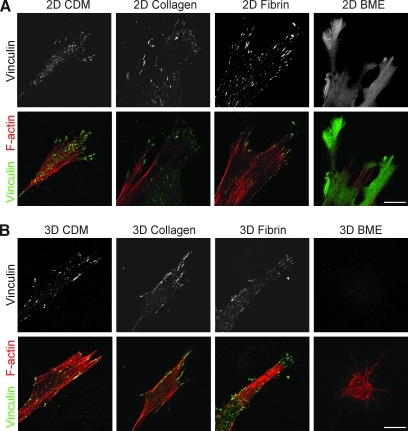
To compare the cell adhesion structures formed by fibroblasts adhering to each of the matrices, antibodies to activated β1 integrin (Fig. 5) and vinculin (Fig. 6) were used. A z stack of confocal images 1 μm apart was collected using a 63 × objective. β1 Integrin expression was observed in cell–matrix adhesions in all four matrices (Fig. 5). Vinculin staining was present in cell adhesions in 3D CDM, collagen, and fibrin (Fig. 6), but was notably diffuse in cells in BME. The cell adhesions on cells in 3D matrices were equally distributed over the cell body, as seen in the z series of adhesions in a fibrin gel in Supplementary Movie 1 (Supplementary Data are available online at www.liebertonline.com/ten). Staining for vinculin in discrete adhesions was weaker than for β1 and was sometimes not detected in collagen gels; a diffuse component of intracytoplasmic staining was observed in the cells stained for vinculin, as described by others. Overall, the fluorescence signal-to-noise ratio was greater for 2D matrices than 3D matrices. Staining for β1 and vinculin on the same cells in 3D matrices confirmed that β1 and vinculin were colocalized at >95% of the adhesions in 3D CDM, collagen, and fibrin (Supplementary Fig. S1). It is theoretically possible that cell adhesions and morphology might differ in cells analyzed in the matrix sandwiches we used for 3D collagen and BME (to ensure that the cells would be suspended in the matrix) compared with cells analyzed in 3D gels poured as a single layer. However, direct comparisons of morphology and integrin-based cell adhesions showed that they were very similar, with no reproducible differences found between cells cultured in sandwiches or a single layer of the 3D matrix (Supplementary Figs. S2 and S3).
FIG. 5.
Distribution of β1 integrin and F-actin in 2D and 3D matrices. Human fibroblasts were cultured on 2D matrix-coated glass coverslips (A) or in 3D matrices (B) and fixed after 24 h of culture. Samples were stained for activated β1 integrin (monoclonal antibody 9EG7, green) and F-actin (phalloidin, red). Images are maximum projections of z slices, unless otherwise indicated. Representative images are shown from three separate experiments. Scale bar represents 10 μm.
FIG. 6.
Distribution of vinculin and F-actin in 2D and 3D matrices. Fibroblasts were cultured on 2D matrix-coated glass coverslips (A) or in 3D matrices (B) and fixed after 24 h of culture. Samples were stained for vinculin (green) and F-actin (phalloidin, red). Images are maximum projections of z slices. Representative images are shown from three separate experiments. Scale bar represents 10 μm.
