Chromatin structure imposed by condensin II at centromeres enables xHJURP-mediated incorporation of CENP-A.
Abstract
Centromeric protein A (CENP-A) is the epigenetic mark of centromeres. CENP-A replenishment is necessary in each cell cycle to compensate for the dilution associated to DNA replication, but how this is achieved mechanistically is largely unknown. We have developed an assay using Xenopus egg extracts that can recapitulate the spatial and temporal specificity of CENP-A deposition observed in human cells, providing us with a robust in vitro system amenable to molecular dissection. Here we show that this deposition depends on Xenopus Holliday junction–recognizing protein (xHJURP), a member of the HJURP/Scm3 family recently identified in yeast and human cells, further supporting the essential role of these chaperones in CENP-A loading. Despite little sequence homology, human HJURP can substitute for xHJURP. We also report that condensin II, but not condensin I, is required for CENP-A assembly and contributes to retention of centromeric CENP-A nucleosomes both in mitosis and interphase. We propose that the chromatin structure imposed by condensin II at centromeres enables CENP-A incorporation initiated by xHJURP.
Introduction
Faithful segregation of the genome requires that chromosomes interact and are pulled by the spindle microtubules during cell division. This is made possible by the kinetochore, a macromolecular assembly built at a single locus in each chromosome known as the centromere (Cleveland et al., 2003; Musacchio and Salmon, 2007). The DNA sequence of this locus is not conserved among eukaryotes and in most cases is even different between the chromosomes of the same organism. What all centromeres have in common are nucleosomes containing a unique histone H3 variant, centromeric protein A (CENP-A; Malik and Henikoff, 2003; Allshire and Karpen, 2008). As any other histone variant, or histone modification, the amount of CENP-A present at centromeres is diluted twofold during DNA replication, and must be replenished to maintain centromere identity. How CENP-A is deposited specifically at centromeres and when this happens has been the subject of many studies in different model organisms over the last decade (Bernad et al., 2009). In budding yeast, a 125-bp sequence present in all chromosomes determines the position in which a single CENP-A–containing nucleosome is assembled (Furuyama and Biggins, 2007). Newly synthesized CENP-A replaces old protein during DNA replication in this organism (Pearson et al., 2001). Fission yeast centromeres are more complex regions of 30–120 kb containing repetitive DNA elements (Clarke et al., 1986). CENP-A is deposited at a 3–5-kb central core element within these regions during S phase and G2, by apparently distinct pathways (Hayashi et al., 2004; Takayama et al., 2008). Plant and metazoan centromeres are embedded in repeated DNA sequences. Loading of CENP-A in Arabidopsis occurs mainly in G2 (Lermontova et al., 2007). A study in Drosophila Kc cells reported that a CENP-A deposition pathway, independent of replication, is active throughout the cell cycle (Ahmad and Henikoff, 2001). In contrast, replenishment of the CENP-A mark occurs in anaphase in the syncytial mitosis of early embryos (Schuh et al., 2007). In HeLa cells, CENP-A deposition takes place in late telophase/early G1, although it requires passage through mitosis (Jansen et al., 2007; Hemmerich et al., 2008). Thus, the timing of CENP-A deposition varies among species and even in different developmental stages.
Another important question is whether a chromatin assembly complex exists for CENP-A, as is the case for other histone H3 variants (Tagami et al., 2004). Histone H3.1, also known as “canonical” histone H3, is deposited throughout chromatin by chromatin assembly factor 1 (CAF-1) complex during DNA replication. At some locations (i.e., active promoters) H3.1 is replaced by H3.3 by a complex containing the histone regulator A (HIRA) protein in a process uncoupled from DNA replication (De Koning et al., 2007; Elsaesser et al., 2010). Two major strategies have been used to identify factors involved in CENP-A incorporation: functional screens in fission yeast, worms, and flies, and biochemical purification of CENP-A–associated proteins in human cells (Hayashi et al., 2004; Obuse et al., 2004; Foltz et al., 2006; Izuta et al., 2006; Okada et al., 2006; Erhardt et al., 2008). Among the factors found in yeast and human cells are Mis18 and Scm3/HJURP (Holliday junction–recognizing protein). Mis18 is present at centromeres at the time of CENP-A incorporation, and some evidences suggest that it could prime centromeric chromatin by modulating its acetylation status (Hayashi et al., 2004; Fujita et al., 2007). Scm3 interacts with CENP-A and facilitates its deposition in budding and fission yeast (Camahort et al., 2007, 2009; Mizuguchi et al., 2007; Stoler et al., 2007; Pidoux et al., 2009; Williams et al., 2009). HJURP, a human protein functionally related to Scm3, associates with predeposited CENP-A, localizes to centromeres in early G1, and is required for CENP-A loading and maintenance (Dunleavy et al., 2009; Foltz et al., 2009; Shuaib et al., 2010). These results strongly suggest that HJURP is a bona fide CENP-A deposition factor. Members of this family of proteins have not been identified in Drosophila or Caenorhabditis elegans, however, implying the existence of alternative pathways for CENP-A assembly (Erhardt et al., 2008; Sánchez-Pulido et al., 2009). Furthermore, the mechanism of action of HJURP and its functional relationship with factors that interact with CENP-A and regulate its deposition in coordination with other major cell cycle events remains to be elucidated.
What directs HJURP to centromeres is also unknown. It is conceivable that centromeric chromatin containing CENP-A and canonical H3 nucleosomes (Blower et al., 2002) adopts a unique three-dimensional structure important not only for kinetochore assembly, but also to be recognized by the CENP-A loading machinery (Bloom, 2007). Factors like condensin, a protein complex that mediates chromosome condensation in mitosis, could contribute to such higher order organization of centromeric chromatin (Bernad et al., 2009). Indeed, previous studies have reported an abnormal morphology of centromeric chromatin and/or kinetochores in mitosis in the absence of condensin in a number of model organisms (Hagstrom et al., 2002; Wignall et al., 2003; Ono et al., 2004; Jäger et al., 2005; Oliveira et al., 2005), and condensin mutants in budding yeast show some loss of CENP-A/Cse4 from centromeres (Yong-Gonzalez et al., 2007). More recently, a defect in de novo loading of CENP-A has been observed after depletion of condensin in HeLa cells by siRNA (Samoshkin et al., 2009). This study does not distinguish between the effects of condensin I and condensin II, the two distinct condensin complexes that exist in vertebrates. These complexes share the SMC2/CAP-E and SMC4/CAP-C core subunits but differ in the specific set of regulatory subunits: CAP-D2, -G, and -H in condensin I and CAP-G2, -D3, and -H2 in condensin II (Hirano, 2005). In somatic cells, condensin I and II are equally abundant. In contrast, condensin I is fivefold more abundant than condensin II in Xenopus egg extracts, and has a predominant role in chromosome condensation (Ono et al., 2003). Intriguingly, condensin II is enriched at centromeres and could therefore have a specific function at this region (Ono et al., 2004).
A robust in vitro system that could recapitulate CENP-A deposition would be an invaluable tool to dissect the requirements of this process in molecular detail. Xenopus egg extracts have proved to be a suitable system for the study of chromatin assembly and to delineate the functions of H3.1 and H3.3 complexes (Quivy et al., 2001; Ray-Gallet et al., 2002). Thus, we set out to develop a quantitative immunofluorescence-based assay to measure CENP-A incorporation in chromosomes assembled in Xenopus egg cell-free extracts. In this experimental system, one can analyze the effect of depleting a factor on a single round of CENP-A loading without accumulation of errors from previous rounds or alteration of cell cycle progression. The latter is especially important given that incorporation of CENP-A appears to be limited to a restricted period of the cell cycle, and thus knocking down a factor that affects cell cycle progression may indirectly affect CENP-A incorporation as well. Finally, the depletion can be rescued by adding back purified proteins or undepleted extract at different times in the course of the assembly reaction. Here we show that the spatial and temporal specificity of CENP-A deposition can be recapitulated on chromosomes assembled in the egg extracts. This reaction requires a functional homologue of HJURP that is present in the extract forming a complex with CENP-A. In addition, we have identified a requirement of condensin II, but no condensin I, for efficient assembly of CENP-A nucleosomes.
