Summary
Coronary artery disease is the number one cause of morbidity and mortality in the Western world. It typically occurs when heart muscle receives inadequate blood supply due to rupture of atherosclerotic plaques. During ischemia, up-regulation of hypoxia inducible factor-1 alpha (HIF-1α) transcriptional factor can activate several downstream angiogenic genes. However, HIF-1α is naturally degraded by prolyl hydroxylase-2 (PHD2) protein. Recently, we cloned the mouse PHD2 gene by comparing the homolog gene in human and rat. The best candidate shRNA sequence for inhibiting PHD2 was inserted behind H1 promoter, followed by a separate hypoxia response element (HRE)-incorporated promoter driving a firefly luciferase (Fluc) reporter gene. This construct allowed us to monitor gene expression noninvasively and was used to test the hypothesis that inhibition of PHD2 by short hairpin RNA interference (shRNA) can lead to significant improvement in angiogenesis and contractility as revealed by in vitro and in vivo experiments.
Keywords: RNA interference, molecular imaging, hypoxia inducible factor (HIF), prolyl hydroxylases (PHD), ischemic heart disease
1. Introduction
Coronary artery disease (CAD) is the leading cause of morbidity and mortality in the Western world (1). Conventional treatment for CAD consists of medical therapy as the first-line strategy, followed by percutaneous coronary intervention (PCI) or coronary artery bypass graft (CABG). However, a significant number of patients will still have refractory angina despite these treatments (2). Over the past decades, a better understanding of the molecular and genetic bases of different diseases has made gene therapy an increasingly viable treatment option (3). With the use of gene transfer techniques, it is now possible to modify somatic cells in ischemic myocardium, to overexpress beneficial or inhibitory pathologic proteins, and to achieve positive therapeutic effects (4). Indeed, several clinical trials evaluating both viral and non-viral delivery of vascular endothelial growth factor (VEGF) and fibroblast growth factor (FGF) have been completed to date (5).
A growing body of evidence suggests that the expression of a single angiogenic factor alone is not sufficient for the functional revascularization of ischemic tissues (6). Newer approaches based on the transcriptional factor HIF-1α may be a more natural choice. HIF-1α is known to control the expression of over 60 genes that affect cell survival and metabolism in adverse conditions, including VEGF (7), insulin-like growth factor (8), erythropoietin (9), nitric oxide synthase (10), among others. However, during normoxia, HIF-1α subunits have an exceptionally short half-life (∼3–5 min) and low steady-state levels (11). This is due to hydroxylation of two prolyl residues (Pro402 and Pro564) by a family of prolyl-4-hydroxylases (PHDs) (12). Hydroxylation of HIF-1α allows recognition by the von Hippel-Lindau (VHL) tumor suppressor, which targets HIF-1α for proteosomal destruction (13). In contrast, increasing the severity of hypoxia retards degradation of HIF-1α subunits, allowing nuclear localization, dimerization with HIF-1β subunits, and formation of a stable DNA-binding HIF complex (14). Thus, the short hairpin RNA (shRNA) plasmid for knockdown of PHD2 (shPHD2) can potentially be used as a novel gene therapy for treatment of ischemic heart disease.
To date, the majority of cardiac gene therapy studies have relied on ex vivo quantification of gene expression (e.g., GFP or lacZ) in small animals or indirect markers (e.g., changes in perfusion or contractility) in clinical trials (15, 16). In order to characterize, visualize, and quantify biological processes at the molecular and cellular levels within intact living organisms, in vivo imaging techniques are needed. Over the past 10 years, molecular imaging has been widely used for oncology studies, but applications in cardiology have been a recent development (17). One such example is the use of reporter genes that can be transferred into cells via a delivery vector and regulated by constitutive, inducible, or tissue-specific promoters.
In this protocol, we outline the procedures used to address the two issues mentioned above--better therapeutic gene and more sophisticated tracking method. We show that the inhibition of HIF-1α degradation via shRNA knockdown of PHD2 in the ischemic heart represents a novel angiogenic therapy approach. At the same time, we track the shRNA vector in vivo through novel molecular imaging technology (18).
2. Material
2.1. Generating the shPHD2 Construct
Mouse ES cell (ATCC). Primary mouse ES cell line at passage 10 from SV129 mouse strain.