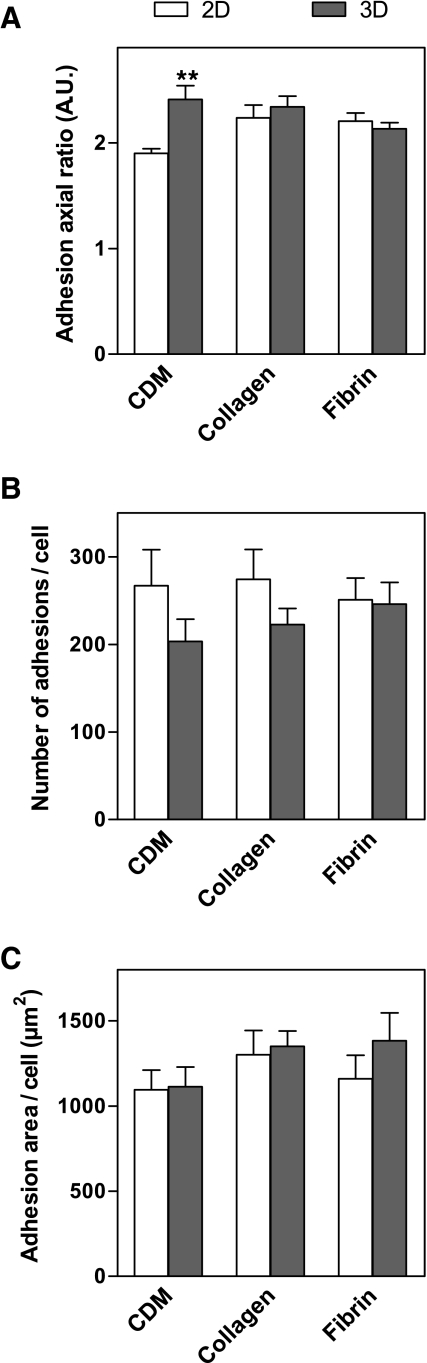
Quantification of cell adhesions
Confocal images of β1 integrin staining of fibroblasts in 2D and 3D matrices were used to calculate the length, width, and area of each cell adhesion structure, the average number of adhesions per cell, and the total summed area of cell adhesions per cell (Fig. 7). β1 Integrin was chosen as the marker for cell adhesions because staining for β1 was stronger than for vinculin, and β1 and vinculin generally colocalized as noted above. Adhesions were quantified in 3D CDM, collagen, and fibrin matrices, the three 3D matrices in which adhesions were observed. The average length of adhesions in 3D matrices was 1.74 ± 0.09 μm, and no significant difference was found between the 3D matrices. Likewise, the average width of adhesions was 0.76 ± 0.02 μm, with no significant difference between the 3D matrices. The length of adhesions in 3D CDM was significantly greater than on 2D CDM, whereas the widths of adhesions in each of the 2D and 3D matrices were not significantly different. Consequently, the axial ratio (length/width) of adhesions in 3D CDM was significantly greater than in 2D CDM (Fig. 7A). The average number of adhesions per cell in 3D matrices was 224 ± 23, and this number did not vary significantly between the 3D matrices (Fig. 7B). In contrast to the substantial differences observed in overall cell morphology and cell spread area in 2D and 3D, the overall total area involving cell–matrix adhesion structures on the cells was not significantly different between any of the paired 2D and 3D matrices (Fig. 7C).
FIG. 7.
Quantification of cell adhesions in 2D and 3D matrices. Z projections of β1 staining of human fibroblasts in 2D and 3D matrices were created. The axial ratio (A) of the cell adhesions were compared for each of the 3D and corresponding 2D matrices. The average number of cell adhesions per cell was also compared (B), along with the total area covered by cell adhesions per cell (C). Statistical analyses were performed using analysis of variance and Tukey posttests (n = 8–13 cells). **p < 0.01.
Discussion
Understanding how cells interact with natural 3D matrices of varying compositions is critical for the rational development of biomimetic scaffolds and microenvironments for tissue engineering. Because little is known about differences in the behavior of a cell in different 3D matrices, we focused on a single cell type—human fibroblasts—as they interacted with four widely used 3D matrices. We directly compared cell morphology, migration, and adhesions to provide fundamental comprehensive information on cellular responses to each type of 2D or 3D matrix.