Results
Loading of CENP-A occurs upon exit from mitosis in Xenopus egg extracts
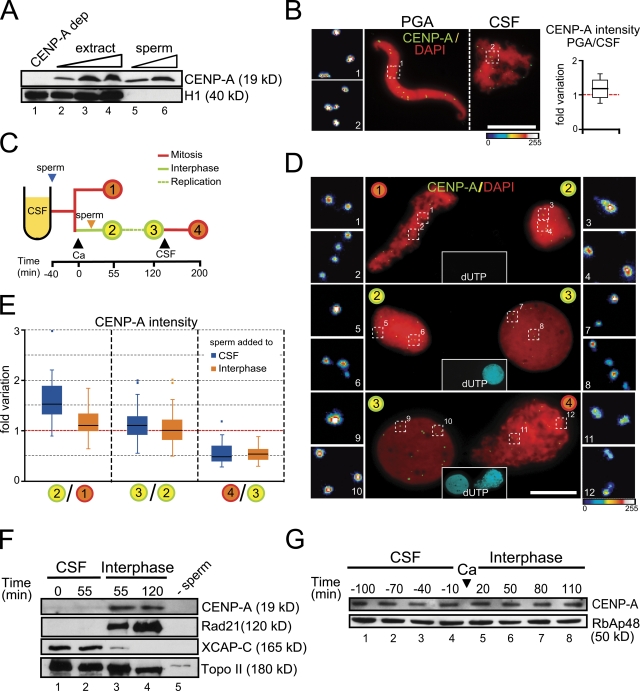
Xenopus egg extracts can recapitulate many cell cycle–specific chromosomal events after addition of demembranated sperm nuclei. These nuclei contain CENP-A that can be detected by immunoblot (Fig. 1 A) and visualized by immunofluorescence upon decondensation of the sperm by incubation with polyglutamic acid (PGA) or in egg extract (Fig. 1 B). The CENP-A antibody detects 10–20 foci per nucleus, consistent with the haploid number of chromosomes in X. laevis, which is 18. These foci colocalize with other centromeric proteins like CENP-T or Bub1, further supporting that they correspond to centromeres and little CENP-A labeling is observed outside them (Fig. S1 A; Rivera and Losada, 2009). Moreover, in vitro–translated myc–CENP-A added to the extract also incorporates to the centromeres labeled by CENP-T, albeit rather inefficiently (Fig. S1 B). For this reason, we decided to assess CENP-A loading by measuring the change in the fluorescent signal of CENP-A at individual centromeres. Nuclei in two different time points of an assembly reaction, or under two different conditions, were spun over the same coverslip, and then processed for immunofluorescence and imaged together. On these images, we quantified the intensity of CENP-A signals and calculated the average increase in CENP-A staining for each pair of nuclei (see Materials and methods for details). We first noticed that the ratio of CENP-A content between the PGA-treated nuclei and the mitotic chromosomes assembled in cytostatic factor (CSF) extract (i.e., prepared from eggs arrested in meiosis II) was 1.2 ± 0.1 (Fig. 1 B, graph). Thus, no incorporation of CENP-A occurred during incubation in this CSF extract. However, 55 min after adding calcium to the CSF assembly mixture to drive degradation of cyclin B and entry into interphase (sample labeled “2” in Fig. 1, C–E), a clear increase of CENP-A could be observed with respect to the signal detected in mitotic chromosomes (sample labeled “1” in Fig. 1, C–E; blue box on the left in the graph in Fig. 1 E). Further incubation in interphase allowed DNA replication to be completed (as indicated by incorporation of a biotinylated nucleotide; insets in Fig. 1 D), but the increase in CENP-A labeling was minor during this period (sample labeled “3” in Fig. 1, C–E; blue box on the middle in Fig. 1 E). These results were confirmed by Western blot analysis of chromatin fractions. The amount of CENP-A present on chromatin after incubation of the sperm in a CSF extract was probably under a detection threshold (Fig. 1 F, lanes 1 and 2), but became visible 55 min after addition of calcium to a sample incubated in CSF for 40 min, and did not increase further (Fig. 1 F, lanes 3 and 4). There is no variation in the levels of soluble CENP-A present in the extract during this assembly reaction (Fig. 1 G). When the nuclei were driven back to mitosis by addition of CSF extract, thereby turning into condensed chromosomes with paired sister chromatids, the intensity of CENP-A labeling was reduced almost twofold due to the resolution of sister centromeres (i.e., each single dot in an interphase nuclei was split into two; sample labeled “4” in Fig. 1, C–E; blue box on the right in Fig. 1 E). Thus, incorporation of CENP-A into sperm chromatin occurs in early interphase and is uncoupled from bulk DNA replication. In fact, the intensity of CENP-A signals in late interphase nuclei assembled in the presence or absence of the DNA replication inhibitor aphidicolin is identical, again indicating that DNA replication is not required for CENP-A loading (Fig. S1 C).
Figure 1.
CENP-A deposition occurs in early interphase in Xenopus egg extracts. (A) Sperm nuclei contain CENP-A. Immunoblot analysis of increasing amounts of egg extract (1, 2, and 3 µl; lanes 2–4), 3 × 105 and 6 × 105 sperm nuclei (lanes 5 and 6), and 1 µl of an extract depleted of CENP-A as control (lane 1). Unlike CENP-A, histone H1 is present in the soluble extract but not in the sperm nuclei. (B) Sperm nuclei were incubated with a buffer containing polyglutamic acid (PGA) or in CSF extract, mixed, centrifuged on the same coverslip, stained with anti-CENP-A (green) and DAPI (red), and imaged together. One representative pair of nuclei is shown. Bar, 10 µm. In the blown-up images on the left the intensity of the CENP-A labeling has been coded with a color gradient going from blue (minimum) to white (maximum). The intensity of CENP-A signals was measured for 15 pairs of nuclei and plotted as fold variation of the average signal for each pair. (C) Outline of the chromatin assembly experiment and the time points at which samples were taken for analysis. The time of calcium addition is considered t = 0. Black arrowheads indicate additions to the extract. The blue and orange arrowheads indicate the time of sperm addition in the two different experiments described in the main text. (D) Representative images of pairs of nuclei from two consecutive time points of the assembly reaction processed and analyzed together. Nuclei were stained with CENP-A (green) and DAPI (red). Replication was visualized by incorporation of biotin-dUTP (cyan, insets in the center). Bar, 10 µm. (E) Graph showing the fold variation of CENP-A signal intensities between two given time points (indicated by the numbers as in panel C) for two different time course experiments: blue boxes for the experiment in which sperm is added to CSF extract and orange boxes for addition of sperm in interphase extract. Data for each time point come from 15 pairs of nuclei in at least two independent experiments. (F) Immunoblot analysis of chromatin fractions obtained after incubation of sperm nuclei in CSF extract (lanes 1 and 2) or incubated in CSF for 40 min and then driven in interphase by addition of calcium (lanes 3 and 4). The times indicated correspond to the scheme in panel C. A mock assembly reaction without sperm DNA is shown as control (lane 5). (G) Immunoblot analysis of samples taken from an egg extract at the indicated times. The time of calcium addition is considered t = 0. The histone chaperone RbAp48 is shown as loading control.
In the time course experiments described in the previous paragraph, centromeres do not get to assemble a full complement of CENP-A. However, if we directly compared the amount of CENP-A between mitotic chromosomes assembled in CSF and late interphase nuclei from the same assembly reaction (corresponding to time points 1 and 3, respectively, in Fig. 1 C), the numbers that we obtained were closer to a twofold increase (1.9 ± 0.1, n = 24 experiments). We also checked CENP-A assembly after exit from the second mitosis, and found similar loading efficiency (not depicted). Henceforth, we performed CENP-A loading assays under different depletion conditions by comparing a late interphase sample and a mitotic sample.
A window of opportunity for CENP-A loading in early interphase
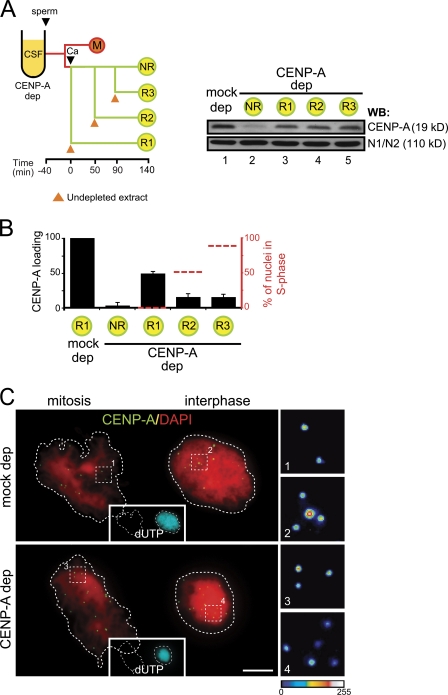
When a CSF extract is first treated with calcium and sperm chromatin is added after 40 min, i.e., once cyclin B has been degraded and the extract is in interphase, incorporation of CENP-A is clearly reduced (Fig. 1 E, orange boxes). This result suggests that previous passage of centromeric chromatin through mitosis could be important for efficient CENP-A incorporation. Alternatively, a factor required for CENP-A loading may be present or exert its function on centromeric chromatin only in early G1. We next asked whether CENP-A can be loaded on chromatin at any time during interphase. For this, we performed the CENP-A loading assay in extracts depleted of CENP-A to which one half volume of undepleted extract was added at different times in interphase (R1, R2, or R3; R is for “rescue”) or not added (NR [no rescue]; Fig. 2 A). As expected, when there is no CENP-A in the extract the loading efficiency relative to a mock-depleted extract is less than 5% (Fig. 2 B, NR condition; and Fig. 2 C). This result validates the assay and indicates that the observed increase in CENP-A staining reflects loading of new CENP-A and is not due to enhanced accessibility of the antibody to centromeres in interphase. Addition of undepleted extract to the CENP-A–depleted extract can partially restore CENP-A loading when added at the same time as calcium (Fig. 2, A and B; 49% efficiency of CENP-A loading in R1 condition). However, if CENP-A availability is restored 50 min and 90 min post-calcium (Fig. 2, A and B; R2 and R3, respectively), when replication is underway in 50% (R2) and 90% (R3) of the nuclei, the efficiency of CENP-A loading drops to 15%. Thus, CENP-A incorporation appears to be restricted to early interphase, before DNA replication starts.