Cell culture medium: DMEM, high glucose with L-glutamine (Gibco-BRL, Grand Island, NY, USA). Store at 4 °C.
Fetal bovine serum (FBS) (Hyclone, City, State, USA). Store at −20 °C.
Penicillin G-Streptomycin: Penicillin 100 U/mL DMEM culture medium and Streptomycin 100 μg/mL DMEM culture medium (Gibco-BRL). Store at −20 °C.
Leukemia inhibitory factor (LIF) (Sigma, St Louis, MO, USA). Store at 4 °C.
Feeder cell. primary fibroblast cells from a 12.5 day-old mouse embryo.
1× Trypsin–EDTA. 0.05% Trypsin, 1 mM EDTA/4Na (Gibco-BRL). Store at 4 °C.
RNA easy kit (Qiagen, Valencia, CA, USA). Store at room temperature.
iScript cDNA synthesis kit (Biorad, Hercules, CA,USA). Store at −20 °C.
pGEM-T (Promega, Madison, WI, USA ). Store at −20 °C.
pSuper shRNA vector (Oligoengine, Seattle, WA,USA). Store at −20 °C.
Five tandem repeats of hypoxia responsive element (5×HRE) (synthesized by Stanford Protein and Nucleic Acid Facility).
Plasmid pGL3 including SV40-Firefly luciferase (Promega). Store at −20 °C.
Restriction endonucleases and T4 ligase. (New England Biolabs, Ipswich, MA,USA ). Store at −20 °C.
S.O.C. medium (Gibco-BRL). Store at room temperature.
Amp selection LB agar plates (100 μg/mL, ampicillin). Store at 4 °C.
Chemical-competent Escherichia coli Top 10 cells (Invitrogen, Carlsbad, CA, USA). Store at −80 °C.
2.2. Testing the shPHD2 Construct In Vitro
Mouse myoblast cell line C2C12 (ATCC). Culturing under 37 °C at 5% CO2.
Lipofectamine 2000 (Invitrogen, Carlsbad, CA,USA). Store at 4 °C.
0.25% Trypsin (Gibco-BRL). Store at 4 °C.
D-Luciferin (Synchem, Elk Grove Village, IL, USA). Store at −20 °C.
We inserted firefly luciferase under the CMV promoter in pcDNA3.1 (Invitrogen) at the SalI restriction enzyme site. Store at −20 °C.
RIPA buffer (Sigma). Store at 4 °C.
Trizol reagent (Invitrogen). Store at 4 °C.
10% SDS-PAGE gel (Biorad). Store at 4 °C.
Hybond-P memberane (GE, Fairfield, CT,USA ).
Nonfat dry milk (Biorad).
Mouse HIF-1 α antibody (Novus, Littleton, CO,USA). Store at 4 °C.
2.3. Evaluation of the shPHD2 Construct In Vivo
Two-month-old FVB female mice (Charles River Laboratories, Wilmington, MA).
Anesthetic reagents and instruments for mouse heart injection: 3% isoflurane + 97% oxygen, sterile forceps and scissors (World Precision Instruments, Inc., Sarasota, FL, USA), needle holders (Accurate Surgical & Scientific Instruments Corp., Westbury, NY, USA), Guthrie double hook retractor (Fine Science Tools, Inc., Foster City, CA, USA), 5-0 Sofsilk suture (Auto Suture Company, Norwalk, CT, USA), 0.5 mL gas-tight Hamilton syringe, and 30 gauge needle (Hamilton Company, Reno, NV, USA).
70% ethanol. Rossville Gold Shield Alcohol, proof 200 grade .
Standard materials for large-scale plasmid preparation.
D-Luciferin (Synchem, Felsberg, Germany). 150 mg/kg body weight for intraperitoneal injection.
Xenogen In Vivo Imaging System (IVIS) 200 small animal imaging system (Calipfer Lise Sciences, Hopkinton, MA).
0.5 mL gas-tight Hamilton syringe and 30-gauge needle (Hamilton Company, Reno, NV).
Siemens-Acuson Sequioa C512 Small Animal Echocardiogram System. This system is used to compare heart function changes between shPHD2 treated group and shScramble control group.
Muscle homogenization solution: 20 mM Na4P2O4, 20 mM NaHPO4, 1 mM MgCl2, 0.5 mM EDTA, and 303 mM sucrose.