Fibroblasts in each type of 3D matrix tended to display a more cylindrical or spindle-shaped phenotype, with fewer protrusions or lamellipodia, as well as less total cell spread area than on a 2D matrix or glass substrate. Except in BME, the cell axial ratio (length vs. width) of fibroblasts in each type of 3D matrix was quite similar. This shape resembles the phenotype of fibroblasts and mesenchymal cells in vivo.1,9,11,51 Fibroblasts on 2D matrices, especially on collagen, fibrin, and glass, had a spread phenotype. The reduced number of cell protrusions and lamellipodia on 2D CDM compared with the other 2D substrates may be due to the fact that 2D CDM is a compressed matrix in which fibers are still present in the X–Y plane, so cells spread along fibers rather than on a uniformly coated substrate; in fact, a recent study has established that narrow linear 1D matrix patterns strongly mimic 3D fibrillar matrix in effects on cell morphology, cytoskeleton, and migration.20
The strikingly rounded, nonmotile phenotype of human fibroblasts in 3D BME differs markedly from the behavior of these cells in the other 3D matrices. It is, however, consistent with the fact that fibroblasts in vivo are predominantly found in the connective tissue stroma and not in the thin basement membrane that separates the epithelium from the connective tissue. Others have also found that fibroblasts do not grow well in the presence of Matrigel/BME.41 In contrast, however, these cells spread very well and migrated particularly rapidly on BME as a 2D substrate, perhaps because it mimics a planar basement membrane. The relatively diffuse distribution of vinculin in adhesions of rapidly migrating fibroblasts on 2D BME is reminiscent of the diffuse contacts observed in rapidly migrating embryonic fibroblasts before they start to form focal adhesions and slow their rates of migration during maturation in cell culture.52 Cell differentiation is known to differ depending on whether cells are cultured on top of or within BME.41 Although the fibroblasts failed to spread in 3D BME, they continued to interact with the BME via small filopodial projections, resembling microglia in the brain that remain stationary while scanning their environment using filopodial protrusions.53
In 2D cell culture, the actin cytoskeleton has been well established to have important roles in cell morphology and migration, as well as linking the cell interior and the ECM through cell adhesions.54–59 In vivo, however, fibroblasts exhibit few actin stress fibers,60 except in tissues that undergo constant large mechanical loading, such as tendon.61 We observed a similar paucity of stress fibers in 3D CDM, collagen, and fibrin, in which only few thin stress fibers were seen near the plasma membrane. Thus, the actin phenotypes in 3D matrices are much more similar to cells in vivo than to cells on 2D matrix-coated coverslips. The observation that numerous stress fibers span the cells in all of the 2D models examined in this study is consistent with higher levels of cytoskeletal stress associated with stiff, noncompliant substrates, which promote the formation of focal adhesions and stress fibers.62–65
Many cells, such as fibroblasts and macrophages, must migrate through ECM to carry out functions such as tissue repair and remodeling. Thus, it is important to characterize and compare cell migration in the various 3D matrices. Our results revealed that the velocity of fibroblasts is dependent on both the dimensionality and the composition of the matrix—it varied not only between the 2D matrices and the 3D matrices of the same compositions, but also between 3D matrices of differing composition. Fibroblasts migrating within 3D CDM had the greatest velocity and directionality, at 30 μm/h and 0.7, respectively, and their velocity was 1.6 times that of the cells in 2D CDM. The latter data are similar to those in a previous study comparing 2D and 3D migration in this CDM model, which reported a migration rate of 30 μm/h and a velocity 1.5 times higher in 3D CDM than on 2D CDM.1 However, to our knowledge, no one has directly compared cell migration between multiple 3D matrices. For inducing maximal rates of migration, the effectiveness of matrices varied substantially in the following order: CDM > collagen > fibrin > BME. Consequently, we conclude that one cannot merely study cell migration or other processes in 3D, but that it is also crucial to consider the molecular composition of the 3D model system being investigated.
The velocities of migration of cells on 2D matrices were different than in 3D matrices, and the relative order of rates also differed, with velocities on BME > CDM > collagen > fibrin. The differences between rates of migration and organization of the actin cytoskeleton in 2D versus 3D environments may result at least in part from the different physical properties of the migratory substrate. It has been suggested that cells often migrate more slowly on a 2D matrix than within a 3D matrix because 2D matrix has a greater density/concentration of ligand on which the cells can make large focal adhesions and remain more firmly anchored to the substrate.66,67 However, a striking exception was found on or within BME, which might result from the nature of normal fibroblasts to remain in the connective tissue and not migrate into the basement membrane. In contrast, malignant cells of the fibroblast lineage (human fibrosarcoma cells) do migrate in 3D BME gels.8
Interestingly, the directional persistence of cell migration often depended on the composition, but not necessarily on the dimensionality, of the matrices. Whether cells migrate with directional persistence or more randomly can be regulated by a number of mechanisms,50,68 to which the present study adds matrix composition. Time-lapse movies of these fibroblasts migrating in 3D matrices revealed that the cells in 3D CDM, collagen, and fibrin migrated by mesenchymal mode. Mesenchymal migration is the standard mode of migration for NIH 3T3 fibroblasts and is characterized by cycles of protrusion, attachment, and retraction69,70 using localized (as opposed to diffuse) adhesions.47,50
Cells interact with ECM through adhesions. Adhesions have been described in detail for cells on 2D matrix, and the best characterized forms are the focal adhesions—mature focal adhesions contain primarily the αvβ3 integrin. In contrast, longer, thinner structures termed fibrillar adhesions have been implicated in matrix assembly and primarily utilize the α5β1 integrin.71,72 A recent study using immortalized tumor cells has challenged the existence of any detectable cell adhesions in a 3D collagen gel, even though the cells retained a requirement for several adhesion components including vinculin.44 Our study addresses this unresolved question about the existence of cell adhesions in 3D environments directly and quantitatively using primary human fibroblasts embedded in a variety of different types of 3D matrix.