Figure 2.
A window of opportunity for CENP-A loading in early interphase. (A) Outline of the chromatin assembly experiment (left) and immunoblot analysis of total extracts to show soluble CENP-A levels under the indicated conditions (right). The histone binding protein N1/N2 is shown as loading control. Sperm was added at t = 0 to CSF extracts (mock depleted or CENP-A depleted). For the mock-depleted extract, one-half volume of undepleted interphase extract was added at the time of calcium addition. For the CENP-A–depleted extract, either no extract was added back (no rescue [NR] condition) or one-half volume of undepleted interphase extract was added at the times indicated (R1, R2, and R3). (B) Bar graph showing the CENP-A loading efficiency under the conditions explained above, with respect to the mock-depleted extract. A small sample of the assembly mixture was taken right before addition of undepleted extract to check the percentage of nuclei showing some incorporation of biotin-dUTP (i.e., undergoing replication, red dotted line) at the time of restoring CENP-A availability, although analysis of CENP-A incorporation was performed comparing CENP-A signals from fully replicated interphase nuclei (taken at t = 140 min) and mitotic chromosomes from images such as those shown in C. At least 15 pairs of nuclei were measured for each condition. Bars represent mean ± SE. (C) Representative images of a mass of mitotic chromosomes next to a replicated interphase nuclei stained with anti-CENP-A (green) and DAPI (red) taken from the mock-depleted (top) and CENP-A–depleted (bottom) samples. Replication was visualized by incorporation of biotin-dUTP (cyan). Bar, 10 µm.
CAF-1 and HIRA are not major CENP-A loaders
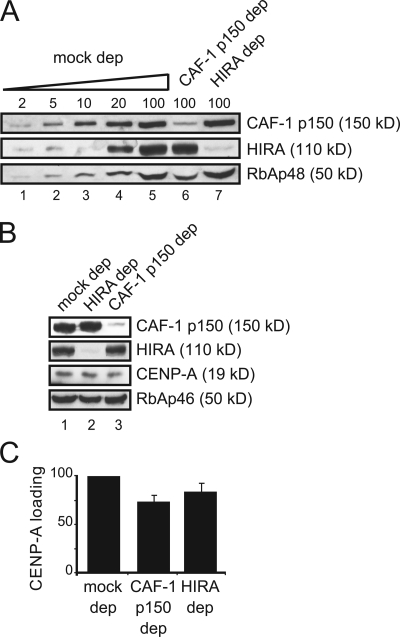
We next tested whether the H3.1 and H3.3 deposition complexes have any role in the deposition of CENP-A. Antibodies against the CAF-1 p150 subunit and HIRA were able to immunodeplete more than 95% of the corresponding protein (Fig. 3 A) without affecting the levels of soluble CENP-A (Fig. 3 B). We then measured the increase in CENP-A staining between interphase nuclei and mitotic chromosomes assembled under each condition (Fig. 3 C). A small decrease in the efficiency of CENP-A loading was observed in the CAF-1–depleted extracts (73± 6%) and in the HIRA-depleted extracts (84± 9%). We noticed that depletion of CAF-1 p150 also decreases the levels of RbAp48 (Fig. 3 A), but not the levels of RbAp46 (Fig. 3 B), histone chaperones present in both H3.1 and H3.3 deposition complexes. Experiments in vitro with purified histones and recombinant CENP-A and RbAp48 showed incorporation of CENP-A nucleosomes on plasmid DNA, prompting the hypothesis that CENP-A could be loaded on chromatin by a passive mechanism involving this chaperone (Furuyama et al., 2006). Also, human cells with reduced levels of RbAp46/48 show defects in CENP-A loading (Hayashi et al., 2004; Dunleavy et al., 2009). Thus, it was possible that the observed decrease in CENP-A loading efficiency upon CAF-1 p150 depletion could be due to co-depletion of RbAp48. However, when we performed the CENP-A incorporation assay after immunodepletion of RbAp48 with a specific antibody, CENP-A loading was not affected (unpublished data). We conclude that neither CAF-1 p150 nor HIRA are the major loaders of CENP-A in Xenopus egg extracts.
Figure 3.
CENP-A incorporation in CAF-1 p150– and HIRA-depleted extracts. (A) Immunoblot analysis of increasing amounts of a mock-depleted extract (expressed as percentage of a 1.5-µl aliquot) and 1.5-µl aliquots of CAF-1 p150– and HIRA-depleted extracts with the indicated antibodies. RbAp48 levels are also shown. (B) Immunoblot analysis of aliquots from a mock-depleted extract, HIRA- and CAF-1 p150–depleted extracts to show the remaining levels of CENP-A. RbAp46 levels are also shown. (C) Bar graph showing the CENP-A loading efficiency in the depleted extracts with respect to the mock. Bars represent mean ± SE from two independent experiments.
Human HJURP promotes CENP-A deposition in Xenopus egg extracts
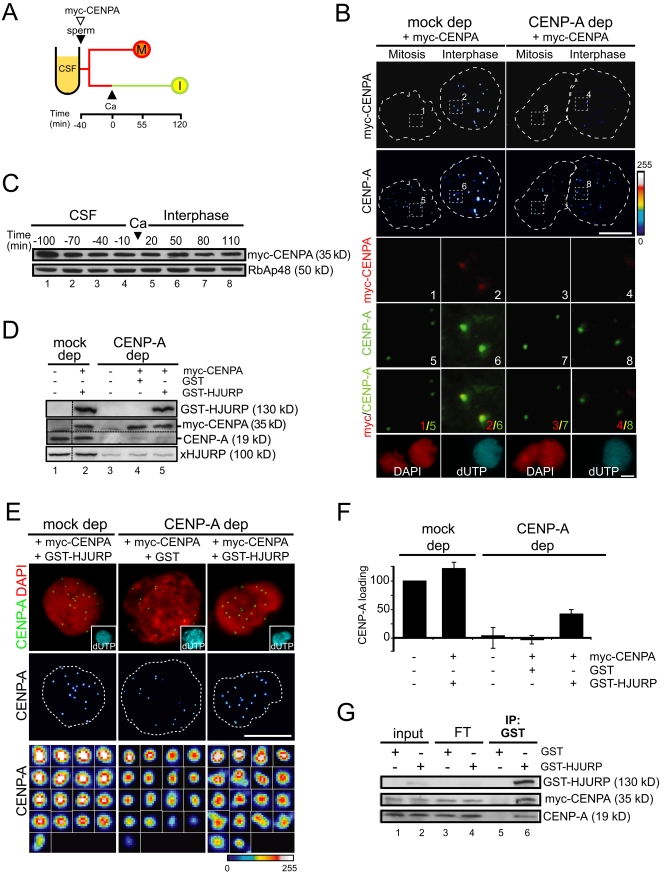
To rescue CENP-A loading, in vitro–translated myc–CENP-A was added to CENP-A–depleted CSF extracts (and mock-depleted extracts as control) along with sperm DNA. After 40 min each mixture was divided in two, calcium was added to one tube, and incubation proceeded for additional 120 min (Fig. 4 A). Only the interphase nuclei from the mock-depleted extract showed centromere-specific myc staining (Fig. 4 B, left panels). This result confirms that incorporation of CENP-A does not take place in CSF but in subsequent interphase and also indicates that the exogenous protein is functional. However, myc–CENP-A does not go to chromatin in the CENP-A–depleted extract (Fig. 4 B, right panels) despite being stable in the extract (Fig. 4 C), and is present at levels similar to endogenous CENP-A (Fig. 4 D, lane 2). We reasoned that depletion of CENP-A might co-deplete an essential component of the loading machinery. Given the recent identification of HJURP as a CENP-A–specific chaperone in human cells (Dunleavy et al., 2009; Foltz et al., 2009), we next tested whether addition of this protein along with myc–CENP-A to a CENP-A–depleted extract could restore CENP-A loading. Indeed, addition of recombinant human HJURP (GST-HJURP), but not GST, allowed CENP-A incorporation (Fig. 4, E and F). Quantification of the increase in CENP-A staining with respect to a mock-depleted extract showed a recovery of 42% in CENP-A loading in the presence of GST-HJURP, but no recovery at all when GST was added instead (Fig. 4 F). Consistent with these results, GST-HJURP was found to coimmunoprecipitate with myc–CENP-A and endogenous CENP-A (Fig. 4 G) Thus, it is likely that a homologue of human HJURP exists in the egg extract and is essential for CENP-A loading.