Muscle microsome preparation wash buffer: 20 mM Tris–HCl (pH 7.0), 60 mM KCl, 303 mM sucrose.
Muscle microsome storage solution: 20 mM Tris–HCl (pH 7.0) and 303 mM sucrose.
Tissue-Tek OCT compound (Sakura Finetek Inc., Torrance, CA, USA).
Rat anti-CD31 (BD Biosciences, San Jose, CA) monoclonal antibody. This antibody binds to endothelial CD31 markers expressed on small blood vessels.
2-methylbutane (Sigma).
Rabbit anti-rat IgG (Sigma, final concentration 0.46 mg/mL).
Papain (Sigma, final concentration 45 μg/mL).
L-cystein (Sigma, final concentration 22 mM).
Iodoacetic acid (Sigma, final concentration 10 mM).
20% rabbit serum (Jackson ImmunoResearch Laboratories, Inc.).
Alexa 488 conjugated rabbit anti-rat antibodies (Molecular Probe).
Alexa 594 conjugated rabbit anti-rat antibodies (Molecular Probe).
KPBS: 356 μM KH2PO4, 1.64 mM K2HPO4, 160 mM NaCl.
Gelatin (Sigma).
Vector Mouse on Mouse (M.O.M) Kits (Vector Laboratories, Inc., Burlingame, CA).
3. Methods
3.1. Generating the shPHD2 Construct
Compare the human and rat PHD2 gene sequences. Because the mouse PHD2 cDNA cannot be found in gene bank, we compared the exact sequence that has been isolated from both. By comparing human and rat PHD2 genes in the Sanger center (www.sanger.ac.uk), we selected the 350 bp fragment sequence in both human and rat PHD2 gene, which is located at the 3’-end of both sequence. We hypothesized this sequence is the same as in mouse genome.
Clone the mouse PHD2 gene from mouse ES cells using PCR method. ~350 bp fragment in 3’ end mouse PHD2 gene.
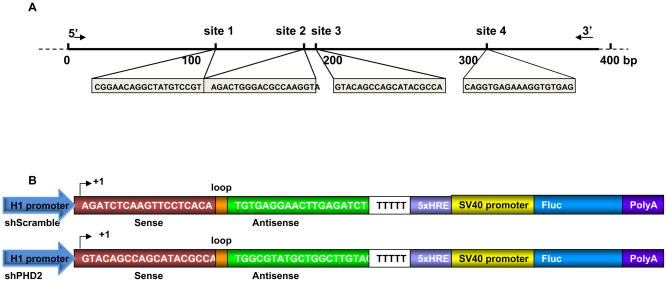
Design RNA interference targeting sequences using a commercially available web-based software (http://www.ambion.com/techlib/misc/siRNA_finder.html). 4 sequences of sites were designed as the shRNA target. The targeting sequences are shown in Fig. 1a. Sequence for the control short hairpin scramble (shScramble) antisense is TGTGAGGAACTTGAGATCT.
Fig. 1. Schema of the shPHD2 knockdown sites and the reporter constructs.
(A) Individual sequences of four small interfering RNA target sites against the PHD2 gene. (B) Schema of a classic hairpin carrying the site-2 sequence (shPHD2) and a control hairpin carrying the scramble sequence (shScramble). The H1 promoter drives the expression of a hairpin structure in which the sense and antisense strands of the small interfering RNA are connected by a 9-bp long loop sequence. In addition, a separate 5xHRE-SV40 promoter driving firefly luciferase (Fluc) is used to track shRNA activity in vitro and in vivo. 5x HRE, 5 repeat of hypoxia response elements; SV40, simian virus 40. (Reproduced from ref. 24 with permission).
3.2. Testing the shPHD2 Construct In Vitro
Clone the four shRNA fragments under H1 promoter. To determine the site that possesses the optimal knock-down efficiency, we cloned the sense and antisense downstream of the H1 promoter by the PCR method (Fig. 1b).
Test the best knocking down efficiency fragment. These four shRNA constructs were used to transfect C2C12 myoblasts in 6-well plates in conjunction with the pCMV-luciferase plasmid, which confirmed equal transfection efficiency. After 48 hours of cell culture, mRNA levels of PHD2 within C2C12 cells were measured by RT-PCR. Construction of the H1 promoter driving sense and antisense was performed as described (19). The No. 2 fragment demonstrated the best efficiency.