When fibroblast adhesions were previously characterized in a complex 3D CDM based heavily on fibronectin, the cell adhesions were termed 3D matrix adhesions; they had a characteristically long, slender phenotype dependent solely on the α5β1 integrin rather than the αvβ3 integrin, which is characteristic of focal adhesions.1 Adhesions have also been reported in 3D collagen73–75 and 3D fibrin,76 but the absence of quantification and direct comparisons of these adhesions under different conditions of matrix composition and directionality has left the extent or even existence of these adhesion structures open to question.44 We found that cell–matrix adhesions were readily detectable in each of the 3D matrices, particularly by examining for prominent aggregates of activated, ligand-occupied integrins. These adhesions were distributed all over the cell surface rather than being located primarily on the ventral (lower) surface, as observed on cells in traditional 2D cell culture. The axial ratio (L/W) of adhesions in 3D CDM was greater than in 2D CDM, but was not significantly different between 2D and 3D matrices of fibrin and collagen. This could be because CDM has long, linear fibers, which contribute to the formation of adhesions with a greater L/W ratio. Notably, however, there were no major differences under 2D versus 3D conditions in the average number of adhesions per cell and the total area of adhesions. However, we did observe in 3D collagen gels that, even though prominent adhesions were readily detectable by immunostaining for integrins, staining for vinculin was variable and at times not detectable (see Supplementary Fig. S2 and data not shown). We speculate that because these collagen gels lack the large organized fibrils of collagen seen in vivo, there may be less force at the cell adhesions, resulting in diminished accumulation of force-dependent cell adhesion components such as vinculin.77 Overall, our results suggest that even though classical focal adhesions per se and stress fibers are not prominent in 3D matrices, the area of the cell surface devoted to interactions mediated by other types of adhesive structures besides focal adhesions with the surrounding matrix is at least as large for 3D as for 2D cell–matrix interactions. Our quantification also establishes that there are no significant differences in total interaction area devoted to defined cell adhesion structures between matrices of different composition.
Overall, our results establish that biochemical composition and dimensionality both play important roles in fibroblast cell–matrix interactions. Of the four common 3D matrices studied here, each matrix has distinct effects on cell morphology and especially on cell migration. Consequently, one cannot simply study cells in 3D. Moreover, the choice of the best 3D molecular microenvironment for 3D studies or biomimetic tissue engineering will depend on the actual in vivo system being studied, mimicked, or repaired. For example, our study establishes that cells can migrate exceptionally efficiently on BME in a flat 2D configuration and yet are completely unable to migrate in the same material in a 3D configuration. This material would consequently be useful for promoting lateral migratory dispersion of cells while blocking penetration into the matrix. The approaches and findings presented in this study should facilitate the design of scaffolds for tissue engineering78 and the analysis of the cellular effects of changing ECM microenvironments in biological processes as diverse as development, wound healing, tumor progression, and remodeling of bioengineered tissues.
Supplementary Material
Acknowledgments
Dr. Marinilce Santos provided valuable advice and assistance with time-lapse microscopy and collagen matrix production. The authors thank Dr. Allison Berrier for numerous helpful discussions, Dr. Angelo Green for generously providing the labeled β1 integrin antibody, and Kazue Matsumoto for support with the CDM and in many other aspects. This work was supported by the HHMI-NIH Research Scholars Program and the Intramural Research Program of the National Institute of Dental and Craniofacial Research, National Institutes of Health.
Disclosure Statement
No competing financial interests exist.