Figure 4.
Human HJURP promotes loading of Xenopus CENP-A. (A) Outline of the chromatin assembly experiment. Mitotic chromosomes (M) and interphase nuclei (I) were assembled in mock- and CENP-A–depleted extracts containing in vitro–translated (IVT) myc-CENPA. (B) Nuclei and chromosomes were stained with antibodies against myc (top) and CENP-A (second from top), both color coded with a gradient as in Fig. 1 B. Insets 1–4 show that myc signals (red) overlap with CENP-A signals (green) in interphase nuclei of mock-depleted extracts (inset 2 and 6), whereas weak myc signals in CENP-A–depleted interphase nuclei (inset 4) are mostly background. DAPI staining (red) and biotin-dUTP (cyan) are shown in the bottom row. Bar, 10 µm. (C) IVT myc-CENP-A was added to a CENP-A–depleted extract along with sperm DNA and samples of the reaction were taken at the indicated times and analyzed by immunoblot with anti-myc and anti-RbAp48 as loading control. The time of calcium addition is considered t = 0. Cell cycle progression was monitored by chromosome morphology after DAPI staining (not depicted). (D) Immunoblot analysis of the extracts used for chromatin assembly in panel E. Antibodies against human and Xenopus HJURP were used to detect GST-HJURP (top) and endogenous xHJURP (bottom), respectively, whereas anti-CENPA was used to simultaneously detect myc-CENPA and endogenous CENP-A (middle). (E) Representative images of interphase nuclei from the indicated extracts stained with anti–CENP-A (green) and DAPI (red). Insets in the images of the first row show biotin-dUTP incorporation (cyan). In the images from the middle and bottom rows, CENP-A labeling has been color coded as in Fig. 1 B. In the bottom row, all centromeres from the nuclei presented above are shown at higher magnification. Bar, 10 µm. (F) Graph showing the loading efficiency of CENP-A in the indicated extracts compared with a mock-depleted extract. Bars represent mean ± SE from two independent experiments. (G) GST-HJURP interacts with myc-CENPA and endogenous CENP-A. Analysis of an immunoprecipitation reaction with anti-GST from an egg extract containing myc-CENPA and either GST alone or GST-HJURP. Aliquots (1.5%) from input and flow-through (FT) fractions were also analyzed.
xHJURP is stored in a complex with CENP-A
A homology database search has recently proposed the existence of an HJURP protein in Xenopus tropicalis (Sánchez-Pulido et al., 2009). A full-length cDNA encoding this protein is not yet available. Maybe not surprisingly given the low sequence conservation between human HJURP (hHJURP) and its putative counterpart in Xenopus, antibodies against the former do not recognize any protein in the immunoprecipitates of CENP-A from egg extracts. We therefore raised new antibodies against a fragment of the putative Xenopus HJURP (xHJURP) and found that they recognize and immunoprecipitate a 100-kD protein (Fig. S2, A and B). CENP-A is present in these immunoprecipitates (Fig. S2 B). Using these antibodies, we confirmed that CENP-A depletion also removes xHJURP from the extract (Fig. 4 D, bottom), thereby explaining why addition of myc–CENP-A alone cannot rescue CENP-A loading in a CENP-A–depleted extract. The levels of other histone chaperones are not affected by the depletion of CENP-A (Fig. S2 C).
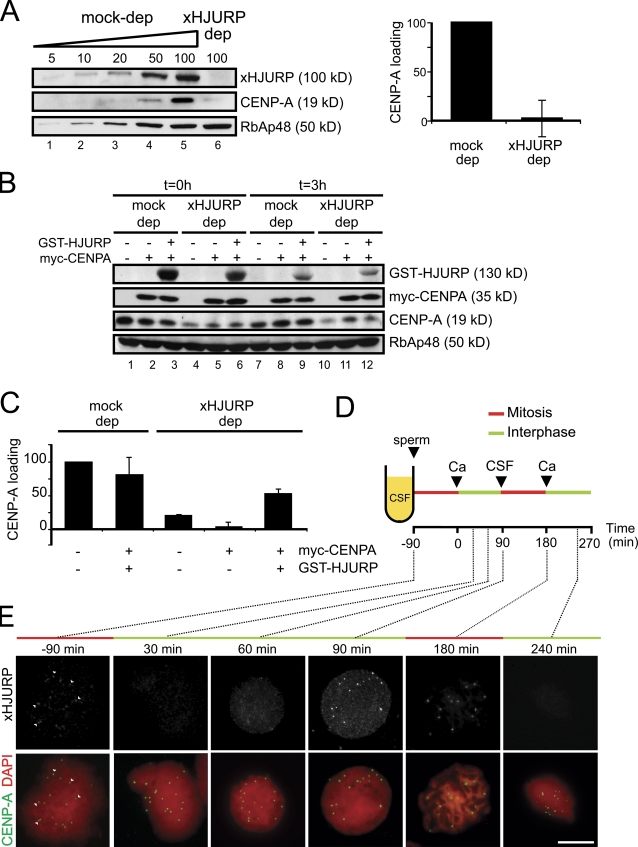
xHJURP is essential for CENP-A deposition and localizes to centromeres
When xHJURP is removed from the extract, CENP-A levels are reduced to 20–50% and loading is abolished (Fig. 5 A). Even if myc–CENP-A is added to the xHJURP-depleted extract to reconstitute CENP-A levels, loading is impaired (Fig. 5 B, lane 5; and Fig. 5 C, fourth bar from the left). Again, we confirmed that myc–CENP-A is stable in the xHJURP-depleted extract throughout the duration of the experiment (Fig. 5 B). Only when hHJURP (GST-HJURP) is also added is loading partially restored (Fig. 5 B, lane 6; and Fig. 5 C, last bar). Thus, the role of HJURP in deposition of the centromeric histone is conserved in Xenopus egg extracts.
Figure 5.
xHJURP is required for CENP-A deposition. (A) Left, immunoblot analysis of increasing amounts of a mock-depleted extract (5–100% of a 1.5-µl aliquot, lanes 1–5) and a 1.5-µl aliquot of an xHJURP-depleted extract (lane 6). Right, graph showing the loading efficiency of CENP-A in an extract depleted of xHJURP. Data come from two different experiments. Bars represent mean ± SE. (B) Immunoblot analysis of mock-depleted and xHJURP-depleted extracts replenished or not with myc-CENP-A and GST-HJURP at the time of sperm addition (t = 0, lanes 1–6) and at the end of the experiment (t = 3 h, lanes 7–12). (C) Loading efficiency of CENP-A in the indicated extracts. Data come from two independent experiments. Bars represent mean ± SE. (D) Outline of the chromatin assembly reaction used to assess the localization of xHJURP throughout the cell cycle reproduced in the extract by successive additions of calcium and CSF extract. Sperm chromatin (800/µl) was added at time 0. The times at which aliquots were taken for analysis are indicated. (E) Immunofluorescent staining with anti-xHJURP (green), CENP-A (red), and DAPI (blue). White arrowheads point to HJURP signals that colocalize with CENP-A signals on CSF assembled chromosomes. Bar, 10 µm.
In human cells, HJURP localizes transiently to centromeres in late telophase/early G1 whereas fission yeast Scm3 remains at centromeres throughout the cell cycle except from prophase to telophase. We performed a time course to examine xHJURP localization in egg extracts (Fig. 5 D). Weak xHJURP staining could be detected at some centromeres on mitotic chromosomes assembled in CSF (Fig. 5 E, white arrowheads in −90-min sample). The signal was lost upon calcium addition (30-min sample). A diffuse staining all over chromatin was observed by 60 min, but 30 min later xHJURP was enriched at all centromeres and stayed there when CSF was added to promote entry in mitosis (180-min sample). Again, calcium addition led to delocalization of xHJURP from centromeres (240-min sample). These observations suggest that HJURP is not an integral part of the centromere, raising the possibility that it acts enzymatically. Supporting this possibility, restoring HJURP levels to 25% of normal levels by adding one-third volume of undepleted extract to an HJURP-depleted extract fully restores CENP-A loading (Fig. S3).