Clone the best fragment into pSuper shRNA vector. The fragment No. 2 knocking down site was inserted after H1 promoter in the vector pSuper as described in the Oligoengine™ manual.
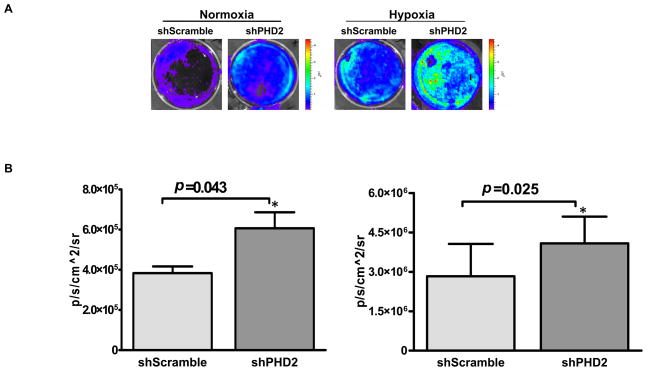
Insert 5×HRE fragment as hypoxia indication. A hypoxia sensing 5xHRE-SV40 promoter driving Fluc cassette was also inserted into the backbone of pSuper vector. The 5 copies of hypoxia response element (5xHRE) derived from the erythropoietin gene are activated through binding of the HIF-1 complex (20), enabling us to monitor the efficacy of the upstream shPHD2 knockdown compared to the upstream shScramble control (Fig. 2a). SpeI site on the pSuper was cut in order to insert the hypoxia response element HRE-SV40-firefly luciferase cassette.
Fig. 2. In vitro characterization of mouse shPHD2.
(A) In vitro imaging results indicate that Fluc signals increased significantly in response to shPHD2 therapy during both normoxia and hypoxia conditions by binding of HIF-1α protein on the 5xHRE binding site. (B) Quantitative analysis of Fluc bioluminescence signals from cell plates. (Reproduced from ref. 24 with permission).
3.3. Evaluation of the shPHD2 Construct In Vitro
Culture mouse C2C12 myoblasts in DMEM medium (high glucose) supplemented with 10% fetal bovine serum as described in the ATCC protocol. Use Lipofectamine 2000 for the transfection. Culture cells for 1 day after shRNA fragment transfection before placing cells in a hypoxia chamber filled with 5% CO2, 1% O2, and 94% N2 at 37°C. Keep cells under hypoxic conditions for 48 hours. At the end of the hypoxic treatment, harvest cells immediately to extract RNA and protein.
Use RT-PCR to compare the expression of antigenic genes (bFGF, tranferin, FLT, KDR, TGF, PAI-1) in transfected cells under nomoxia versus hypoxia. Prepare total RNA from C2C12 cells with Trizol reagent. The primer sets used in the amplification reaction are shown in Table 1. Separate PCR products in 1% agarose gel and quantify the intensity of the bands with Labworks 4.6 image acquisition and analysis software.
To confirm the shPHD2 knocking down efficiency, isolate nuclear extracts and detect HIF-1α protein by western blot. After 48 hrs of hypoxia culture, wash the C2C12 cells with phosphate buffered saline (PBS) and homogenize cells with 200 μL of homogenization buffer (RIPA). Fractionate the supernatants on SDS-PAGE and blot to a Hybond-P membrane. Block the membranes with 5% nonfat dry milk in 1 x TBS containing 0.05% Tween 20. Incubate with the antibodies against HIF-1α. Detect with secondary HRP-conjugated antibodies and the ECL detection system. For the in vivo western blot, take heart samples at days 1, 4, 7, 14, and 28 after plasmid injection (see Note 1).