References
- 1.Cukierman E. Pankov R. Stevens D.R. Yamada K.M. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 2.Roskelley C.D. Desprez P.Y. Bissell M.J. Extracellular matrix-dependent tissue-specific gene expression in mammary epithelial cells requires both physical and biochemical signal transduction. Proc Natl Acad Sci USA. 1994;91:12378. doi: 10.1073/pnas.91.26.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Streuli C.H. Bailey N. Bissell M.J. Control of mammary epithelial differentiation: basement membrane induces tissue-specific gene expression in the absence of cell-cell interaction and morphological polarity. J Cell Biol. 1991;115:1383. doi: 10.1083/jcb.115.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson C.M. Bissell M.J. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2006;22:287. doi: 10.1146/annurev.cellbio.22.010305.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun T. Jackson S. Haycock J.W. MacNeil S. Culture of skin cells in 3D rather than 2D improves their ability to survive exposure to cytotoxic agents. J Biotechnol. 2006;122:372. doi: 10.1016/j.jbiotec.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 6.Cukierman E. Pankov R. Yamada K.M. Cell interactions with three-dimensional matrices. Curr Opin Cell Biol. 2002;14:633. doi: 10.1016/s0955-0674(02)00364-2. [DOI] [PubMed] [Google Scholar]
- 7.Stegemann J.P. Nerem R.M. Altered response of vascular smooth muscle cells to exogenous biochemical stimulation in two- and three-dimensional culture. Exp Cell Res. 2003;283:146. doi: 10.1016/s0014-4827(02)00041-1. [DOI] [PubMed] [Google Scholar]
- 8.Zaman M.H. Trapani L.M. Sieminski A.L. Mackellar D. Gong H. Kamm R.D., et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci USA. 2006;103:10889. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elsdale T. Bard J. Collagen substrata for studies on cell behavior. J Cell Biol. 1972;54:626. doi: 10.1083/jcb.54.3.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Even-Ram S. Yamada K.M. Cell migration in 3D matrix. Curr Opin Cell Biol. 2005;17:524. doi: 10.1016/j.ceb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 11.Friedl P. Brocker E.B. The biology of cell locomotion within three-dimensional extracellular matrix. Cell Mol Life Sci. 2000;57:41. doi: 10.1007/s000180050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abbott A. Cell culture: biology's new dimension. Nature. 2003;424:870. doi: 10.1038/424870a. [DOI] [PubMed] [Google Scholar]
- 13.Yamada K.M. Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130:601. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Rangarajan R. Zaman M.H. Modeling cell migration in 3D: status and challenges. Cell Adh Migr. 2008;2:106. doi: 10.4161/cam.2.2.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ravanti L. Heino J. Lopez-Otin C. Kahari V.M. Induction of collagenase-3 (MMP-13) expression in human skin fibroblasts by three-dimensional collagen is mediated by p38 mitogen-activated protein kinase. J Biol Chem. 1999;274:2446. doi: 10.1074/jbc.274.4.2446. [DOI] [PubMed] [Google Scholar]
- 16.Koike T. Vernon R.B. Hamner M.A. Sadoun E. Reed M.J. MT1-MMP, but not secreted MMPs, influences the migration of human microvascular endothelial cells in 3-dimensional collagen gels. J Cell Biochem. 2002;86:748. doi: 10.1002/jcb.10257. [DOI] [PubMed] [Google Scholar]
- 17.Fisher K.E. Pop A. Koh W. Anthis N.J. Saunders W.B. Davis G.E. Tumor cell invasion of collagen matrices requires coordinate lipid agonist-induced G-protein and membrane-type matrix metalloproteinase-1-dependent signaling. Mol Cancer. 2006;5:69. doi: 10.1186/1476-4598-5-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berrier A.L. Yamada K.M. Cell-matrix adhesion. J Cell Physiol. 2007;213:565. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 19.Daley W.P. Peters S.B. Larsen M. Extracellular matrix dynamics in development and regenerative medicine. J Cell Sci. 2008;121:255. doi: 10.1242/jcs.006064. [DOI] [PubMed] [Google Scholar]