Condensin II prevents CENP-A eviction
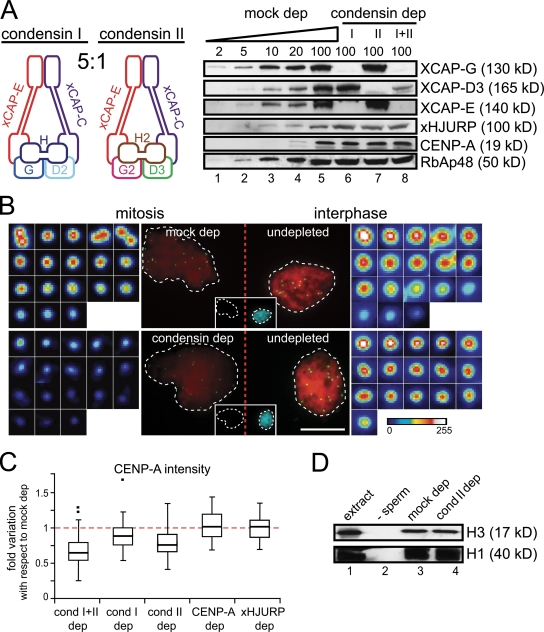
We next investigated whether condensin contributes to CENP-A assembly. As mentioned before, there are two distinct condensin complexes in Xenopus egg extracts that can be depleted separately or simultaneously without affecting CENP-A levels in the soluble extracts (Fig. 6 A). Targeting of each complex to chromatin are independent events, and in particular, centromeric enrichment of condensin II does not require condensin I (Fig. S4). Quantitation of the CENP-A signals present in mitotic chromosomes assembled in CSF extracts depleted of both condensins with respect to control samples revealed a significant reduction in the amount of CENP-A (Fig. 6, B and C). Because there is no loading of CENP-A during mitosis, this result implies that CENP-A present in the sperm chromatin added to the extract is being lost or evicted. Removal of condensin II is largely responsible for this effect, although condensin I may also contribute (Fig. 6 C). Depletion of CENP-A or depletion of xHJURP does not lead to eviction. Unlike CENP-A nucleosomes, the stability of H3 nucleosomes is not altered by the absence of condensin II, at least as judged by immunoblot analysis of chromatin fractions (Fig. 6 D).
Figure 6.
Condensin II prevents eviction of CENP-A nucleosomes from centromeres. (A) Left, subunit composition of the two condensin complexes. Right, immunoblot analysis of increasing amounts of a mock-depleted extract (expressed as percentage of a 1.5-µl aliquot) and extracts depleted of condensin I, condensin II, or both with the indicated antibodies. RbAp48 is shown as loading control. (B) Sperm chromatin was added to mock-depleted (top) and condensin-depleted (bottom) CSF extracts and incubated for 90 min. To serve as reference, sperm chromatin was added to an undepleted CSF extract, incubated for 40 min, and then calcium was added and incubation continued for 90 min. The latter samples (replicated interphase nuclei, as evidenced by the incorporation of biotin dUTP) were mixed with the former ones (mitotic chromosomes), centrifuged over coverslips, and stained with anti-CENP-A (green), DAPI (red), and streptavidin (cyan). Representative images of the photographed pairs are shown, with corresponding blown-up images of each individual centromere. (C) CENP-A signals in chromosomes assembled in depleted CSF extracts were measured in comparison with a reference sample, as described in B. The resulting numbers are expressed and plotted as fold variation relative to the average value obtained for the mock-depleted extract, arbitrarily set to 1 (red dotted line). Data from at least two independent experiments are shown for the depletion of condensins and from a single experiment in the case of CENP-A and xHJURP depletions. (D) Immunoblot analysis of chromatin fractions from assembly reactions in mock- and condensin II–depleted extracts. An aliquot of extract (lane 1) and a chromatin fraction from a mock assembly reaction with no sperm (lane 2) were also analyzed.
To check whether the lack of condensin also results in eviction of CENP-A in interphase, we compared the CENP-A signals in nuclei assembled by adding sperm to an interphase extract, without previous incubation in CSF. As shown in Fig. 1 E (orange squares), the nuclei assembled under this condition load little maternal CENP-A. We observed a reduction in CENP-A signal intensities in the absence of condensin II (Fig. S5). Together, these results indicate that condensin II contributes to retention of CENP-A nucleosomes already present in centromeric chromatin, both in mitosis and interphase.
Condensin II is required for proper loading of CENP-A
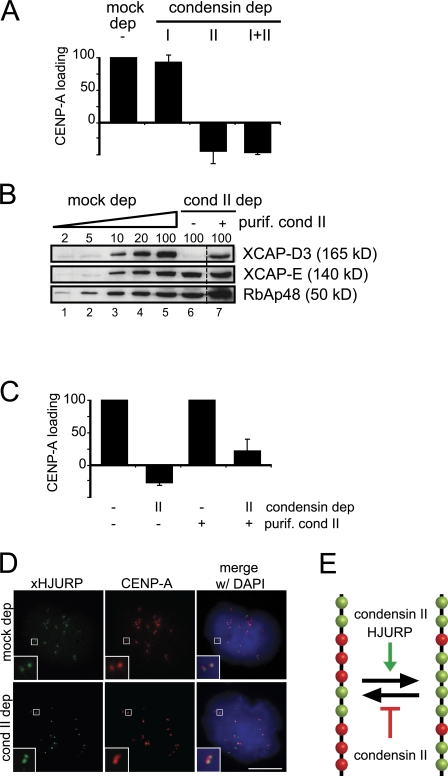
We next assayed CENP-A loading in the absence of each condensin complex. Nuclei assembled in extracts depleted of condensin I loaded CENP-A almost as well as those present in mock-depleted extracts. In contrast, depletion of condensin II alone or double depletion of both condensin I and II led to negative CENP-A loading values suggesting that, in addition to the eviction problem described in the previous paragraph, de novo loading of CENP-A was severely impaired (Fig. 7 A). Immunodepletion of condensin II was performed with an antibody against XCAP-D3, whereas an antibody against XCAP-E was used to immunodeplete both condensins simultaneously. Because the same defect in CENP-A loading is observed using two distinct antibodies, it is unlikely that it is due to unspecific removal of a factor other than condensin II. Nevertheless, to further test the specificity of the defect, affinity-purified condensin II was added back to a condensin II–depleted extract. We found that restoration of the condensin II level to around 30% of the level in a mock-depleted extract (Fig. 7 B, lane 7) led to partial recovery of CENP-A loading (23%, Fig. 7 C). We conclude that condensin II is specifically required for proper CENP-A nucleosome assembly into centromeres.
Figure 7.
Condensin II is required for efficient loading of CENP-A. (A) Bar graph showing the CENP-A loading efficiency in the indicated extracts in two independent experiments. Bars represent mean ± SE. (B) Immunoblot analysis of increasing amounts of a mock-depleted extract (expressed as percentage of a 1.5-µl aliquot, lanes 1–5) and an extract depleted with anti-XCAP-D3 without (lane 6) or with 1:10 volume of affinity-purified condensin II (lane 7). (C) Bar graph showing the CENP-A loading efficiency in mock- and condensin II–depleted extracts with and without adding back purified condensin II. (D) Immunofluorescent staining of mitotic chromosomes assembled in mock- and condensin II–depleted extracts with antibodies against xHJURP (green) and CENP-A (red). DNA was counterstained with DAPI (blue). Bar, 10 µm. (E) Model to explain the role of xHJURP and condensin II in CENP-A dynamics. CENP-A and H3 nucleosomes are shown in green and red, respectively. Net incorporation of CENP-A will depend on the balance between addition of new CENP-A nucleosomes, a process in which xHJURP has a key role, and eviction of these nucleosomes. Condensin II prevents eviction of CENP-A nucleosomes but it is also required for their incorporation.
Depletion of condensin II did not affect the levels of xHJURP in the soluble extract (Fig. 6 A). xHJURP targeting to centromeres, best observed by late interphase and in subsequent mitosis, was also normal (Fig. 7 D). Thus, the defect in CENP-A loading is not due to HJURP being unable to recognize the centromere region in the absence of condensin II.