Table 1.
| bFGF | Forward | 5’—CTTCAAGGACCCCAAGCGGGCTCTA—3’ |
| Reverse | 5’—CGAGTTTATACTGCCCAGTT—3’ | |
| Transferrin | Forward | 5’—CTTCAAGGACCCCAAGCGGCTCTAC—3’ |
| Reverse | 5’—GTTCGTTTCAGTGCCACATACCAAC—3’ | |
| FLT | Forward | 5’—TGAAGTCTGCTCGCTATTTGGTA—3’ |
| Reverse | 5’—CTATGGTGCATGGTTCTGTTGTT—3’ | |
| KDR | Forward | 5’—GAAGCTACTGCCGTCCGATTGAG—3’ |
| Reverse | 5’—TGCTGGCTTTGGTGAGGTTTGAT—3’ | |
| TGF | Forward | 5’—AAATTCGACATGATCCAGGGACT—3’ |
| Reverse | 5’—TGCACTTACACGACTTCACCACC—3’ | |
| PAI | Forward | 5’—ATGGCTCAGAGCAACAAGTTCAA—3’ |
| Reverse | 5’—GACAAAGGCTGTGGAGGAAGACG—3’ |
3.4. Testing the shPHD2 Efficacy In Vivo
-
1
Get approval from the Animal Research Committee of the institute.
-
2
Ligate the mid left anterior descending (LAD) artery of adult female FVB mice. Anesthetize mice using isoflurane inhalation. Place a tube through the mouth into the trachea and connect the tube to a Harvard rodent ventilator. After stabilizing the respiratory status of the animal, a make a thoracotomy incision in the fourth intercostal space and use a surgical retractor to expose the heart. Ligate the LAD permanently with a 7-0 non-absorbable surgical suture. Confirm myocardial infarction by myocardial blanching and EKG changes. Wait for 10 minutes. Inject 25 μg (1 μg/μL) of shRNA plasmid at the peri-infarct zone or 25 μg (1 μg/μL) of shScramble plasmid as control using a 30-gauge Hamilton needle (see Notes 2 and 3).
-
3
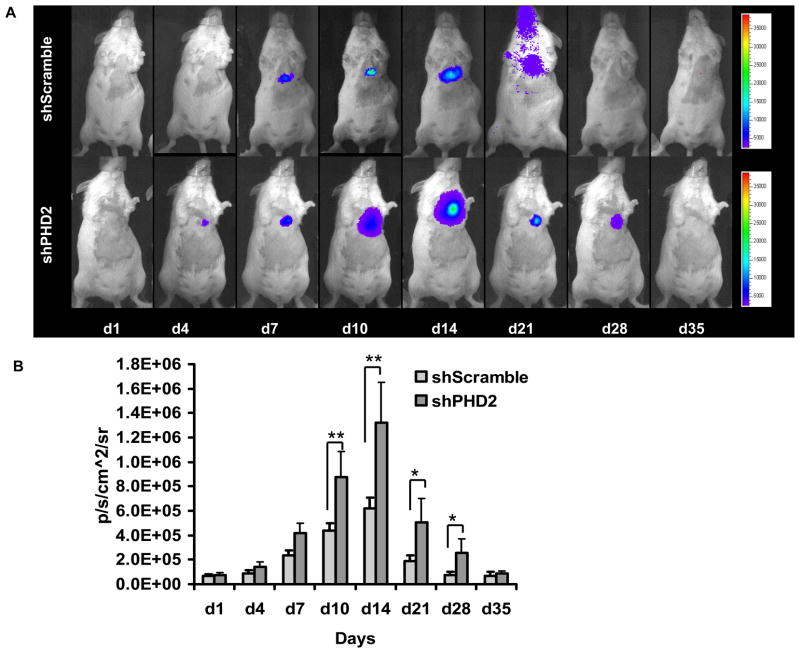
Perform cardiac bioluminescence imaging with the Xenogen IVIS. After intraperitoneal injection of the reporter probe D-luciferin (150 mg/kg body weight), image animals for 1–10 minutes. Scan the same mice were repetitively for 4 weeks according to the specific study design (Fig. 3). Quantify bioluminescence signals in units of maximum photons per second per centimeter squared per steradian (p/s/cm2/sr) using the built-in software on the IVIS system (21, 22).
-
3
Perform echocardiography before (day −7) and after (week 2, week 4, week 8) the LAD ligation with the Siemens-Acuson Sequioa C512 system equipped with a multi-frequency (8–14 MHZ) 15L8 transducer. Investigators are blinded to group designation. Analyze M-mode images using the Siemens built-in software. Measure left ventricular end diastolic diameter (EDD) and end-systolic diameter (ESD) and calculate left ventricular fractional shortening by the formula: LVFS = [EDD−ESD]/EDD.