- 20.Doyle A.D. Wang F.W. Matsumoto K. Yamada K.M. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184:481. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grinnell F. Fibroblast-collagen-matrix contraction: growth-factor signalling and mechanical loading. Trends Cell Biol. 2000;10:362. doi: 10.1016/s0962-8924(00)01802-x. [DOI] [PubMed] [Google Scholar]
- 22.Rhee S. Grinnell F. Fibroblast mechanics in 3D collagen matrices. Adv Drug Deliv Rev. 2007;59:1299. doi: 10.1016/j.addr.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandulache V.C. Parekh A. Dohar J.E. Hebda P.A. Fetal dermal fibroblasts retain a hyperactive migratory and contractile phenotype under 2-and 3-dimensional constraints compared to normal adult fibroblasts. Tissue Eng. 2007;13:2791. doi: 10.1089/ten.2006.0412. [DOI] [PubMed] [Google Scholar]
- 24.Karamichos D. Brown R.A. Mudera V. Complex dependence of substrate stiffness and serum concentration on cell-force generation. J Biomed Mater Res A. 2006;78:407. doi: 10.1002/jbm.a.30814. [DOI] [PubMed] [Google Scholar]
- 25.Maaser K. Wolf K. Klein C.E. Niggemann B. Zanker K.S. Brocker E.B., et al. Functional hierarchy of simultaneously expressed adhesion receptors: integrin alpha2beta1 but not CD44 mediates MV3 melanoma cell migration and matrix reorganization within three-dimensional hyaluronan-containing collagen matrices. Mol Biol Cell. 1999;10:3067. doi: 10.1091/mbc.10.10.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grinnell F. Petroll W.M. Cell motility and mechanics in three-dimensional collagen matrices. Annu Rev Cell Dev Biol. 2010;26:335. doi: 10.1146/annurev.cellbio.042308.113318. [DOI] [PubMed] [Google Scholar]
- 27.Hartmann A. Boukamp P. Friedl P. Confocal reflection imaging of 3D fibrin polymers. Blood Cells Mol Dis. 2006;36:191. doi: 10.1016/j.bcmd.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 28.Sun T. Haycock J. Macneil S. In situ image analysis of interactions between normal human keratinocytes and fibroblasts cultured in three-dimensional fibrin gels. Biomaterials. 2006;27:3459. doi: 10.1016/j.biomaterials.2006.01.058. [DOI] [PubMed] [Google Scholar]
- 29.Radisky D.C. Levy D.D. Littlepage L.E. Liu H. Nelson C.M. Fata J.E., et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436:123. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedl P. Dynamic imaging of cellular interactions with extracellular matrix. Histochem Cell Biol. 2004;122:183. doi: 10.1007/s00418-004-0682-0. [DOI] [PubMed] [Google Scholar]
- 31.Friedl P. Maaser K. Klein C.E. Niggemann B. Krohne G. Zanker K.S. Migration of highly aggressive MV3 melanoma cells in 3-dimensional collagen lattices results in local matrix reorganization and shedding of alpha2 and beta1 integrins and CD44. Cancer Res. 1997;57:2061. [PubMed] [Google Scholar]
- 32.Yamada K.M. Pankov R. Cukierman E. Dimensions and dynamics in integrin function. Braz J Med Biol Res. 2003;36:959. doi: 10.1590/s0100-879x2003000800001. [DOI] [PubMed] [Google Scholar]
- 33.Clark R.A. Lanigan J.M. DellaPelle P. Manseau E. Dvorak H.F. Colvin R.B. Fibronectin and fibrin provide a provisional matrix for epidermal cell migration during wound reepithelialization. J Invest Dermatol. 1982;79:264. doi: 10.1111/1523-1747.ep12500075. [DOI] [PubMed] [Google Scholar]
- 34.Mosesson M.W. Introduction: fibrinogen as a determinant of the metastatic potential of tumor cells. Blood. 2000;96:3301. [PubMed] [Google Scholar]
- 35.Roeder B.A. Kokini K. Sturgis J.E. Robinson J.P. Voytik-Harbin S.L. Tensile mechanical properties of three-dimensional type I collagen extracellular matrices with varied microstructure. J Biomech Eng. 2002;124:214. doi: 10.1115/1.1449904. [DOI] [PubMed] [Google Scholar]
- 36.Horch R.E. Bannasch H. Kopp J. Andree C. Stark G.B. Single-cell suspensions of cultured human keratinocytes in fibrin-glue reconstitute the epidermis. Cell Transplant. 1998;7:309. doi: 10.1177/096368979800700309. [DOI] [PubMed] [Google Scholar]
- 37.Alavi A. Stupack D.G. Cell survival in a three-dimensional matrix. Methods Enzymol. 2007;426:85. doi: 10.1016/S0076-6879(07)26005-7. [DOI] [PubMed] [Google Scholar]