Discussion
The mechanism of centromere specification and propagation has remained elusive for decades. The realization that CENP-A is the major marker of this locus has led to the identification of a number of factors that play a role in the deposition of the centromeric histone (Allshire and Karpen, 2008; Bernad et al., 2009; Torras-Llort et al., 2009). Despite major advances, a molecular understanding of this process and its cell cycle regulation is still lacking. To tackle this issue we have developed an immunofluorescence-based assay that measures CENP-A incorporation at centromeres of chromosomes assembled in the Xenopus egg cell-free system. Using this assay, we demonstrate that CENP-A assembly occurs in early interphase and is uncoupled from DNA replication. The H3.1 and H3.3 deposition complexes containing p150 CAF-1 and HIRA, respectively, previously shown to be critical for efficient assembly of their respective nucleosomes in Xenopus egg extracts, are not major loaders of the centromeric histone variant in this system (Quivy et al., 2001; Ray-Gallet et al., 2002; Tagami et al., 2004). Instead, we have found that more than half of the CENP-A in the egg extract is present in a complex with xHJURP, a member of the recently identified Scm3/HJURP family of proteins proposed to be CENP-A–specific chaperones from yeast to human (Camahort et al., 2007, 2009; Mizuguchi et al., 2007; Stoler et al., 2007; Dunleavy et al., 2009; Foltz et al., 2009; Pidoux et al., 2009; Sánchez-Pulido et al., 2009; Williams et al., 2009; Shuaib et al., 2010). Exogenously added CENP-A competes poorly with endogenous CENP-A for the binding to HJURP, which explains why myc-CENPA incorporates into centromeres rather inefficiently and is not useful to measure CENP-A loading (Fig. S1 B; unpublished data). Remarkably, we found that hHJURP is able to interact and load xCENP-A in an extract depleted of the endogenous xHJURP despite the little sequence conservation between human and frog HJURP. The region of human HJURP that binds to CENP-A has been recently mapped to the N-terminal domain (Shuaib et al., 2010), consistent with the identification within this region of a relatively conserved domain (the Scm3 domain) in all HJURP/Scm3-related proteins (Sánchez-Pulido et al., 2009). Conversely, HJURP recognizes the CATD domain of CENP-A, and this domain is highly conserved among CENP-A proteins from different species. How CENP-A nucleosomes are actually assembled into chromatin remains to be elucidated. The action of remodeling complexes like FACT and RSF, identified in the CENP-A immunoprecipitates, could be required in concert with that of HJURP (Obuse et al., 2004; Foltz et al., 2006; Okada et al., 2009; Perpelescu et al., 2009). In any case, our results not only support the evolutionary conservation of the CENP-A deposition process and its epigenetic nature, but further substantiate the key role of HJURP.
Timing issues in CENP-A assembly
In human cells, CENP-A levels are low in late G1-S and increase in G2-M, and this regulation likely contributes to the temporal and spatial specificity of CENP-A loading (Shelby et al., 1997, 2000). It has been proposed that one function of HJURP may be to protect CENP-A from degradation (Dunleavy et al., 2009). In yeast, proteolysis of Cse4 is also important to restrict Cse4/CENP-A localization (Collins et al., 2004). In contrast, CENP-A levels are constant during the chromatin assembly experiments described here. It is therefore possible that the CENP-A degradation machinery is not active in the embryonic cycles. Additional mechanisms must exist to prevent incorporation of CENP-A at noncentromeric regions. Remarkably, we observed that addition of undepleted extracts to a CENP-A–depleted extract restores loading only if added at the time of calcium addition. We speculate that efficient targeting or incorporation of CENP-A requires a post-translational modification of HJURP/CENP-A or of the centromeric chromatin that occurs in mitosis or early G1. Mis18 function could be involved in this priming. At least in human cells, its binding to centromeres appears to precede the binding of HJURP (Fujita et al., 2007; Foltz et al., 2009). Nuclear envelope formation or DNA replication may prevent loading of CENP-A at later time points in interphase.
The role of condensin II in CENP-A assembly
Our data show that condensin II is specifically required for proper CENP-A nucleosome dynamics (Fig. 7 E). It has been proposed that the easier disassembly of CENP-A nucleosomes compared with H3 nucleosomes could facilitate the clearance of those promiscuously incorporated into euchromatic regions (Moreno-Moreno et al., 2006; Conde e Silva et al., 2007) or deposited at sites of DNA damage (Zeitlin et al., 2009). In turn, this disassembly would have to be counterbalanced by some property of centromeric chromatin, and we speculate that this property is linked to the action of condensin II in this region both in mitosis and interphase. It will be of great interest to determine if other factors shown to be critical for CENP-A assembly may play a role in preventing eviction.
Condensin II likely contributes to the three-dimensional organization of the alternating blocks of CENP-A nucleosomes and H3 nucleosomes present in centromeric chromatin (Blower et al., 2002; Marshall et al., 2008; Santaguida and Musacchio, 2009). In turn, this organization dictates the assembly of the kinetochore and may also facilitate the targeting of the CENP-A assembly machinery (Silva and Jansen, 2009). CENP-C and proteins of the constitutive centromere-associated network (CCAN) bind this centromeric chromatin: CENP-T/CENP-W associate with H3 nucleosomes, whereas CENP-N and CENP-C interact with CENP-A (Hori et al., 2008; Carroll et al., 2009, 2010). CENP-C is absent from sperm DNA but assembles onto centromeres upon incubation in the CSF extract (Milks et al., 2009). Future studies will have to determine whether this is also the case for other components of CCAN—not yet described in Xenopus—and how the targeting of these factors is affected by the absence of condensin II. We are currently developing antibodies toward some of these proteins. Although we found that HJURP can still target the centromeres in the absence of condensin II, we speculate that its action, or the action of other chromatin-remodeling factors acting downstream of HJURP to accomplish CENP-A incorporation, may be ineffective under this condition.
In summary, our results indicate that CENP-A deposition in the Xenopus egg extract in vitro system is subject to the same spatial and temporal restrictions that apply in human cells, and that the major players involved are highly conserved. We have demonstrated that xHJURP is necessary for the loading of CENP-A and have shown that condensin II affects CENP-A dynamics, most likely by promoting proper organization of centromeric chromatin both in mitosis and interphase. We are confident that this assay will be a very powerful tool to further understand the molecular mechanisms underlying the process of the specification and propagation of centromeric chromatin.
Materials and methods
Antibodies
A fragment of recombinant Xenopus HJURP was amplified from IMAGE clone 5157296 with the primers 5′-CACCATGATTCCTTGCCAACATAG-3′ and 5′-TCAGTTTGCACTCCGGTGTC-3′, cloned in pENTR/D-TOPO and transferred to pDEST-17 (Invitrogen) to produce a His-tagged polypeptide in Escherichia coli that was purified and injected in rabbits for antibody production. The embryonic form of histone H1, known as B4, was purified fused to GST from a pGEX plasmid and also injected into rabbits. The following peptides were coupled to KLH and used as antigens to produce rabbit polyclonal antibodies against: XCAP-E (CSKTKERRNRMEVDK), XCAP-G (CEKTKKNLSKLLNEEAN), XCAP-D3 (CRQRISGKAPLKPSN), Asf1 (CVGSASEEYDQVLDS), xCENP-T (NLLIERHLPMEYRRC), N1/N2 (CSHLVRKKRKTEEES), RbAp46 (CEPDIPASELEAQGS), and RbAp48 (CEDTEGGVDPEGQGS). Other antibodies used in this study were against Xenopus CENP-A (Rivera and Losada, 2009), Rad21 (Losada et al., 1998), CAF-1 p150 (Quivy et al., 2001), HIRA (Ray-Gallet et al., 2002), nucleoplasmin (PA3C5, a generous gift from S. Dilworth, Imperial College Faculty of Medicine, London, UK; [Dilworth et al., 1987]), human HJURP (Sigma-Aldrich), GST (CNIO Monoclonal Antibodies Unit), myc (a rabbit polyclonal antibody for Western blot and a mouse monoclonal [clone 9B11] for immunofluorescence, both from Cell Signaling Technology), and histone H3 (ab1791; Abcam). A fraction of the CENP-A antibody was labeled with DyLight 549 (DyLight Microscale Labeling kit; Thermo Fisher Scientific).
Preparation and use of Xenopus egg extracts
Cytostatic factor (CSF)–arrested low speed supernatants of Xenopus eggs were prepared in XBE2 buffer (10 mM K-Hepes, pH 7.7, 0.1 M KCl, 2 mM MgCl2, 0.1 mM CaCl2, 5 mM EGTA, and 50 mM sucrose) and driven to interphase by addition of 100 µg/ml cycloheximide and 0.5 mM CaCl2. Aphidicolin (Sigma-Aldrich) was used at 25 µg/ml and biotin-16-dUTP (Roche) at 10 µM. Chromosome assembly reactions containing 800 sperm nuclei/µl were processed for immunofluorescence as described previously (Rivera and Losada, 2009) and mounted using Mowiol. Isolation of chromatin fractions for immunoblot analysis for Fig. 6 D was performed as described previously (Rivera and Losada 2009). For Fig. 1 F the protocol was modified: 400 µl of assembly reactions containing 800 sperm nuclei/µl extract were used and two rounds of centrifugation on sucrose cushions were required to achieve purity. Immunoprecipitation reactions were performed on 10 µl of protein A agarose beads using 100 µl of extract and 2–5 µg of antibody.