-
4
Embed the explanted hearts into OCT compound. Process 5 μm frozen sections for immunostaining. Use a rat anti-CD31 to detect microvascular density in the peri-infarct area. Count the number of capillary vessels by a blinded investigator in ten randomly selected areas using a light microscope (x 200 magnification). Use additional samples to examine the infarction size by Masson’s trichrome staining.
Fig. 3. Molecular imaging of shRNA plasmid fate after intramyocardial delivery.
(A) Following myocardial infarction, activation of HIF-1α protein binds to 5xHRE site to activate Fluc expression. Infarcted mice injected with shPHD2 (bottom row) had more robust Fluc signals compared to infarcted mice injected with shScramble (top row) due to knocking down of PHD2, which result in more HIF-1α protein binding to 5 x HRE site. Peak expression occurred within week 1–2 as reflected by the Fluc imaging signals. (B) Detailed quantitative analysis of Fluc signals from all animals injected with shPHD2 or shScramble plasmid with LAD ligation. Signal activity is expressed as p/sec/cm2/sr. (Reproduced from ref. 24 with permission).
Acknowledgments
The authors would like to thank Dr. Robert Robbins and his staff for providing the surgical service. This work was supported in part by grants from the NIH HL095571 (JCW), NIH HL093172 (JCW), and AHA Western Postdoctoral Fellowship (MH).
Footnotes
We harvest the heart tissue under microscopy to isolate the peri-infarct and infarct area tissues.
To obtain the optimal quality of plasmids for in vivo injection, we recommend preparing the plasmids by CsCl2-ethidium bromide equilibrium centrifugation.
To obtain the large yield of shPHD2 plasmid, we select DH5 alpha and fresh E. coli solution to do the isolation.
References
- 1.Rosamond W, Flegal K, Friday G, Furie K, Go A, Greenlund K, Haase N, Ho M, Howard V, Kissela B, Kittner S, Lloyd-Jones D, McDermott M, Meigs J, Moy C, Nichol G, O'Donnell CJ, Roger V, Rumsfeld J, Sorlie P, Steinberger J, Thom T, Wasserthiel-Smoller S, Hong Y. Heart disease and stroke statistics--2007 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2007;115:e69–171. doi: 10.1161/CIRCULATIONAHA.106.179918. [DOI] [PubMed] [Google Scholar]
- 2.Kim MC, Kini A, Sharma SK. Refractory angina pectoris: mechanism and therapeutic options. J Am Coll Cardiol. 2002;39:923–934. doi: 10.1016/s0735-1097(02)01716-3. [DOI] [PubMed] [Google Scholar]
- 3.Nabel GJ. Genetic, cellular and immune approaches to disease therapy: past and future. Nat Med. 2004;10:135–141. doi: 10.1038/nm990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yla-Herttuala S, Alitalo K. Gene transfer as a tool to induce therapeutic vascular growth. Nat Med. 2003;9:694–701. doi: 10.1038/nm0603-694. [DOI] [PubMed] [Google Scholar]
- 5.Isner JM. Myocardial gene therapy. Nature. 2002;415:234–239. doi: 10.1038/415234a. [DOI] [PubMed] [Google Scholar]
- 6.Luttun A, Tjwa M, Moons L, Wu Y, Angelillo-Scherrer A, Liao F, Nagy JA, Hooper A, Priller J, De Klerck B, Compernolle V, Daci E, Bohlen P, Dewerchin M, Herbert JM, Fava R, Matthys P, Carmeliet G, Collen D, Dvorak HF, Hicklin DJ, Carmeliet P. Revascularization of ischemic tissues by PlGF treatment, and inhibition of tumor angiogenesis, arthritis and atherosclerosis by anti-Flt1. Nat Med. 2002;8:831–840. doi: 10.1038/nm731. [DOI] [PubMed] [Google Scholar]
- 7.Vincent KA, Feron O, Kelly RA. Harnessing the response to tissue hypoxia: HIF-1 alpha and therapeutic angiogenesis. Trends Cardiovasc Med. 2002;12:362–367. doi: 10.1016/s1050-1738(02)00186-x. [DOI] [PubMed] [Google Scholar]