- 38.Kleinman H.K. McGarvey M.L. Hassell J.R. Star V.L. Cannon F.B. Laurie G.W., et al. Basement membrane complexes with biological activity. Biochemistry. 1986;25:312. doi: 10.1021/bi00350a005. [DOI] [PubMed] [Google Scholar]
- 39.Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat Rev Cancer. 2003;3:422. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- 40.Timpl R. Brown J.C. Supramolecular assembly of basement membranes. Bioessays. 1996;18:123. doi: 10.1002/bies.950180208. [DOI] [PubMed] [Google Scholar]
- 41.Kleinman H.K. Preparation of gelled substrates. Curr Protoc Cell Biol. May, 2001. Chapter 10: Unit 10.3. [DOI] [PubMed]
- 42.White E.S. Thannickal V.J. Carskadon S.L. Dickie E.G. Livant D.L. Markwart S., et al. Integrin alpha4beta1 regulates migration across basement membranes by lung fibroblasts: a role for phosphatase and tensin homologue deleted on chromosome 10. Am J Respir Crit Care Med. 2003;168:436. doi: 10.1164/rccm.200301-041OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan K.T. Cortesio C.L. Huttenlocher A. Integrins in cell migration. Methods Enzymol. 2007;426:47. doi: 10.1016/S0076-6879(07)26003-3. [DOI] [PubMed] [Google Scholar]
- 44.Fraley S.I. Feng Y. Krishnamurthy R. Kim D.H. Celedon A. Longmore G.D., et al. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat Cell Biol. 2010;12:598. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cukierman E. Preparation of extracellular matrices produced by cultured fibroblasts. Curr Protoc Cell Biol. Jan, 2007. Chapter 10: Unit 10.9. [DOI] [PubMed]
- 46.Hooper S. Marshall J.F. Sahai E. Tumor cell migration in three dimensions. Methods Enzymol. 2006;406:625. doi: 10.1016/S0076-6879(06)06049-6. [DOI] [PubMed] [Google Scholar]
- 47.Wolf K. Mazo I. Leung H. Engelke K. von Andrian U.H. Deryugina E.I., et al. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160:267. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trevigen. Protocol 3430–005-01. www.trevigen.com/angiocell/cultrexbme.php. www.trevigen.com/angiocell/cultrexbme.php
- 49.Ridley A.J. Schwartz M.A. Burridge K. Firtel R.A. Ginsberg M.H. Borisy G., et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 50.Friedl P. Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2010;188:11. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beningo K.A. Dembo M. Wang Y.L. Responses of fibroblasts to anchorage of dorsal extracellular matrix receptors. Proc Natl Acad Sci USA. 2004;101:18024. doi: 10.1073/pnas.0405747102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Couchman J.R. Rees D.A. The behaviour of fibroblasts migrating from chick heart explants: changes in adhesion, locomotion and growth, and in the distribution of actomyosin and fibronectin. J Cell Sci. 1979;39:149. doi: 10.1242/jcs.39.1.149. [DOI] [PubMed] [Google Scholar]
- 53.Nimmerjahn A. Kirchhoff F. Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 54.Chen C.S. Mrksich M. Huang S. Whitesides G.M. Ingber D.E. Geometric control of cell life and death. Science. 1997;276:1425. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 55.Vicente-Manzanares M. Choi C.K. Horwitz A.R. Integrins in cell migration—the actin connection. J Cell Sci. 2009;122:199. doi: 10.1242/jcs.018564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Clainche C. Carlier M.F. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 57.Lock J.G. Wehrle-Haller B. Stromblad S. Cell-matrix adhesion complexes: master control machinery of cell migration. Semin Cancer Biol. 2008;18:65. doi: 10.1016/j.semcancer.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 58.Yamaguchi H. Wyckoff J. Condeelis J. Cell migration in tumors. Curr Opin Cell Biol. 2005;17:559. doi: 10.1016/j.ceb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 59.Gardel M.L. Schneider I.C. Aratyn-Schaus Y. Waterman C. Integration of actin and adhesion dynamics in cell migration. Annu Rev Cell Dev Biol. 2010;26:315. doi: 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hinz B. Gabbiani G. Mechanisms of force generation and transmission by myofibroblasts. Curr Opin Biotechnol. 2003;14:538. doi: 10.1016/j.copbio.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 61.Ralphs J.R. Waggett A.D. Benjamin M. Actin stress fibres and cell-cell adhesion molecules in tendons: organisation in vivo and response to mechanical loading of tendon cells in vitro. Matrix Biol. 2002;21:67. doi: 10.1016/s0945-053x(01)00179-2. [DOI] [PubMed] [Google Scholar]