Immunodepletion and add-back
Immunodepletions were performed using Affi-prep Protein A support (Bio-Rad Laboratories) as described previously (Losada et al., 1998). In brief, depletion of 100 µl of extract required one (CENP-A) or two (xHJURP, condensin) rounds of incubation with 50 µl of beads bound to 30 µg of antibody. Condensin I was depleted with anti-XCAP-G, condensin II with anti-XCAP-D3, and both condensins together either with anti-XCAP-E. In the case of CAF-1 p150 and HIRA, 33 and 10 µl of crude sera, respectively, were bound to 50 µl of beads. Antibodies were cross-linked to the beads using dimethyl pimelimidate (Sigma-Aldrich). For add-back experiments, myc-CENPA was synthesized in vitro using the TNT Quick Coupled Transcription/Translation system (Promega) and diluted in the extract 1:10 to 1:20 so that myc-CENPA was one- to fivefold more concentrated than endogenous CENP-A (Fig. 5, lane 1). X. laevis CENP-A (IMAGE clone 6327326) was cloned into plasmid pCS+MT (kindly provided by D. Turner, University of Michigan, Ann Arbor, MI) to produce a protein with six copies of the myc tag in the N terminus. Recombinant human HJURP-GST was purified from E. coli and 0.3–0.4 ng/µl was added to the extracts along with myc-CENPA (Dunleavy et al., 2009). For affinity purification of condensin II, 90 µg of anti-XCAP-D3 antibody cross-linked to protein A–magnetic beads was incubated with 1 ml of egg extract for 1 h. After extensive washing, the bound complex was eluted with 100 µl of 1mg/ml D3 peptide in XBE2 buffer for 1 h at room temperature, concentrated in a Centricon unit (Millipore), and stored at −80°C. Two distinct XCAP-D3 antibodies raised against the same antigen were used for depletion of condensin II and for affinity purification of the complex.
Image analysis for the CENP-A loading assay
Relative measurements of CENP-A content were obtained by processing and imaging together samples from consecutive time points in the assembly reaction. The sample from the first time point was kept on ice until required. In most cases, two 15-µl aliquots of the assembly reactions (e.g., CSF-assembled chromosomes and interphase nuclei) were mixed, and immediately fixed and spun over the same coverslip. For imaging, chromatin states were elucidated by morphology (condensed chromosomes vs. interphase nuclei) and/or dUTP incorporation (replicated vs. nonreplicated nuclei). In most experiments, undepleted CSF-assembled chromosomes were used as standard reference. Images were taken using a microscope (model DM6000; Leica), with a HCX PlanApochromat 63×/1.4 oil immersion objective. Quantification and processing were done with ImageJ software (http://rsb.info.nih.gov/ij). Images of CENP-A staining were set to binary using the Threshold tool, and then individual centromeres were selected as regions of interest (ROIs) applying the Analyze Particles function. When required, the Watershed function was used to separate centromeres in close vicinity. Alternatively, centromeric regions were selected manually using drawing tools. Using ROI Manager, ROI lists were created for individual nuclei. The average Integrated Density (ID = average pixel intensity × area) was first calculated from the IDs of centromeres within each nucleus and then a ratio between the IDs of each imaged pair of nuclei was obtained. Finally, the average ID ratio (IDr) of at least 15 pairs of nuclei was calculated. The relative CENP-A loading efficiency of a depleted extract (Δ) with respect to a mock-depleted, control extract (C) is calculated as (IDrΔ − 1)/(IDrC − 1). In this case, we imaged together and quantified the CENP-A signals of nuclei assembled in CSF and late interphase, corresponding to point “1” and “3” of Fig. 1 C, respectively. To measure CENP-A eviction, the average integrated density of CENP-A signals in chromosomes assembled in depleted CSF extracts (Fig. 6 C) or in late interphase nuclei (Fig. S5), a reference sample was prepared in undepleted interphase and CSF extract, respectively. The resulting values are expressed as a ratio relative to the average value obtained for the mock-depleted extract.
Online supplemental material
Fig. S1 shows that CENP-A incorporation occurs at centromeres and is independent of ongoing DNA replication. Fig. S2 shows that xHJURP associates with CENP-A in the egg extract and that other known histone chaperones are not affected by CENP-A depletion. Fig. S3 shows that substoichiometric amounts of xHJURP can restore full CENP-A loading. Fig. S4 shows that condensin I and condensin II are targeted to chromosomes independently of each other. Fig. S5 shows that condensin prevents eviction of CENP-A also in interphase. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201005136/DC1.
Acknowledgments
We are grateful to D. Turner and S. Dilworth for reagents. We are also indebted to members of the Chromosome Dynamics and DNA Replication groups of the CNIO, and in particular J. Méndez, for helpful discussions.
This research has been supported by the Spanish Ministry of Science and Innovation (grants BFU2007-66627 and CSD-Inesgen to A. Losada, a Juan de la Cierva contract for R. Bernad, and FPI fellowships to P. Sánchez and T. Rivera); the European Commission Epigenome Network of Excellence (LSHG-CT-2004-503433) to G. Almouzni and A. Losada; MIRG-31126 to A. Losada; and EMBO fellowship ALTF 77-2007 for R. Bernad. Further support for the group of G. Almouzni comes from La Ligue Nationale contre le Cancer (Equipe labelisée Ligue 2010); PIC Programs (“Retinoblastome” and “Replication, Instabilite chromosomique et cancer”); European Commission ITN FP7-PEOPLE-2007 “Image DDR” and FP7-PEOPLE-2008 “Nucleosome 4D”; ACI-2007-Cancéropôle IdF “Breast cancer and Epigenetics”; ANR “FaRC” PCV06_142302 and ANR “ECenS” ANR-09-BLAN-0257-01; and INCa “GepiG”.
Footnotes
Abbreviations used in this paper:
- CAF-1
- chromatin assembly factor 1
- CENP-A
- centromeric protein A
- CSF
- cytostatic factor
- HIRA
- histone regulator A
- HJURP
- Holliday junction–recognizing protein
References
- Ahmad K., Henikoff S. 2001. Centromeres are specialized replication domains in heterochromatin. J. Cell Biol. 153:101–110 10.1083/jcb.153.1.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire R.C., Karpen G.H. 2008. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet. 9:923–937 10.1038/nrg2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad R., Sánchez P., Losada A. 2009. Epigenetic specification of centromeres by CENP-A. Exp. Cell Res. 315:3233–3241 10.1016/j.yexcr.2009.07.023 [DOI] [PubMed] [Google Scholar]
- Bloom K. 2007. Centromere dynamics. Curr. Opin. Genet. Dev. 17:151–156 10.1016/j.gde.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Blower M.D., Sullivan B.A., Karpen G.H. 2002. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell. 2:319–330 10.1016/S1534-5807(02)00135-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R., Li B., Florens L., Swanson S.K., Washburn M.P., Gerton J.L. 2007. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell. 26:853–865 10.1016/j.molcel.2007.05.013 [DOI] [PubMed] [Google Scholar]
- Camahort R., Shivaraju M., Mattingly M., Li B., Nakanishi S., Zhu D., Shilatifard A., Workman J.L., Gerton J.L. 2009. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell. 35:794–805 10.1016/j.molcel.2009.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Silva M.C., Godek K.M., Jansen L.E., Straight A.F. 2009. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 11:896–902 10.1038/ncb1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Milks K.J., Straight A.F. 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189:1143–1155 10.1083/jcb.201001013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Amstutz H., Fishel B., Carbon J. 1986. Analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA. 83:8253–8257 10.1073/pnas.83.21.8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D.W., Mao Y., Sullivan K.F. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 112:407–421 10.1016/S0092-8674(03)00115-6 [DOI] [PubMed] [Google Scholar]
- Collins K.A., Furuyama S., Biggins S. 2004. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 14:1968–1972 10.1016/j.cub.2004.10.024 [DOI] [PubMed] [Google Scholar]
- Conde e Silva N., Black B.E., Sivolob A., Filipski J., Cleveland D.W., Prunell A. 2007. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J. Mol. Biol. 370:555–573 10.1016/j.jmb.2007.04.064 [DOI] [PubMed] [Google Scholar]
- De Koning L., Corpet A., Haber J.E., Almouzni G. 2007. Histone chaperones: an escort network regulating histone traffic. Nat. Struct. Mol. Biol. 14:997–1007 10.1038/nsmb1318 [DOI] [PubMed] [Google Scholar]
- Dilworth S.M., Black S.J., Laskey R.A. 1987. Two complexes that contain histones are required for nucleosome assembly in vitro: role of nucleoplasmin and N1 in Xenopus egg extracts. Cell. 51:1009–1018 10.1016/0092-8674(87)90587-3 [DOI] [PubMed] [Google Scholar]
- Dunleavy E.M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., Almouzni-Pettinotti G. 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 137:485–497 10.1016/j.cell.2009.02.040 [DOI] [PubMed] [Google Scholar]
- Elsaesser S.J., Goldberg A.D., Allis C.D. 2010. New functions for an old variant: no substitute for histone H3.3. Curr. Opin. Genet. Dev. 20:110–117 10.1016/j.gde.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt S., Mellone B.G., Betts C.M., Zhang W., Karpen G.H., Straight A.F. 2008. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J. Cell Biol. 183:805–818 10.1083/jcb.200806038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R., III, Cleveland D.W. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469 10.1038/ncb1397 [DOI] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Bailey A.O., Yates J.R., III, Bassett E.A., Wood S., Black B.E., Cleveland D.W. 2009. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 137:472–484 10.1016/j.cell.2009.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., Obuse C., Yanagida M. 2007. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell. 12:17–30 10.1016/j.devcel.2006.11.002 [DOI] [PubMed] [Google Scholar]