- 8.Fukuda R, Hirota K, Fan F, Jung YD, Ellis LM, Semenza GL. Insulin- like growth factor 1 induces hypoxia-inducible factor 1-mediated vascular endothelial growth factor expression, which is dependent on MAP kinase and phosphatidylinositol 3-kinase signaling in colon cancer cells. J Biol Chem. 2002;277:38205–38211. doi: 10.1074/jbc.M203781200. [DOI] [PubMed] [Google Scholar]
- 9.Stolze I, Berchner-Pfannschmidt U, Freitag P, Wotzlaw C, Rossler J, Frede S, Acker H, Fandrey J. Hypoxia-inducible erythropoietin gene expression in human neuroblastoma cells. Blood. 2002;100:2623–2628. doi: 10.1182/blood-2001-12-0169. [DOI] [PubMed] [Google Scholar]
- 10.Sandau KB, Fandrey J, Brune B. Accumulation of HIF-1alpha under the influence of nitric oxide. Blood. 2001;97:1009–1015. doi: 10.1182/blood.v97.4.1009. [DOI] [PubMed] [Google Scholar]
- 11.Jewell UR, Kvietikova I, Scheid A, Bauer C, Wenger RH, Gassmann M. Induction of HIF-1alpha in response to hypoxia is instantaneous. Faseb J. 2001;15:1312–1314. [PubMed] [Google Scholar]
- 12.Chan DA, Sutphin PD, Denko NC, Giaccia AJ. Role of prolyl hydroxylation in oncogenically stabilized hypoxia-inducible factor-1alpha. J Biol Chem. 2002;277:40112–40117. doi: 10.1074/jbc.M206922200. [DOI] [PubMed] [Google Scholar]
- 13.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 14.Jiang BH, Semenza GL, Bauer C, Marti HH. Hypoxia-inducible factor 1 levels vary exponentially over a physiologically relevant range of O2 tension. Am J Physiol. 1996;271:C1172–1180. doi: 10.1152/ajpcell.1996.271.4.C1172. [DOI] [PubMed] [Google Scholar]
- 15.Pislaru S, Janssens SP, Gersh BJ, Simari RD. Defining gene transfer before expecting gene therapy: putting the horse before the cart. Circulation. 2002;106:631–636. doi: 10.1161/01.cir.0000019621.18368.b7. [DOI] [PubMed] [Google Scholar]
- 16.Yla-Herttuala S, Markkanen JE, Rissanen TT. Gene therapy for ischemic cardiovascular diseases: some lessons learned from the first clinical trials. Trends Cardiovasc Med. 2004;14:295–300. doi: 10.1016/j.tcm.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Wu JC, Yla-Herttuala S. Human gene therapy and imaging: cardiology. Eur J Nucl Med Mol Imaging. 2005;32(Suppl 2):S346–357. doi: 10.1007/s00259-005-1897-6. [DOI] [PubMed] [Google Scholar]
- 18.Huang M, Chan DA, Jia F, Xie X, Li Z, Hoyt G, Robbins RC, Chen X, Giaccia AJ, Wu JC. Short hairpin RNA interference therapy for ischemic heart disease. Circulation. 2008;118:S226–233. doi: 10.1161/CIRCULATIONAHA.107.760785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyagishi M, Taira K. U6 promoter-driven siRNAs with four uridine 3' overhangs efficiently suppress targeted gene expression in mammalian cells. Nature biotechnology. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- 20.Ruan H, Su H, Hu L, Lamborn KR, Kan YW, Deen DF. A hypoxia-regulated adeno-associated virus vector for cancer-specific gene therapy. Neoplasia. 2001;3:255–263. doi: 10.1038/sj.neo.7900157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao F, Lin S, Xie X, Ray P, Patel M, Zhang X, Drukker M, Dylla SJ, Connolly AJ, Chen X, Weissman IL, Gambhir SS, Wu JC. In vivo visualization of embryonic stem cell survival, proliferation, and migration after cardiac delivery. Circulation. 2006;113:1005–1014. doi: 10.1161/CIRCULATIONAHA.105.588954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang M, Chan D, Jia F, Xie X, Li Z, Hoyt G, Robbins R, Chen X, Giaccia A, Wu J. Short hairpin RNA interference therapy for ischemic heart disease. Circulation. 2008;118:S226–233. doi: 10.1161/CIRCULATIONAHA.107.760785. [DOI] [PMC free article] [PubMed] [Google Scholar]