- 62.Discher D.E. Janmey P. Wang Y.L. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 63.Dubash A.D. Menold M.M. Samson T. Boulter E. Garcia-Mata R. Doughman R., et al. Chapter 1. Focal adhesions: new angles on an old structure. Int Rev Cell Mol Biol. 2009;277:1. doi: 10.1016/S1937-6448(09)77001-7. [DOI] [PubMed] [Google Scholar]
- 64.Bershadsky A.D. Balaban N.Q. Geiger B. Adhesion-dependent cell mechanosensitivity. Annu Rev Cell Dev Biol. 2003;19:677. doi: 10.1146/annurev.cellbio.19.111301.153011. [DOI] [PubMed] [Google Scholar]
- 65.Doyle A.D. Yamada K.M. Cellular mechanotransduction: interactions with the extracellular matrix. In: Mofrad M.R.K., editor; Kamm R.D., editor. Cellular Mechanotransduction. New York, NY: Cambridge University Press; 2010. p. 120. [Google Scholar]
- 66.DiMilla P.A. Stone J.A. Quinn J.A. Albelda S.M. Lauffenburger D.A. Maximal migration of human smooth muscle cells on fibronectin and type IV collagen occurs at an intermediate attachment strength. J Cell Biol. 1993;122:729. doi: 10.1083/jcb.122.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodman S.L. Risse G. von der Mark K. The E8 subfragment of laminin promotes locomotion of myoblasts over extracellular matrix. J Cell Biol. 1989;109:799. doi: 10.1083/jcb.109.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petrie R.J. Doyle A.D. Yamada K.M. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10:538. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Palecek S.P. Loftus J.C. Ginsberg M.H. Lauffenburger D.A. Horwitz A.F. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature. 1997;385:537. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 70.Lauffenburger D.A. Horwitz A.F. Cell migration: a physically integrated molecular process. Cell. 1996;84:359. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 71.Pankov R. Cukierman E. Katz B.Z. Matsumoto K. Lin D.C. Lin S., et al. Integrin dynamics and matrix assembly: tensin-dependent translocation of alpha(5)beta(1) integrins promotes early fibronectin fibrillogenesis. J Cell Biol. 2000;148:1075. doi: 10.1083/jcb.148.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zamir E. Katz M. Posen Y. Erez N. Yamada K.M. Katz B.Z., et al. Dynamics and segregation of cell-matrix adhesions in cultured fibroblasts. Nat Cell Biol. 2000;2:191. doi: 10.1038/35008607. [DOI] [PubMed] [Google Scholar]
- 73.Tamariz E. Grinnell F. Modulation of fibroblast morphology and adhesion during collagen matrix remodeling. Mol Biol Cell. 2002;13:3915. doi: 10.1091/mbc.E02-05-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wozniak M.A. Desai R. Solski P.A. Der C.J. Keely P.J. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J Cell Biol. 2003;163:583. doi: 10.1083/jcb.200305010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y.K. Wang Y.H. Wang C.Z. Sung J.M. Chiu W.T. Lin S.H., et al. Rigidity of collagen fibrils controls collagen gel-induced down-regulation of focal adhesion complex proteins mediated by alpha2beta1 integrin. J Biol Chem. 2003;278:21886. doi: 10.1074/jbc.M300092200. [DOI] [PubMed] [Google Scholar]
- 76.Zhou X. Rowe R.G. Hiraoka N. George J.P. Wirtz D. Mosher D.F., et al. Fibronectin fibrillogenesis regulates three-dimensional neovessel formation. Genes Dev. 2008;22:1231. doi: 10.1101/gad.1643308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Galbraith C.G. Yamada K.M. Sheetz M.P. The relationship between force and focal complex development. J Cell Biol. 2002;159:695. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huebsch N. Mooney D.J. Inspiration and application in the evolution of biomaterials. Nature. 2009;462:426. doi: 10.1038/nature08601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.