- Furuyama S., Biggins S. 2007. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. USA. 104:14706–14711 10.1073/pnas.0706985104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T., Dalal Y., Henikoff S. 2006. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc. Natl. Acad. Sci. USA. 103:6172–6177 10.1073/pnas.0601686103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K.A., Holmes V.F., Cozzarelli N.R., Meyer B.J. 2002. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 16:729–742 10.1101/gad.968302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M. 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 118:715–729 10.1016/j.cell.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Hemmerich P., Weidtkamp-Peters S., Hoischen C., Schmiedeberg L., Erliandri I., Diekmann S. 2008. Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 180:1101–1114 10.1083/jcb.200710052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. 2005. Condensins: organizing and segregating the genome. Curr. Biol. 15:R265–R275 10.1016/j.cub.2005.03.037 [DOI] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.H., Suzuki E., Okawa K., et al. 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 135:1039–1052 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- Izuta H., Ikeno M., Suzuki N., Tomonaga T., Nozaki N., Obuse C., Kisu Y., Goshima N., Nomura F., Nomura N., Yoda K. 2006. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 11:673–684 10.1111/j.1365-2443.2006.00969.x [DOI] [PubMed] [Google Scholar]
- Jäger H., Rauch M., Heidmann S. 2005. The Drosophila melanogaster condensin subunit Cap-G interacts with the centromere-specific histone H3 variant CID. Chromosoma. 113:350–361 10.1007/s00412-004-0322-4 [DOI] [PubMed] [Google Scholar]
- Jansen L.E., Black B.E., Foltz D.R., Cleveland D.W. 2007. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176:795–805 10.1083/jcb.200701066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I., Fuchs J., Schubert V., Schubert I. 2007. Loading time of the centromeric histone H3 variant differs between plants and animals. Chromosoma. 116:507–510 10.1007/s00412-007-0122-8 [DOI] [PubMed] [Google Scholar]
- Losada A., Hirano M., Hirano T. 1998. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 12:1986–1997 10.1101/gad.12.13.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik H.S., Henikoff S. 2003. Phylogenomics of the nucleosome. Nat. Struct. Biol. 10:882–891 10.1038/nsb996 [DOI] [PubMed] [Google Scholar]
- Marshall O.J., Marshall A.T., Choo K.H. 2008. Three-dimensional localization of CENP-A suggests a complex higher order structure of centromeric chromatin. J. Cell Biol. 183:1193–1202 10.1083/jcb.200804078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milks K.J., Moree B., Straight A.F. 2009. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol. Biol. Cell. 20:4246–4255 10.1091/mbc.E09-05-0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G., Xiao H., Wisniewski J., Smith M.M., Wu C. 2007. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 129:1153–1164 10.1016/j.cell.2007.04.026 [DOI] [PubMed] [Google Scholar]
- Moreno-Moreno O., Torras-Llort M., Azorín F. 2006. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res. 34:6247–6255 10.1093/nar/gkl902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Obuse C., Yang H., Nozaki N., Goto S., Okazaki T., Yoda K. 2004. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 9:105–120 10.1111/j.1365-2443.2004.00705.x [DOI] [PubMed] [Google Scholar]
- Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R., III, Desai A., Fukagawa T. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446–457 10.1038/ncb1396 [DOI] [PubMed] [Google Scholar]
- Okada M., Okawa K., Isobe T., Fukagawa T. 2009. CENP-H-containing complex facilitates centromere deposition of CENP-A in cooperation with FACT and CHD1. Mol. Biol. Cell. 20:3986–3995 10.1091/mbc.E09-01-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira R.A., Coelho P.A., Sunkel C.E. 2005. The condensin I subunit Barren/CAP-H is essential for the structural integrity of centromeric heterochromatin during mitosis. Mol. Cell. Biol. 25:8971–8984 10.1128/MCB.25.20.8971-8984.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T., Losada A., Hirano M., Myers M.P., Neuwald A.F., Hirano T. 2003. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 115:109–121 10.1016/S0092-8674(03)00724-4 [DOI] [PubMed] [Google Scholar]
- Ono T., Fang Y., Spector D.L., Hirano T. 2004. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol. Biol. Cell. 15:3296–3308 10.1091/mbc.E04-03-0242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C.G., Maddox P.S., Salmon E.D., Bloom K. 2001. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 152:1255–1266 10.1083/jcb.152.6.1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perpelescu M., Nozaki N., Obuse C., Yang H., Yoda K. 2009. Active establishment of centromeric CENP-A chromatin by RSF complex. J. Cell Biol. 185:397–407 10.1083/jcb.200903088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux A.L., Choi E.S., Abbott J.K., Liu X., Kagansky A., Castillo A.G., Hamilton G.L., Richardson W., Rappsilber J., He X., Allshire R.C. 2009. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell. 33:299–311 10.1016/j.molcel.2009.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy J.P., Grandi P., Almouzni G. 2001. Dimerization of the largest subunit of chromatin assembly factor 1: importance in vitro and during Xenopus early development. EMBO J. 20:2015–2027 10.1093/emboj/20.8.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Gallet D., Quivy J.P., Scamps C., Martini E.M., Lipinski M., Almouzni G. 2002. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell. 9:1091–1100 10.1016/S1097-2765(02)00526-9 [DOI] [PubMed] [Google Scholar]
- Rivera T., Losada A. 2009. Shugoshin regulates cohesion by driving relocalization of PP2A in Xenopus extracts. Chromosoma. 118:223–233 10.1007/s00412-008-0190-4 [DOI] [PubMed] [Google Scholar]
- Samoshkin A., Arnaoutov A., Jansen L.E., Ouspenski I., Dye L., Karpova T., McNally J., Dasso M., Cleveland D.W., Strunnikov A. 2009. Human condensin function is essential for centromeric chromatin assembly and proper sister kinetochore orientation. PLoS ONE. 4:e6831 10.1371/journal.pone.0006831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Pulido L., Pidoux A.L., Ponting C.P., Allshire R.C. 2009. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 137:1173–1174 10.1016/j.cell.2009.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Musacchio A. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M., Lehner C.F., Heidmann S. 2007. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 17:237–243 10.1016/j.cub.2006.11.051 [DOI] [PubMed] [Google Scholar]
- Shelby R.D., Vafa O., Sullivan K.F. 1997. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 136:501–513 10.1083/jcb.136.3.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby R.D., Monier K., Sullivan K.F. 2000. Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 151:1113–1118 10.1083/jcb.151.5.1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M., Ouararhni K., Dimitrov S., Hamiche A. 2010. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc. Natl. Acad. Sci. USA. 107:1349–1354 10.1073/pnas.0913709107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M.C., Jansen L.E. 2009. At the right place at the right time: novel CENP-A binding proteins shed light on centromere assembly. Chromosoma. 118:567–574 10.1007/s00412-009-0227-3 [DOI] [PubMed] [Google Scholar]
- Stoler S., Rogers K., Weitze S., Morey L., Fitzgerald-Hayes M., Baker R.E. 2007. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl. Acad. Sci. USA. 104:10571–10576 10.1073/pnas.0703178104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 116:51–61 10.1016/S0092-8674(03)01064-X [DOI] [PubMed] [Google Scholar]
- Takayama Y., Sato H., Saitoh S., Ogiyama Y., Masuda F., Takahashi K. 2008. Biphasic incorporation of centromeric histone CENP-A in fission yeast. Mol. Biol. Cell. 19:682–690 10.1091/mbc.E07-05-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torras-Llort M., Moreno-Moreno O., Azorín F. 2009. Focus on the centre: the role of chromatin on the regulation of centromere identity and function. EMBO J. 28:2337–2348 10.1038/emboj.2009.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wignall S.M., Deehan R., Maresca T.J., Heald R. 2003. The condensin complex is required for proper spindle assembly and chromosome segregation in Xenopus egg extracts. J. Cell Biol. 161:1041–1051 10.1083/jcb.200303185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.S., Hayashi T., Yanagida M., Russell P. 2009. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol. Cell. 33:287–298 10.1016/j.molcel.2009.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong-Gonzalez V., Wang B.D., Butylin P., Ouspenski I., Strunnikov A. 2007. Condensin function at centromere chromatin facilitates proper kinetochore tension and ensures correct mitotic segregation of sister chromatids. Genes Cells. 12:1075–1090 10.1111/j.1365-2443.2007.01109.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S.G., Baker N.M., Chapados B.R., Soutoglou E., Wang J.Y., Berns M.W., Cleveland D.W. 2009. Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc. Natl. Acad. Sci. USA. 106:15762–15767 10.1073/pnas.0908233106 [DOI] [PMC free article] [PubMed] [Google Scholar]